Open Access Article
Open Access ArticleChanges in degrading ability, populations and metabolism of microbes in activated sludge in the treatment of phenol wastewater†
Rui Chen,
Long-Fei Ren,
Jiahui Shao,
Yiliang He and
Xiaofan Zhang *
*
School of Environmental Science and Engineering, Shanghai Jiao Tong University, 800 Dongchuan Road, 200240 Shanghai, PR China. E-mail: xf_zhang@sjtu.edu.cn; Tel: +86-21-5474563
First published on 15th November 2017
Abstract
Herein, changes in the degrading ability, populations and metabolism of microbes in activated sludge exposed to 60–350 mg L−1 phenol are thoroughly investigated. A phenol degradation experiment is conducted using activated sludge as inoculum over 140 days. The results suggest that the sludge efficiently degrades 250 mg L−1 phenol; however, it is unable to remove 350 mg L−1 phenol completely in two days, which might be caused by the decreased activities of catechol 1,2 dioxygenase (C12O) and catechol 2,3 dioxygenase (C23O). The specific oxygen uptake rate (SOUR) of the sludge and extracellular polymeric substances (EPS) generation are inhibited at the beginning of phenol degradation and then increase with phenol loading. A large amount of humic acid (HA) is produced during the degradation of 350 mg L−1 phenol due to cell decomposition. Illumina-MiSeq sequencing indicates that denitrifiers are competitive clusters at high phenol concentrations. The present study provides a comprehensive understanding of mechanisms of microbial responses to toxic compounds.
1. Introduction
Phenolic compounds are one of the most hazardous and widespread pollutants in the industrial field. They are recognized to be hematotoxic and hepatotoxic, provoking mutagenesis and carcinogenesis toward living organisms.1 Large quantities of phenol are used in the production of polycarbonate resins, explosives, textiles, drugs, and disinfectants.2 The concentration of phenol in discharge from these industries varies from 0.1 to over 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mg L−1.3,4 Without appropriate treatments, phenol and other phenolic compounds are prone to accumulate in sediments, natural waters, and animals due to their recalcitrant nature, which poses a huge threat to the environment and humans.5
000 mg L−1.3,4 Without appropriate treatments, phenol and other phenolic compounds are prone to accumulate in sediments, natural waters, and animals due to their recalcitrant nature, which poses a huge threat to the environment and humans.5
Conventional treatments of phenol wastewater include adsorption, solvent extraction, membrane separation, chemical oxidation and biological processes.6,7 Since other methods often cause secondary pollution, biological processes have received more and more attention due to their low cost and complete mineralization. Various biodegradation approaches have been developed recently, and activated sludge remains a primary approach in the removal of phenol. Furthermore, its effectiveness in absorbing and degrading organic compounds makes it a competitive candidate in practice.8 Besides, activated sludge is more likely to remain stable compared with pure strains with a fluctuation in influent loading.9 Thus, great efforts have been devoted to phenol removal via the activated sludge process. It is demonstrated that up to 2000 mg L−1 phenol could be successfully degraded by activated sludge with mixed liquor suspended solids (MLSS) of 2850 mg L−1 at 25 °C and pH of 7 ± 0.5.8 Nevertheless, previous studies also suggest that even though acclimated sludge could withstand phenol of high concentrations, the predominant population and bioactivity of sludge are significantly influenced by phenol.10,11
Microbial characteristics can effectively elucidate the degradation mechanism and reflect the performance of biological processes since biodegradation relies heavily on the functions and activity of microorganisms. Many studies have investigated the influence of phenol on certain characteristics of activated sludge, which mainly focus on microbial communities, metabolism and surface properties.9,12 Phenol removal is accomplished by microorganisms that are capable of employing phenol as a carbon and energy source. The Proteobacteria phylum is able to utilize a wide range of aromatic compounds, and is also detected as the most abundant group in microbial communities for phenol degradation.12 Other species such as Firmicutes, Chloroflexi, Actinobacteria and Bacteroides could coexist with Proteobacteria to form a symbiotic colony.6,12 These microorganisms are usually enveloped by a matrix of molecular polymer, known as extracellular polymeric substances (EPS). The production of EPS is affected by many factors, such as active secretion, cell lysis, and adsorption from the environment.13 An immediate increase in EPS was detected in a study where the carbon source was changed from acetate to phenol.14 This was a direct consequence of the decomposition of dead cells and could be one of the self-protection strategies for sludge in a phenol environment. EPS also play an important role in storing organic substrates and adsorbing pollutants in wastewater.15 The absorbed phenol is degraded through oxidization and then ring cleavage occurs. There are two cleavage pathways in aerobic condition, one is via the ortho pathway catalyzed by catechol 1,2 dioxygenase (C12O) and the other is via meta cleavage catalyzed by catechol 2,3 dioxygenase (C23O).16 meta cleavage is usually detected throughout the phenol degradation process, and coexistence of meta and ortho cleavage is found at a relatively higher phenol concentration.9 The shift in these two pathways is achieved by microbial regulation on enzyme activities in different operating conditions. Most metabolic activities take place in the presence of oxygen, and thus specific oxygen uptake rate (SOUR) could represent the bioactivity of microbes in the degradation. It is reported that the SOUR of sludge was significantly inhibited when the phenol concentration increased to 400 mg L−1, which contributed to the incomplete removal of phenol.11 Undoubtedly, several researchers independently studied the influence of phenol on a certain characteristic of activated sludge, whereas the coherence of these characteristics is hardly known. Since biodegradation is accomplished by a series of biochemical reactions, it is of great value to explore multiple changes in sludge characteristics in a consecutive process. Thus, a comprehensive investigation is still needed to further explain the phenol degradation mechanisms.
This study is designed to determine several aspects of the microbial dynamics during phenol degradation. Activated sludge is cultured and acclimated to degrade phenol with a concentration ranging from 60 to 350 mg L−1. The total DNA of the sludge is extracted to reveal the population succession and its impact on microbial metabolism. Metabolic activities such as SOUR, enzyme activities and EPS are studied to analyze their relationship with phenol removal ability. To the best of our knowledge, this is the first time that the phenol degradation process is elucidated by combining microbial degrading ability, populations and metabolic activities.
