Open Access Article
Open Access ArticleSynthesis of a poly(Gd(III)-DOTA)–PNA conjugate as a potential MRI contrast agent via post-synthetic click chemistry functionalization†
Xiaoxiao Wanga,
Mark Milnea,
Francisco Martínezb,
Timothy J. Schollb and
Robert H. E. Hudson *a
*a
aDepartment of Chemistry, The University of Western Ontario, 1151 Richmond St., London, Ontario N6A 5B7, Canada. E-mail: rhhudson@uwo.ca
bDepartment of Medical Biophysics, The Robarts Research Institute, The University of Western Ontario, 1151 Richmond St., London, Ontario N6A 5B7, Canada
First published on 22nd September 2017
Abstract
Herein we present a method for the simultaneous, multiple conjugation of alkynyl-(Gd(III)-DOTA) moieties to a PNA oligomer possessing a domain of azide residues. This is achieved in a single Huisgen type copper-catalyzed azide alkyne cycloaddition (CuAAC) reaction after the oligomer assembly is completed. A triplex formed between the labelled PNA and riboadenylic acid, [(Gd(III)-DOTA)4–PNA]2:poly(rA), displayed higher relaxivities at any field strength compared to the single-stranded probe at the same concentration of Gd3+ ions as determined by nuclear magnetic resonance dispersion (NMRD) studies.
Over the past two decades, molecular imaging has been pursued utilizing various modalities including magnetic resonance imaging (MRI), single photon emission tomography (SPECT), positron emission tomography (PET), fluorescence and bioluminescence imaging. Although SPECT/PET techniques are very sensitive, the requirement for radioactive agents and the attendant risks limits their use to justifiably serious applications and not routine screening/diagnoses. Fluorescence and bioluminescence imaging have been widely employed, but are limited by fundamental drawbacks, such as low efficiency of light transmission through tissues, difficulty in quantification, and signal interference from light absorption by haemoglobin and other biomolecules.1 Therefore, by comparison, MRI has the advantage of being a non-invasive imaging technique that provides high spatial resolution, deep tissue penetration, an unsurpassed ability to distinguish soft tissues and the ability to image subjects repeatedly. With these aforementioned advantages, MRI has proved highly useful in clinical diagnoses; however, the need for development of new contrast agents (CAs) to improve specificity and enhance sensitivity remains.2,3
To date, small molecular Gd3+ chelates are routinely used for clinical imaging to enhance image contrast by shortening the relaxation times of proximal water protons. Modifications of Gd3+-based CAs have been made in attempts to improve the low molar relaxivity (r1) of chelated gadolinium. For example, clinically used small molecule contrast agents have been conjugated to high molecular weight macromolecules including proteins,4 polymers,5 self-assembled peptide amphiphile nanofibers6 and oligonucleotides.7 These efforts have resulted in the significant enhancement of r1. Additionally, the development of targeted MR contrast agents is of increasing importance, fuelled by the continual discovery of novel molecular targets such as DNA or RNA sequences, or proteins/enzymes involved in specific pathologies.
We were interested to investigate the possibility of producing a generically targeted probe that could report on global mRNA anabolism. Since the 3′-ends of all fully processed eukaryotic mRNAs, with the exception of most histone genes, have a polyadenylic acid [poly(rA)] tail, this was envisioned as a target that could serve as a scaffold for assembling a relatively high density of labels via triplex formation. The poly(rA) of mRNA consists of 200–250 adenosines initially in the nucleus, and is shortened to ca. 50 adenosines in the cytosol. The tail plays a significant role in mRNA stability, translation and transport.8–12 The polyadenylation of mRNA is catalysed by the enzyme poly(rA) polymerase (PAP) and by neo-PAP, a recently identified human poly(rA) polymerase, that is significantly over-expressed in human cancer cells.13,14 This suggests the polyadenylated RNA as a potential malignancy selective target for the development of hybridization probes.15 Some small molecules, like the protoberberine alkaloids, have been found to bind to poly(rA) with high affinity leading to self-structure formation which has been proposed as a method to interfere with the normal biological functioning of mRNA.15–17
Using peptide nucleic acid (PNA) as a targeting agent offers the advantages of high affinity, specificity, biological stability. There are a limited number of examples of PNA–DOTA chelator conjugates for use in imaging by targeting mRNA.18–21 While gene specific agents have appeared,19 a poly(rA) targeted agent has not. Thus, in this work, a PNA hybridization probe containing and integral, chelated Gd3+ as a CA for targeting poly(rA) has been prepared and characterized.