2. Material and methods
2.1 Reactor operation
Activated sludge was cultured in a lab-made reactor with a working volume of 3 L to investigate the removal of phenol (60–350 mg L−1). Synthetic nutrient solution was added to the bottom of the reactor using a peristaltic pump every two days (BJ100-2J, LongerPump). Phenol was the sole carbon source in the nutrient solution and its concentration was increased stepwise to 60 mg L−1, 120 mg L−1, 250 mg L−1 and 350 mg L−1. The composition of the nutrient medium is listed in Table 1. The hydraulic retention time (HRT) was 48 h. The temperature and pH of the reactor were kept constant at 25 °C and 7, respectively. Effluent from the upper part of the reactor was settled thoroughly and added back to the reactor. An air pump provided dissolved oxygen (3 mg L−1) for microbial growth. Activated sludge was sampled to analyse the SOUR, enzyme activities of C12O and C23O, EPS, hydrophobicity and populations until phenol was totally degraded in two days. All measurements were conducted in triplicate and then averaged.| Compound | Concentration |
|---|---|
| NH4Cl | 200 mg L−1 |
| K2HPO4 | 700 mg L−1 |
| KH2PO4 | 550 mg L−1 |
| CaCl2 | 2.5 mg L−1 |
| MgSO4·7H2O | 60 mg L−1 |
| FeCl3·6H2O | 5 mg L−1 |
| EDTA | 3.75 μg L−1 |
| MnSO4·H2O | 1.25 μg L−1 |
| CoCl2·6H2O | 0.25 μg L−1 |
| CaCl2 | 0.25 μg L−1 |
| ZnSO4·7H2O | 0.25 μg L−1 |
| CuSO4·5H2O | 0.025 μg L−1 |
| H3BO3 | 0.025 μg L−1 |
| Na2MoO4·2H2O | 0.025 μg L−1 |
| Phenol | 60–350 mg L−1 |
2.2 Seed biomass
Activated sludge was collected from the aerobic tank in a water purification plant in Shanghai, China. The sludge was washed with tap water and sieved with a screen mesh. Then, it was added to the reactor, and the initial MLSS was 3000 mg L−1 in the reactor.2.3 Chemical analysis
The concentrations of phenol and ammonium in the effluent of the reactor were measured every two days. The effluent was first filtered through a water phase needle filter (13 mm, 0.45 μm, SCAA-101, ANPEL Scientific Instrument Co. Ltd) for the measurement. The concentration of phenol was determined using a UV-vis spectrophotometer (UV-1800, MAPADA Instruments Co. Ltd) at 269 nm. Ammonium and MLSS were determined on the basis of standard methods.17 The total organic carbon (TOC) in the influent and effluent was measured using a TOC Analyzer (Multi N/C 3100, Analytik Jena Co. Ltd).2.4 Sludge morphology and hydrophobicity
Sludge morphology was observed via scanning electron microscopy (SEM, JSM-6700F, JEOL). Samples were washed with 0.2 mol L−1 phosphate buffer three times and allowed to settle down naturally. Then, the settled sludge was immersed in 10% methanal for 4 hours for fixation. Then, the sludge was dehydrated with 40%, 60%, 80% and 100% ethanol for 20 min. After freeze-drying, the sample was observed via SEM.18The hydrophobicity of the sludge was reflected by its contact angle. The same amount of sludge was retained on an aqueous filtering membrane (50 mm, 0.45 μm, SCBB-206 ANPEL Scientific Instrument Co. Ltd) and then dried naturally at room temperature; its water contact angle was measured using a Dropmeter A-200 contact angle system (MAIST Vision Inspection and Measurement).
2.5 DNA analysis
Sludge DNA was extracted using a soil DNA extraction kit (OMEGA Biotech) following the instructions from the manufacturer. The DNA of raw sludge and sludge adapted to 120 mg L−1, 250 mg L−1 and 350 mg L−1 phenol was marked as S_I, S_II, S_III and S_IV, respectively. Polymerase chain reaction (PCR) amplification, Illumina-MiSeq sequencing and population analysis were carried out according to previous descriptions.192.6 EPS measurements
Activated sludge was collected and centrifuged at 8000 rpm for 5 min to extract EPS. The supernatant was discarded and 0.05% NaCl was added to the sludge residue, and it was placed in a 60 °C water bath for 30 minutes. After centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min, the supernatant was filtered through a water phase needle filter (13 mm, 0.45 μm, SCAA-101, ANPEL Scientific Instrument Co. Ltd) and collected for EPS analysis. The content of proteins (PN) and humic acid (HA) was measured by a modified Lowry method using bovine serum albumin as the standard.20 The content of polysaccharides (PS) was measured by the anthrone–sulfuric method using glucose as the standard.21 Three-dimension excitation–emission matrix (3D-EEM) fluorescence spectra were obtained using a fluorescence spectrophotometer (F-7000, HITACHI). The range of the emission wavelength was set from 220 nm to 520 nm, and the range of the excitation wavelength was from 220 nm to 420 nm. The interval of emission and excitation was 5 nm with a scanning speed of 12
000 rpm for 15 min, the supernatant was filtered through a water phase needle filter (13 mm, 0.45 μm, SCAA-101, ANPEL Scientific Instrument Co. Ltd) and collected for EPS analysis. The content of proteins (PN) and humic acid (HA) was measured by a modified Lowry method using bovine serum albumin as the standard.20 The content of polysaccharides (PS) was measured by the anthrone–sulfuric method using glucose as the standard.21 Three-dimension excitation–emission matrix (3D-EEM) fluorescence spectra were obtained using a fluorescence spectrophotometer (F-7000, HITACHI). The range of the emission wavelength was set from 220 nm to 520 nm, and the range of the excitation wavelength was from 220 nm to 420 nm. The interval of emission and excitation was 5 nm with a scanning speed of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nm min−1.
000 nm min−1.
2.7 Enzyme assays and intermediate detection
Activated sludge was washed twice with phosphate buffer (0.1 mol L−1, pH 7.5), and then the sludge was disrupted on ice in 5 s treatments at 5 s intervals for 10 min using an ultrasonic cell crusher (XINYI Ultrasonic Equipment). The crude cell extract was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 20 min at 4 °C; then, the supernatant was subjected to C12O and C23O assays immediately as previously described.22,23 The C12O and C23O activities were measured on a Cary 60 UV-vis spectrophotometer (AGILENT Technologies) at 260 nm and 375 nm, respectively. The absorbance was auto-recorded every 30 s for 6 min at 25 °C. The specific activities of C12O and C23O were expressed as nanomoles catechol converted per minute per milligram protein.
000 rpm for 20 min at 4 °C; then, the supernatant was subjected to C12O and C23O assays immediately as previously described.22,23 The C12O and C23O activities were measured on a Cary 60 UV-vis spectrophotometer (AGILENT Technologies) at 260 nm and 375 nm, respectively. The absorbance was auto-recorded every 30 s for 6 min at 25 °C. The specific activities of C12O and C23O were expressed as nanomoles catechol converted per minute per milligram protein.