Generally, there are two strategies for labelling of PNA with Gd3+ chelates currently in use, these being either incorporation of a DOTA monomer during the synthesis, often as a side-chain modified lysine, or as a terminal residue.22 Additionally, the attachment of a Gd(III)-DOTA-containing monomer may be done during solid-phase oligomerization or, alternatively, in a post-synthetic labelling step that can take place either in solution or on the solid-phase resin. As an example of the first approach, the use of a Fmoc-Lys(DOTA)-OH was used during the solid-phase synthesis of a PNA peptide chimera. The resulting DOTA-labelled PNA was then radio-metalled.20 Alternatively, a post-synthetic labelling of PNA with chelators whilst on solid-phase can be done as exemplified by the work of Wickstrom.18 N-terminal labelling on solid support was achieved using DOTA-(t-OBu)3COOH. After removal from the resin, a Gd3+ chelation step was required to produce a labelled probe. All of these procedures perform the metalation step after the complete assembly of the PNA-chelator conjugate. While the chelation can be quite efficient, the conditions can be somewhat forcing in terms of temperature and pH, in some instances, and preclude the use of sensitive functionalities. Therefore, an approach that is chemoselective, robust, and tolerant to functionalities on PNA is still needed.
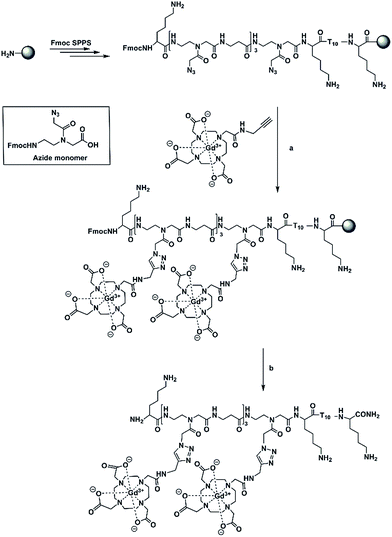
Based on our previous work on the post-synthetic functionalization of PNA,23 we have pursued a labelling strategy utilizing copper-catalysed click chemistry on polymer supported targets.24 Although most examples of CuAAC applied to the derivatization of PNA are performed in solution, we have focused our attention on polymer-supported CuAAC because it combines advantages of click chemistry and that of SPPS in which an excess of reactants can be used and easily washed away from the solid support. As well, the post-synthetic functionalization strategy reduces the overall synthetic effort as well as introducing a step that is amenable to structural diversification. In order to pursue this approach, an Fmoc-azide PNA monomer (Az) in which a nucleobase was replaced with an azide group (Scheme 1), has been synthesized.25
Accumulation of Gd3+-containing probes at the target site is usually a main concern owing to the limited concentration of the specific mRNAs in cancer cells. This has been addressed in Wickstrom's study in which multiple chelators have been conjugated to a PNA–peptide dendrimer.18,26 They demonstrated that the relaxivity per probe rose as the number of Gd3+ per probe rose.
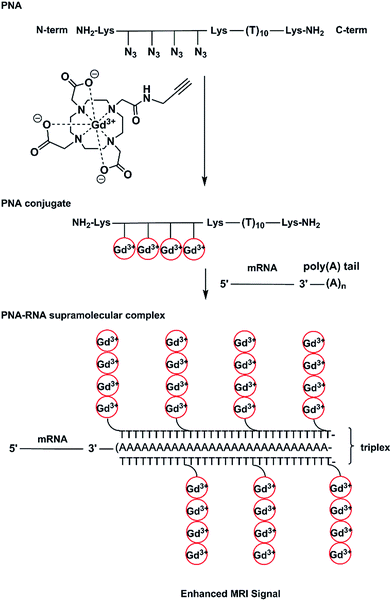
In our work, we have prepared a PNA sequence including four azide monomers which forms the labelling domain and a thymine decamer which forms a poly(rA) targeting domain.27–29 The azide units are spaced by β-alanine residues to avoid steric interactions between chelators. Alkynyl-(Gd(III)-DOTA) chelates have been attached to PNA via on-resin CuAAC to yield a hybridization probe, (Gd(III)-DOTA)4–PNA (Scheme 1). The PNA was designed to bind to poly(rA) sequences and form a comb-like triplex structure that could also serve to concentrate Gd3+ ions in the microenvironment of the target.