At the end of the experiment, the mixture in the reactor was sampled for intermediate identification by gas chromatography-mass spectrometry (GCMS-QP2010, SHIMAZDU) following a previously described procedure.24
2.8 SOUR test
Sludge was sampled and incubated in phenol solutions that were aerated thoroughly until saturated with dissolved oxygen (DO). A DO meter was used to record the change in DO concentration at 30 s intervals, and SOUR was calculated by dividing the slope of the DO decline curve with the MLSS.3. Results and discussion
3.1 Operation performance
Phenol and ammonium served as the only carbon and nitrogen sources for cell growth, and their removal efficiencies are depicted in Fig. 1(i). The periods with 60 mg L−1, 120 mg L−1, 250 mg L−1 and 350 mg L−1 phenol are marked as stage 1, 2, 3 and 4, respectively. It can be recognized that sludge was capable of completely degrading 60 mg L−1 and 120 mg L−1 phenol. However, when the phenol concentration increased to 250 mg L−1, the removal efficiency reduced significantly. Moreover, the settleability of the sludge became poor and overgrowth of filamentous microorganisms was further observed, as reflected in Fig. 1(ii). It is hypothesized that sludge bulking causes accumulation of phenol in the effluent. In order to reduce the filamentous bacteria, DO was adjusted to 1 mg L−1. Insufficient oxygen influenced the removal of phenol by the microorganisms, and led to the fluctuation shown in Fig. 1(i). Filamentous bacteria were gradually controlled, and 250 mg L−1 phenol was degraded completely in two days. The synthetic phenol was increased to 350 mg L−1 at day 78; thereafter, the removal efficiency of phenol in two days decreased. Similar result was obtained when un-acclimated activated sludge was fed with 300 mg L−1 phenol, where the system suffered severe sludge washout, which led to poor phenol removal.25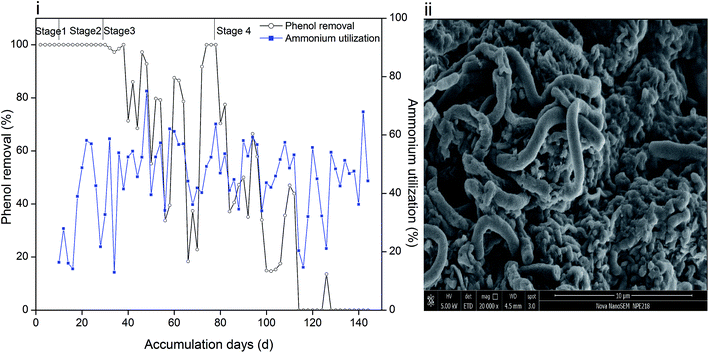 | ||
| Fig. 1 Removal efficiency of phenol and utilization of ammonium (i) and surface morphology of sludge (ii). | ||
The utilization efficiency of ammonium in stage 1 was about 20–30%, and then it increased to 40–60% and was maintained within the same range. As a toxic compound, phenol might inhibit the activity of ammonia oxidizing bacteria and lead to a decrease in ammonium oxidation efficiency. However, the inhibition could be overcome by acclimating sludge on the basis that a phenol concentration below 100 mg L−1 is acceptable for microorganisms.26 Ammonium utilization changed in the same trend with phenol removal in stage 3 and early period of stage 4, and the mole ratio of utilized phenol/ammonium was steady as observed from the calculations (ESI Fig. S1†). At the end of the operation, although phenol could not be degraded, ammonium oxidation could still provide energy for heterotrophic nitrifiers and denitrifiers and even act as a nutrient source for ammonium oxidizing bacteria. However, a high phenol loading severely inhibited the microbial activity and resulted in the fluctuation of ammonium utilization efficiency.
The TOC removal performance in each stage is summarized in Table 2. In stage 1–3, the measured TOC in the effluent was below 4 mg L−1, which indicates that the sludge oxidized phenol and its derivatives almost completely. However, 350 mg L−1 influent phenol caused severe organic pollution in the effluent, as reflected by the low TOC removal.
| Phenol concentration (mg L−1) | 60 | 120 | 250 | 350 |
| TOC in influent (mg L−1) | 49.6 | 101.5 | 199.8 | 280.4 |
| TOC in effluent (mg L−1) | 0.6 | 2.6 | 4 | 253.6 |
| TOC removal (%) | 98.8 | 97.7 | 98 | 10.6 |
3.2 Sludge metagenomic
Illumina-MiSeq sequencing was employed to detect the succession of microbe populations during the operation. After sequencing, 153![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 881 quality sequences (average length 395.45 bp) in total were obtained from 4 sludge samples. The RDP classifier was implemented to assign these sequences into 1040 OTUs with a 97% similarity limit. A summary of the sequencing details and microbe diversity estimators is presented in Table 3. The Good's coverage of all the samples was above 99%, which indicates that the sequencing process could represent the actual bacterial community compositions. The Ace and Chao indexes decreased largely with the degradation, which is indicative of reduced richness in biomass. There were higher Shannon values for S_I and S_II than those of S_III and S_IV and lower Simpson values for S_I and S_II than those of S_III and S_IV, which implies that the former samples were more diverse than the latter samples. It is believed that communities with diverse and sufficient microorganisms are more effective in biodegradation.27 On the contrary, simple communities lack the tolerance of utilizing toxic compounds particularly at high concentrations. In stage 1–2, the phenol concentration (less than 120 mg L−1) was acceptable to sludge, and the intermediates were catabolized by microorganisms immediately. Different microorganisms could utilize phenol and its intermediates and thus release inhibition of substrates. In stage 4, the incomplete phenol degradation might cause the accumulation of intermediates such as pyruvate, succinate, and acetate, and the accumulation of these intermediates also slowed down the phenol oxidation process. Therefore, a high phenol loading not only reduced the richness of some abundant populations, but also led to reduced diversity.
881 quality sequences (average length 395.45 bp) in total were obtained from 4 sludge samples. The RDP classifier was implemented to assign these sequences into 1040 OTUs with a 97% similarity limit. A summary of the sequencing details and microbe diversity estimators is presented in Table 3. The Good's coverage of all the samples was above 99%, which indicates that the sequencing process could represent the actual bacterial community compositions. The Ace and Chao indexes decreased largely with the degradation, which is indicative of reduced richness in biomass. There were higher Shannon values for S_I and S_II than those of S_III and S_IV and lower Simpson values for S_I and S_II than those of S_III and S_IV, which implies that the former samples were more diverse than the latter samples. It is believed that communities with diverse and sufficient microorganisms are more effective in biodegradation.27 On the contrary, simple communities lack the tolerance of utilizing toxic compounds particularly at high concentrations. In stage 1–2, the phenol concentration (less than 120 mg L−1) was acceptable to sludge, and the intermediates were catabolized by microorganisms immediately. Different microorganisms could utilize phenol and its intermediates and thus release inhibition of substrates. In stage 4, the incomplete phenol degradation might cause the accumulation of intermediates such as pyruvate, succinate, and acetate, and the accumulation of these intermediates also slowed down the phenol oxidation process. Therefore, a high phenol loading not only reduced the richness of some abundant populations, but also led to reduced diversity.
| Sample | Sequence | OTU | Ace | Chao | Shannon | Simpson | Coverage |
|---|---|---|---|---|---|---|---|
| S_I | 38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 655 655 |
678 | 785.3 | 793.5 | 5.3 | 0.0143 | 0.9980 |
| S_II | 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 552 552 |
768 | 745.3 | 755.7 | 4.8 | 0.0233 | 0.9959 |
| S_III | 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 583 583 |
248 | 322.5 | 330.5 | 2.4 | 0.2511 | 0.9972 |
| S_IV | 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 091 091 |
273 | 287.3 | 284.3 | 3.0 | 0.1710 | 0.9990 |
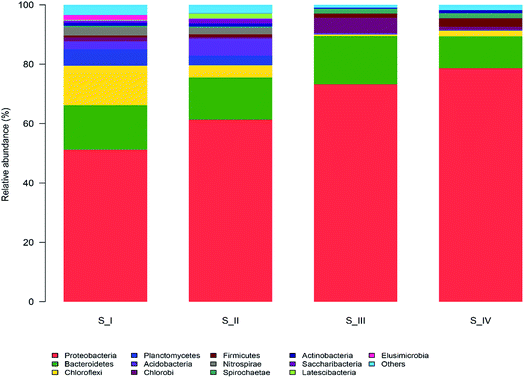
The taxonomic compositions of the microbial communities at the phylum level are illustrated in Fig. 2. There were 36 phyla identified and 13 of them with relatively higher abundance (more than 1%) were marked. The most abundant groups in the entire process were Proteobacteria (51.2–78.7%) and Bacteroidetes (10.7–16.3%). Proteobacteria was found to be the dominant member in several different activated sludge processes.6,12 Ammonium was the only nitrogen source provided and hence, microorganisms that can efficiently utilize ammonium would be more likely to survive in this condition. Many ammonia oxidizing bacteria (AOB) belong to the subclass of Proteobacteria, and this might be the reason why Proteobacteria was the largest group. Bacteroidaceae is often found to be the primary group in biogas digesters, trash ports, and contaminated soils. Reports suggest that Bacteroidetes are able to degrade a range of complicated carbon compounds, which made it possible for them to consume phenol in our reactor.27 There were also obvious proportional changes in the microflora among the four samples. Planctomycetes (3.3–5.6%), Acidobacteria (2.5–5.6%) and Nitrospirae (2.2–2.7%) were abundant in S_I and S_II, whereas these three groups diminished to less than 0.04% when phenol was increased to 250 mg L−1. Both Planctomycetes and Acidobacteria are major groups in traditional wastewater treatment plants; however, they failed to survive in the phenol environment. Nitrospirae is a group of bacteria that oxidizes nitrite, and the decrease of Nitrospirae is supposed to be related to the difficulty in nitrosation under high phenol concentration.28 The competition of electron acceptor between phenol and nitrite oxidation may give rise to the loss of nitrite oxidizing bacteria (NOB). On the contrary, some phyla seemed to be more suitable in high phenol conditions. Firmicutes and Spirochaetae only accounted for 0.6% and 0.5% in the sample of raw sludge, and they accounted for 2.8% and 1.7% of the total populations in 350 mg L−1 phenol, respectively. Bacteria belonging to the Firmicutes phylum have been reported to endure harsh environments due to the function of spores.29 Spirochaetae is seldom studied or discovered in the degradation of aromatic hydrocarbons; however, it took up 5% in a microcosm of petroleum refinery sludge, which is consistent with our results.30
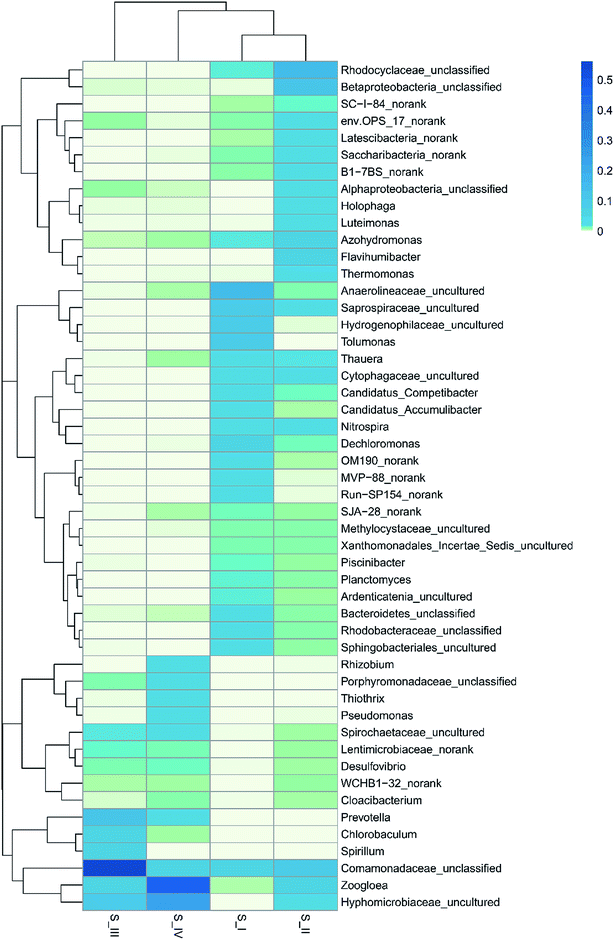
In order to deepen the understanding on microbe dynamics, a hierarchical heatmap analysis was performed based on microbial community profiles at the genus level. According to Fig. 3, the compositional successions in the microbial community structure in the four samples are distinct. Identification of Anaerolineaceae_uncultured (15.4%), Hydrogenophilaceae_uncultured (10.4%), Dechloromonas (5.4%), Saprospiraceae_uncultured (9.0%) and Planctomyces (1.6%) demonstrated a close agreement with reported genera in traditional activated sludge processes.31 Alphaproteobacteria_unclassified (2.7%), Betaproteobacteria_unclassified (11.0%), Rhodocyclaceae_unclassified (13.7%) and Nitrospira (3.2%) were identified to be the functional bacteria in S_II, and they also played an important role in nitrification. There were 2508 sequences belonging to Anaerolineaceae_uncultured in the raw sludge; however, after phenol acclimation, only 10 sequences remained. This might be because phenol cleavage took place in the presence of oxygen, and the aerobic condition was unfavorable for the growth of Anaerolineaceae bacteria. It was noticeable that Rhizobium (2.2%), Thiothrix (3.5%) and Pseudomonas (3.1%) genera with sequences of 491, 765 and 673, respectively, were largely produced in 350 mg L−1 phenol. Moreover, Zoogloea (45.1%) and Hyphomicrobiaceae_uncultured (24.7%) were the most dominant genera. Almost every genus that survived was linked to the nitrogen cycle except Rhizobium and Thiothrix. Rhizobia is a group of fungi that are capable of degrading phenol according to several literature focusing on phenol contaminated sites.32 In addition, Thiothrix was shown to be versatile in anaerobic conditions with or without nitrate.31 Since ammonium was the only nitrogen source in the reactor, nitrate reduction, ammonium and nitrite oxidation were associated with phenol degradation. Pseudomonas is one of the most common microorganisms that is effective in the biodegradation