Using this approach a relatively simple and small construct can form a large supramolecular complex (Fig. 1).
In order to optimize the derivatization chemistry, we first synthesized a test sequence, Fmoc-TTT-Az-K (PNA1, K = Lys = L-lysine), containing one azide unit (Az, Scheme 1). Without purification, the crude resin-bound PNA1 was placed in a manual peptide synthesis vessel and alkynyl-(Gd(III)-DOTA)30–32 (30 equiv.), CuSO4·5H2O (60 equiv.), sodium ascorbate (240 equiv.), and the ligand tris(3-hydroxypropyltriazolylmethyl)amine (THPTA) (60 eq.) were added and isopropanol/H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v) was used as the solvent. After the resin was shaken at room temperature (12 h), the resin was washed and treated under standard cleavage/deprotection conditions. Analysis of the residue LC/MS methods indicated successful attachment of Gd(III)-DOTA moieties and complete consumption of azide-possessing PNA1 to produce a conjugate with mass of 973.4175 [M + 2H]2+ and 648.9204 [M + 3H]3+ (calculated 973.5574 [M + 2H]2+ and 649.3742 [M + 3H]3+). We then proceeded to prepare a PNA containing four azide inserts, PNA2 (Fmoc-K-Az-β-Az-β-Az-β-Az-K-TTTTTTTTTT-K) (Scheme 1). A small portion of PNA was cleaved off the resin and characterization was performed by reversed phase HPLC and HRMS (ESI-TOF). The observed ions at mass values of 1411.6052 [M + 3H]3+ and 1058.9668 [M + 4H]4+ agree well with the calculated masses of 1411.7679 [M + 3H]3+ and 1059.0779 [M + 4H]4+. Proceeding without purification, the resin-bound PNA2 was subjected to CuAAC in the presence of excess alkynyl-(Gd(III)-DOTA) to yield Fmoc(Gd(III)-DOTA)4–PNA (Fmoc-K-Gd-β-Gd-β-Gd-β-Gd-K-TTTTTTTTTT-K) in quantitative conjugation yield. The excess reagents, solvents, and catalyst were easily washed from the resin, whereas the workup to remove these materials using solution phase methods can be laborious, in our experience. HRMS (ESI-TOF) analysis of the Fmoc(Gd(III)-DOTA)4–PNA conjugate showed ions with masses of 1324.0796 [M + 5H]5+ and 1103.4063 [M + 6H]6+ which are in accordance with the calculated masses of 1324.0326 [M + 5H]5+ and 1103.5285 [M + 6H]6+. After Fmoc deprotection and cleavage of the oligomer from the resin, only the usual reversed phase HPLC purification was required. Characterization of purified (Gd(III)-DOTA)4–PNA (K-Gd-β-Gd-β-Gd-β-Gd-K-TTTTTTTTTT-K) via HRMS (ESI-TOF) showed masses at 914.5258 [M + 7H]7+ and 710.6509 [M + 9H]9+ corresponding to the calculated masses of 914.2765 (M + 7H)7+ and 711.3279 [M + 9H]9+.
2, v/v) was used as the solvent. After the resin was shaken at room temperature (12 h), the resin was washed and treated under standard cleavage/deprotection conditions. Analysis of the residue LC/MS methods indicated successful attachment of Gd(III)-DOTA moieties and complete consumption of azide-possessing PNA1 to produce a conjugate with mass of 973.4175 [M + 2H]2+ and 648.9204 [M + 3H]3+ (calculated 973.5574 [M + 2H]2+ and 649.3742 [M + 3H]3+). We then proceeded to prepare a PNA containing four azide inserts, PNA2 (Fmoc-K-Az-β-Az-β-Az-β-Az-K-TTTTTTTTTT-K) (Scheme 1). A small portion of PNA was cleaved off the resin and characterization was performed by reversed phase HPLC and HRMS (ESI-TOF). The observed ions at mass values of 1411.6052 [M + 3H]3+ and 1058.9668 [M + 4H]4+ agree well with the calculated masses of 1411.7679 [M + 3H]3+ and 1059.0779 [M + 4H]4+. Proceeding without purification, the resin-bound PNA2 was subjected to CuAAC in the presence of excess alkynyl-(Gd(III)-DOTA) to yield Fmoc(Gd(III)-DOTA)4–PNA (Fmoc-K-Gd-β-Gd-β-Gd-β-Gd-K-TTTTTTTTTT-K) in quantitative conjugation yield. The excess reagents, solvents, and catalyst were easily washed from the resin, whereas the workup to remove these materials using solution phase methods can be laborious, in our experience. HRMS (ESI-TOF) analysis of the Fmoc(Gd(III)-DOTA)4–PNA conjugate showed ions with masses of 1324.0796 [M + 5H]5+ and 1103.4063 [M + 6H]6+ which are in accordance with the calculated masses of 1324.0326 [M + 5H]5+ and 1103.5285 [M + 6H]6+. After Fmoc deprotection and cleavage of the oligomer from the resin, only the usual reversed phase HPLC purification was required. Characterization of purified (Gd(III)-DOTA)4–PNA (K-Gd-β-Gd-β-Gd-β-Gd-K-TTTTTTTTTT-K) via HRMS (ESI-TOF) showed masses at 914.5258 [M + 7H]7+ and 710.6509 [M + 9H]9+ corresponding to the calculated masses of 914.2765 (M + 7H)7+ and 711.3279 [M + 9H]9+.