of phenolic substances; besides, it could realize biodenitrification of nitrate. It was found that Pseudomonas could utilize phenol and catechol simultaneously, and it also grew well on substrates such as formic acid.33,34 A widely accepted idea is that the family Hyphomicrobiaceae is related to Alphaproteobacteria, which is dominant and competitive under low-nutrient concentrations.35 Zoogloea plays a major part in traditional wastewater treatment owing to its strong ability to adsorb and oxidize organic matter in activated sludge and biofilm. Moreover, some strains belonging to Zoogloea were found out to be denitrifying bacteria, which means that they are able to consume carbohydrates and ammonium as elector donors in denitrification.36 It is believed that phenol degradation and denitrification cooperated until phenol concentration was increased to 350 mg L−1, and thus contributed to the analogous trend in the removal of ammonium and phenol. However, the denitrification showed superiority over phenol degradation at the end of the operation, which is probably related to the alternative carbon source and low DO. Phenol toxicity poses a huge threat to cell activity, and decomposed sludge could be used as inner carbon for denitrification. Besides, the limited DO was more favorable for denitrification than phenol oxidation. Some researchers believe that ammonium removal and phenol degradation can be accomplished simultaneously in AOB and NOB, but opinions are divided since these two processes might also compete with each other over electron acceptor when oxygen is insufficient.37 In our results, denitrification and phenol degradation were achieved simultaneously until phenol concentration was increased to 350 mg L−1; however, high phenol concentration (350 mg L−1) poses a huge threat to sludge activity and phenol oxidation.
3.3 EPS content
The variation in sludge EPS under different phenol concentrations is presented in Fig. 4(i). Proteins (PN), polysaccharides (PS) and humic acid (HA) were the main components in the EPS, and their content and proportions changed with phenol concentration. The PN content increased from 7.0 mg g−1 MLSS to 9.9 mg g−1 MLSS when phenol concentration increased from zero to 120 mg L−1 and then decreased to 6.4 mg g−1 MLSS at the end of the operation with 350 mg L−1 phenol. This is in accordance with other studies suggesting that the PN content is reduced when sludge is exposed to toxic compounds such as 4-chlorophenol and 2,4,6-trichlorophenol.38,39 The PS content decreased from 17.0 mg g−1 MLSS to 10.4 mg g−1 MLSS with the initial addition of 60 mg L−1 phenol. In the following days, the release of PS increased to 18.1 mg g−1 MLSS in 250 mg L−1 phenol and then exhibited an obvious decline to 1.0 mg g−1 MLSS in 350 mg L−1 phenol. PS is important for providing internal carbon in EPS, and the large consumption of PS finally manifested the inability of sludge to use 350 mg L−1 phenol.40 The content of HA in raw sludge was 12.0 mg g−1 MLSS, which increased to 20.6 mg g−1 MLSS when sludge was fed with 250 mg L−1 phenol. The dramatic growth was intensified in the last stage of the operation, where the HA content increased to 95.8 mg g−1 MLSS. As complicated organic matter was probably produced from dead substrates, HA was probably accumulated from the high decomposition of microorganisms in 350 mg L−1 phenol.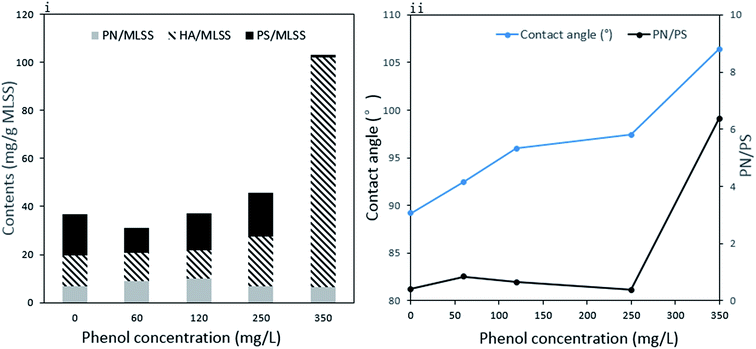 | ||
| Fig. 4 Compositions of extracellular polymeric substances (i) and contact angle of sludge and ratio of PN to PS in EPS (ii). | ||
The total EPS content showed a slight decrease from 36.8 mg g−1 MLSS to 31.1 mg g−1 MLSS with the addition 60 mg L−1 phenol and then gradually increased to 103.2 mg g−1 MLSS in 350 mg L−1 phenol. It is clear that the decrease in total EPS in 60 mg L−1 phenol was mainly caused by reduced PS. The difficulty in utilizing external toxic compounds at the beginning might compel microorganisms to consume their inner sources. EPS, which are carbon and energy reserves, could be utilized by microorganisms when the sludge was initially fed with phenol. Our result is similar to other reports concluding that the immediate replacement of an ordinary carbon source to phenol would lead to the reduced production of EPS.39,41 The release of PS could be recovered when the sludge was adapted to low concentrations of phenol. However, 350 mg L−1 phenol severely inhibited the sludge activity, and even caused significant decomposition of bacteria. The large amount of HA produced from cell decomposition contributed to the accelerated release of EPS at the end of the operation. External substances are first absorbed by EPS and then degraded. Thus, EPS play an important role in adsorbing pollutants and developing a protective shield under harsh environments.42 The increased EPS could provide a more protective barrier around the microorganisms and relieve phenol toxicity to their cells, which is important for the survival of microorganisms at high phenol loading.
In addition, EPS have key influence on the hydrophobicity of microbes.43 To the best of our knowledge, amino acids with hydrophobic parts mostly come from PN in EPS, whereas PS contains some hydrophilic functional groups, such as carboxyl and hydroxyl. As the sludge was surrounded with EPS, there is a positive correlation between cell hydrophobicity and the ratio of PN to PS in EPS. The variations in the contact angle and PN/PS are expressed in Fig. 4(ii). The raw sludge possessed a slightly hydrophilic property and became more and more hydrophobic as the degradation proceeded. An obvious increase in contact angle was detected from 89.2° to 106.4°, whereas PN/PS showed the same tendency. Hydrophobicity plays a major role in the adhesion of organic compounds to cells and cell aggregation. Proper hydrophobic property benefits interactions between cells and the liquid phase. In our study, the change in surface property made it difficult for phenol to attach to cells, which is one of the microbial responses to improve tolerance against toxicity. However, the slight adsorption of phenol by the microorganisms resulted in the high phenol effluent and significant inhibition of the phenol degradation process. A comparable conclusion was drawn from a study using phenol and mono-chlorophenols to feed Pseudomonas putida CP1, where an increase in hydrophobicity prompted cell aggregation under high substrate toxicity.44
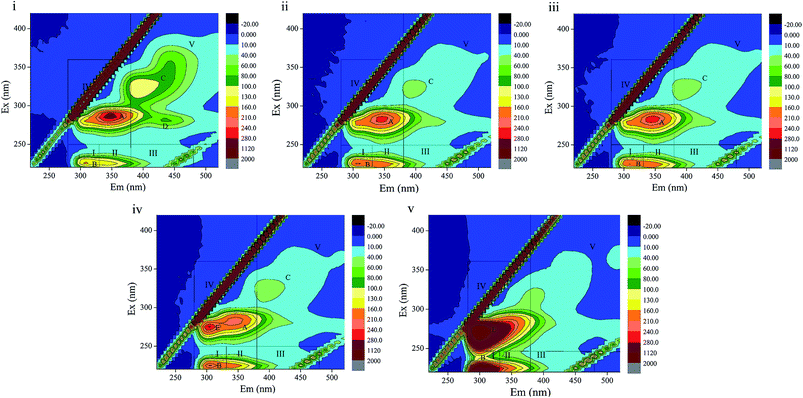
3.4 3D-EEM fluorescence spectra
3D-EEM is a feasible and common method to visualize fluorescence variations in EPS contents. The fluorescence spectra and peak information of sludge EPS from the different stages are presented in Fig. 5 and Table 4. There are four peaks, A, B, C and D, in EEM without phenol (Fig. 5(i)), which are assigned to soluble microbial by-product-like matter (peak A), tyrosine protein-like matter (peak B) and humic acid-like matter (peak C and D). In Fig. 5(ii), peak D located at the excitation/emission wavelengths (Ex/Em) of 280/440 nm disappeared when phenol was added. In the following days, a new peak E, which was observed at the Ex/Em of 275/305 nm occurred with the addition of 250 mg L−1 phenol and enhanced in 350 mg L−1 phenol. This indicates the generation of tyrosine protein-like matter in a higher phenol concentration. Peak A and C disappeared in 350 mg L−1 phenol, which reveals that parts of the humic acid-like matter and soluble microbial by-product-like matter were substituted by tryptophan protein-like matter. Regions in the EEM spectra are utilized to represent the biodegradable (tyrosine-like and soluble microbial by-product-like) property and non-biodegradable (tryptophan-like, fulvic acid-like, and humic acid-like) property of substrates in EPS.19 The quenching effect of 350 mg L−1 phenol on soluble microbial by-product-like matter supports the opinion that EPS were consumed by sludge during starvation. In addition, the generation of tryptophan protein-like matter indicates difficulty in phenol degradation.| Phenol influent (mg L−1) | Peaks | Ex/Em | Composition |
|---|---|---|---|
| 0 | A | 285/345 | Soluble microbial by-product-like matter |
| B | 225/310 | Tyrosine protein-like matter | |
| C | 325/395 | Humic acid-like matter | |
| D | 280/400 | Humic acid-like matter | |
| 60 | A | 285/345 | Soluble microbial by-product-like matter |
| B | 225/305 | Tyrosine protein-like matter | |
| C | 320/395 | Humic acid-like matter | |
| 120 | A | 285/345 | Soluble microbial by-product-like matter |
| B | 225/310 | Tyrosine protein-like matter | |
| C | 325/395 | Humic acid-like matter | |
| 250 | A | 285/345 | Soluble microbial by-product-like matter |
| B | 225/305 | Tyrosine protein-like matter | |
| C | 325/395 | Humic acid-like matter | |
| E | 275/305 | Tryptophan protein-like matter | |
| 350 | B | 220/300 | Tyrosine protein-like matter |
| E | 275/300 | Tryptophan protein-like matter |
The peak locations in EEM varied within 5 nm either in emission wavelength or excitation wavelength. Peak B displayed a blue shift in 60 mg L−1 phenol compared with that of the raw sludge, which evidences the break-up of some structures such as aromatic rings and conjugated bonds. Then peak B displayed a red shift when phenol was increased to 120 mg L−1, which manifests the presence of functional groups such as amino, carbonyl and hydroxyl.45,46 An identical observation was also noted in for humic acid-like matter (peak C). In summary, the fluorescence spectra variation clearly indicates that the increase in phenol had a profound influence on component formations in EPS.
3.5 Enzyme activities
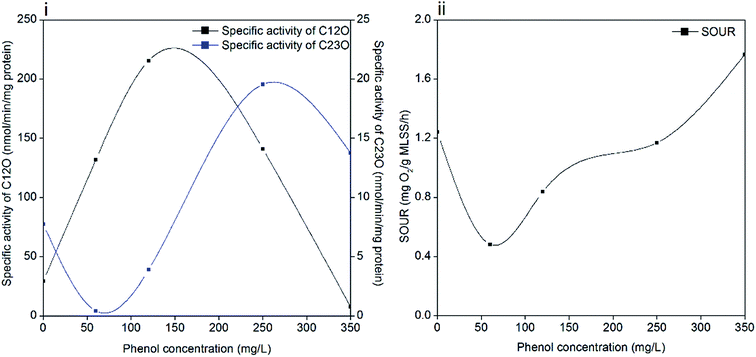
Phenol degradation is accomplished by a series of biochemical reactions catalyzed by different enzymes. Catechol is formed in the first step and subsequently, it is catalyzed to cis,cis-muconic acid by C12O (ortho pathway) or 2-hydroxymuconic semialdehyde (2-HMSA) by C23O (meta pathway).47 In the ortho pathway, succinate and acetyl-coenzyme A are produced in the primary assimilatory steps, and then oxidized completely through the tricarboxylic acid cycle (TCA). While the meta pathway leads to the formation of acetaldehyde, pyruvate and formate during primary assimilation, and then the central carbon precursor 3-phosphoglycerate (PGA) is generated through the glyoxylate cycle with sufficient reduction equivalents.16 The activities of C12O and C23O during the entire degradation process are illustrated in Fig. 6(i). The dioxygenases activities were significantly influenced by the increased phenol loading. The specific activity of C12O increased to the maximum value of 215.5 nmol per min per mg protein when the phenol concentration was 120 mg L−1, followed by a decline afterwards. The specific activity of C23O was barely detected until the phenol concentration was increased to 250 mg L−1, and it reached the maximum value of 19.55 nmol per min per mg protein at the time. In stage 4, the specific activity of C23O decreased to 13.8 nmol per min per mg protein and the specific activity of C12O was 7.6 nmol per min per mg protein. Pyruvic acid and succinic acid were found in the identification of phenol degradation intermediates (ESI Fig. S2†), which validates the meta and ortho pathways.It is found that some strains degrade phenol through the meta and ortho pathways, while others only through one pathway. A number of species of Pseudomonas possess these two enzyme-coding genes. The Rhodococcus sp. strain PNAN5 was identified to express C12O activities, whereas Rhodococcus opacus R7 showed C23O activities in phenol degradation.32 Millions of microorganisms from different species in activated sludge make it possible for the coexistence of two dioxygenases. Besides, the shifting of meta and ortho cleavage is in accordance with the opinion that the degradation pathways can change with a variation in initial substrate concentrations. A study suggested that Pseudomonas putida ATCC 49451 degraded catechol mainly through the ortho pathway, and simultaneous activation of meta and ortho cleavages was observed at sodium benzoate concentrations of 300 mg L−1.48 In our study, ortho cleavage prevailed until the phenol concentration increased to 350 mg L−1, and then meta cleavage showed superiority over ortho cleavage. These two pathways are related with the different energy production capabilities of microbes under different conditions. With excess phenol, sludge can shift energy production from complete oxidization to incomplete oxidization to produce a higher growth rate, which is achieved by shifting ortho cleavage to meta cleavage. This regulation was effective in maintaining microbial activity but inefficient in phenol catabolism. As a result, the degradation process was inhibited significantly in 350 mg L−1 phenol, as confirmed by the poor removal performance. Another reason is also hypothesized to be linked with the DO concentration in the latter stage of operation. Oxygen served as an electron acceptor in aerobic degradation; however, DO decreased to prevent sludge bulking when the phenol concentration increased to 250 mg L−1 and afterwards. Insufficient oxygen restrained phenol degradation process while ammonium degradation was less affected.
3.6 Specific oxygen uptake rate
Phenol degrading microorganisms mainly live in aerobic environments, therefore, SOUR can further represent the specific activities of sludge in the reactor. According to Fig. 6(ii), SOUR declined to 0.48 mg O2 per g MLSS per h when phenol was initially added, which was one-third of the original value (1.24 O2 per g MLSS per h). Then SOUR gradually recovered to 1.76 mg O2 per g MLSS per h in the last stage of operation with 350 mg L−1 phenol.The decrease in SOUR was consistent with the decrease in EPS in 60 mg L−1 phenol, which indicates that phenol addition inhibited cell activity. This observation is also in accordance with studies concluding that even in phenol concentrations less than 100 mg L−1, the SOUR of sludge was inhibited because of phenol toxicity.49,50 The subsequent increase in SOUR reflects the effectiveness of the acclimation process in maintaining sludge activity. It is noteworthy that a further increase in phenol loading did not weaken the bioactivity of sludge, which might be attributed to the survival of denitrifying bacteria and protective shield formed by EPS. The improved tolerance of sludge to phenol was also supported by the finding that sludge adapted to even low phenol concentrations is less susceptible to the phenol loading.9,51 However, the sludge with high bioactivity was unable to remove 350 mg L−1 phenol completely in two days, which is partly due the lack of oxygen. Without enough oxygen, both phenol degrading microorganisms and denitrifying bacteria are unable to oxidize phenol, as suggested in the poor removal efficiency of 350 mg L−1 phenol and low activities of C12O and C23O. Although the SOUR test implies that the sludge activity was high at this stage, the degradation of phenol was actually inhibited.
4. Conclusions
The response mechanisms of microorganisms to phenol were comprehensively investigated in this work. Phenol concentration affected the populations, metabolic activities and surface hydrophobicity of activated sludge, and thus affected the utilization of phenol. The maximum of 250 mg L−1 phenol could be degraded completely in two days after acclimation; however, more time is needed for the total removal of 350 mg L−1 phenol. The result of bacterial community succession suggests that Proteobacteria (51.2–78.7%) and Bacteroidetes (10.7–16.3%) were dominant groups in the degradation. Zoogloea, Hyphomicrobiaceae and other denitrifying bacteria were gradually accumulated with an increase in phenol; however, their ability to oxidize phenol was significantly inhibited under high phenol loading. This was concluded from the low dioxygenases activities and high SOUR value in the last stage of operation. The toxicity of phenol also caused a distinct decrease in the diversity and richness of the microbial communities, which is attributed to the accelerated generation of HA in EPS. Cell hydrophobicity increased with phenol concentration and might have caused substrate adsorption in sludge to be difficult. Meanwhile, the biodegradable soluble microbial by-product-like matter in EPS was gradually replaced by non-biodegradable tryptophan protein-like matter.Conflicts of interest
The authors have no conflicts of interest to declare.Acknowledgements
This work received support from the National Natural Science Foundation of China (No. 21577089), the Sinopec Zhenhai refining and chemical company (314059), and the Major Science and Technology Program for Water Pollution Control and Treatment (2014ZX07206001).References
- J. Michałowicz and W. Duda, Pol. J. Environ. Stud., 2007, 16, 347–362 Search PubMed.
- P. M. van Schie and L. Y. Young, Biorem. J., 2000, 4, 1–18 CrossRef CAS.
- P. M. Fedorak, S. E. Hrudey and D. L. Wise, Biotreatment Systems, 1988 Search PubMed.
- L. Yotova, I. Tzibranska, F. Tileva, G. H. Markx and N. Georgieva, J. Ind. Microbiol. Biotechnol., 2009, 36, 367 CrossRef CAS PubMed.
- L. Gianfreda and J.-M. Bollag, Isolated enzymes for the transformation and detoxification of organic pollutants, Marcel Dekker, New York, 2002 Search PubMed.
- Y. Jiang, L. Wei, H. Zhang, K. Yang and H. Wang, Bioresour. Technol., 2016, 218, 146 CrossRef CAS PubMed.
- J. Rueyshin, W. C. Huang and H. Yahan, J. Hazard. Mater., 2009, 164, 46–52 CrossRef PubMed.
- S. Y. Lee, Y. N. Chun and S. I. Kim, J. Ind. Eng. Chem., 2009, 15, 323–327 CrossRef CAS.
- J. W. Lim, C. E. Seng, P. E. Lim, S. L. Ng, K. C. Tan and S. L. Kew, Water SA, 2012, 39, 695–700 CrossRef.
- A. P. A. Alves, P. S. Lima, M. Dezotti and J. P. Bassin, Int. Biodeterior. Biodegrad., 2017, 123, 146–155 CrossRef CAS.
- M. L. Leong, K. M. Lee, S. O. Lai and B. S. Ooi, Desalination, 2011, 270, 181–187 CrossRef CAS.
- D. R. Joshi, Y. Zhang, Z. Tian, Y. Gao and M. Yang, Appl. Microbiol. Biotechnol., 2016, 1–12 CAS.
- J. Wingender, T. R. Neu and H. C. Flemming, Microb. Extracell. Polym. Subst., 1999, 45–53 Search PubMed.
- C. Wu, Y. Zhou and J. Song, Water Sci. Technol., 2016, 73, 2324 CrossRef CAS PubMed.
- G. P. Sheng, H. Q. Yu and X. Y. Li, Biotechnol. Adv., 2010, 28, 882 CrossRef CAS PubMed.
- B. Kiesel and R. H. Müller, Acta Biotechnol., 2002, 22, 221–234 CrossRef CAS.
- APHA, Standard Methods for the Examination of Water and Wastewater, American Public Health Association, Washington, DC, USA, 21st edn, 2005 Search PubMed.
- Y. P. Moy, J. H. Tay, S. K. Toh, Y. Liu and T. L. Tay, Lett. Appl. Microbiol., 2002, 34, 407–412 CrossRef PubMed.
- Y. Ding, Y. Tian, J. Liu, N. Li, J. Zhang, W. Zuo and Z. Li, Chem. Eng. J., 2016, 286, 198–207 CrossRef CAS.
- B. Frølund, T. Griebe and P. H. Nielsen, Appl. Microbiol. Biotechnol., 1995, 43, 755–761 CrossRef.
- J. Wu, H.-m. Zhou, H.-z. Li, P.-c. Zhang and J. Jiang, Water Res., 2009, 43, 3029–3036 CrossRef CAS PubMed.
- T. Nakazawa and A. Nakazawa, Methods Enzymol., 1970, 17, 518–522 Search PubMed.
- M. Nozaki, Methods Enzymol., 1970, 17, 522–525 Search PubMed.
- P. N. Polymenakou and E. G. Stephanou, Biodegradation, 2005, 16, 403–413 CrossRef CAS PubMed.
- T. L. Tay, Y. P. Moy, A. M. Maszenan and J. H. Tay, Appl. Microbiol. Biotechnol., 2005, 67, 708–713 CrossRef PubMed.
- C. Petronela, R. M. Hlihor, L. C. Apostol, D. Mariana, M. O. Pogacean and M. Gavrilescu, Environ. Eng. Manage. J., 2012, 11, 2053–2058 Search PubMed.
- A. S. Burns, C. W. Pugh, Y. T. Segid, P. T. Behum, L. Lefticariu and K. S. Bender, Biodegradation, 2012, 23, 415–429 CrossRef CAS PubMed.
- R. Yao, H. Yang, M. Yu, Y. Liu and H. Shi, RSC Adv., 2016, 6, 113959–113966 RSC.
- X. Zhuang, Z. Han, Z. Bai, G. Zhuang and H. Shim, Environ. Pollut., 2010, 158, 1119–1126 CrossRef CAS PubMed.
- J. Sarkar, S. K. Kazy, A. Gupta, A. Dutta, B. Mohapatra, A. Roy, P. Bera, A. Mitra and P. Sar, Front. Microbiol., 2016, 7, 1407 Search PubMed.
- M. Wagner, A. Loy, R. Nogueira, U. Purkhold, N. Lee and H. Daims, Antonie van Leeuwenhoek, 2002, 81, 665–680 CrossRef CAS PubMed.
- H. Chen, Y. Y. Fang, X. H. Yin and X. D. Wei, J. Anhui Agric. Sci., 2015, 43, 201–205 Search PubMed.
- A. Kumar, S. Kumar and S. Kumar, Biochem. Eng. J., 2005, 22, 151–159 CrossRef CAS.
- M. M. B. Martín, J. A. S. Pérez, F. G. A. Fernández, J. L. G. Sánchez, J. L. C. López and S. M. Rodríguez, J. Hazard. Mater., 2008, 151, 780–788 CrossRef PubMed.
- K. Lautenschlager, C. Hwang, F. Ling, W. T. Liu, N. Boon, O. Köster, T. Egli and F. Hammes, Water Res., 2014, 62, 40 CrossRef CAS PubMed.
- T. Lukow and H. Diekmann, Biotechnol. Lett., 1997, 19, 1157–1159 CrossRef CAS.
- J. Lu, Q. Jin, Y. He, X. He and J. Zhao, Water, Air, Soil Pollut., 2014, 225, 1–10 CAS.
- Z. Bao, X. Chen, J. Zhao, F. Lin, J. Li and Y. Zhang, Int. Biodeterior. Biodegrad., 2016, 110, 24–31 CrossRef CAS.
- M. S. Miao, Y. J. Zhang, L. Shu, J. Zhang, Q. Kong and N. Li, Int. Biodeterior. Biodegrad., 2014, 95, 61–66 CrossRef CAS.
- Z. W. Wang, Y. Liu and J. H. Tay, Appl. Microbiol. Biotechnol., 2005, 69, 469–473 CrossRef CAS PubMed.
- D. Onbasli and B. Aslim, J. Hazard. Mater., 2009, 168, 64–67 CrossRef CAS PubMed.
- C. S. Laspidou and B. E. Rittmann, Water Res., 2002, 36, 2711–2720 CrossRef CAS PubMed.
- B. M. Wilén, B. Jin and P. Lant, Water Res., 2003, 37, 2127–2139 CrossRef.
- A. Farrell and B. Quilty, J. Ind. Microbiol. Biotechnol., 2002, 28, 316–324 CrossRef CAS PubMed.
- W. Chen, P. Westerhoff, J. A. Leenheer and K. Booksh, Environ. Sci. Technol., 2003, 37, 5701–5710 CrossRef CAS PubMed.
- L. F. Ren, S. Q. Ni, C. Liu, S. Liang, B. Zhang, Q. Kong and N. Guo, Environ. Sci. Pollut. Res., 2015, 22, 2925–2934 CrossRef CAS PubMed.
- R. H. Müller and W. Babel, Appl. Microbiol. Biotechnol., 1996, 46, 156–162 CrossRef.
- K. C. Loh and S. S. Chua, Enzym. Microb. Technol., 2002, 30, 620–626 CrossRef CAS.
- V. J. Inglezakis, S. Malamis, A. Omirkhan, J. Nauruzbayeva, Z. Makhtayeva, T. Seidakhmetov and A. Kudarova, J. Environ. Manage., 2016, 203, 825–830 CrossRef PubMed.
- N. A. Yusoff, S. A. Ong, L. N. Ho, Y. S. Wong, W. F. Khalik and S. L. Lee, Biochem. Eng. J., 2016, 115, 14–22 CrossRef CAS.
- J. W. Lim, J. Z. Tan and C. E. Seng, Appl. Water Sci., 2013, 3, 515–525 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra09225c |
| This journal is © The Royal Society of Chemistry 2017 |