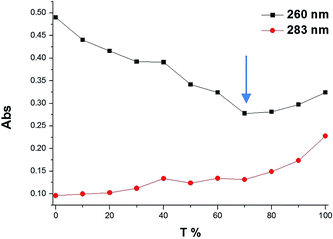
To determine the stoichiometry of binding of the (Gd(III)-DOTA)4–PNA conjugate with a complementary nucleic acid, a Job plot from UV-vis data was constructed. For simplicity and accuracy of the measurement, DNA-A10 was used as the target for the PNA probe. The results at two wavelengths (260 and 283 nm),4 indicated a binding stoichiometry of two (Gd(III)-DOTA)4–PNA to one DNA-dA10, which is consistent with triplex formation (Fig. 2).29 Since it has been shown that homothymidine PNAs form triple helices with oligoriboadenylic acid, specifically (PNA-T8)2:poly(rA40),27 we believe that (Gd(III)-DOTA)4–PNA similarly is capable of triplex formation with poly(rA), as illustrated in Scheme 1.
Temperature dependent UV studies were performed at 2 μM individual strand concentration for [(Gd(III)-DOTA)4–PNA]2:poly(rA). Experiments were repeated thrice and the Tm values were measured from the first derivative plots (Fig. S3†). The melt curves shows a biphasic profile with Tm values of 56 °C and 85 °C, respectively, which is consistent with the presence of a triplex, and supported by the Job plot studies.
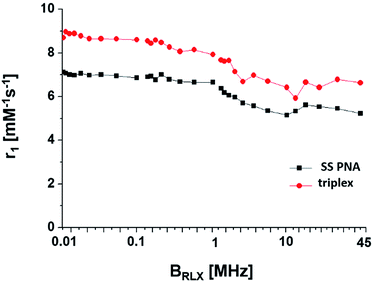
To evaluate the (Gd(III)-DOTA)4–PNA as an MRI probe, its NMRD profile at a series of temperatures were collected. A value of 5.6 mM−1 s−1 was observed at 20 MHz and 25 °C per Gd3+ ion (Fig. 3). The NMRD profile of the triplex, [(Gd(III)-DOTA)4–PNA]2:poly(rA), at 25 °C possessed a r1 of 6.6 mM−1 s−1 at 20 MHz. These findings are in good agreement with other DO3A monoamide compounds (4–5 mM−1 s−1) under similar conditions.33 Evaluation over the entire profile gives some indication of the solution structure of our probe. An increase in relaxivity is seen at ∼20 MHz for both (Gd(III)-DOTA)4–PNA and the triplex, [(Gd(III)-DOTA)4–PNA]2:poly(rA) at 298 K. This increase has been attributed to the slowing of molecular rotation for the probe (Gd(III)-DOTA)4–PNA,34 and ascribed to triplex formation when in the presence of poly(rA). This is further seen in the temperature dependent studies where the increase in relaxivity at 20 MHz is not present for higher temperatures, most likely due destabilization of the triple–helical complex (Fig. S4†).
It is worth noting that concentrations of 20 μM or greater were easily achieved with this PNA conjugate despite the perceived poor solubility of PNAs. In this study, the solubility of the PNA probe benefits from its relatively short (10-mer) length and that it possesses a lysine on each end of this sequence as well as being conjugated to the Gd3+DOTA complexes which add a significant polar component.
It is also noteworthy that triplex formation of this probe with an average full length poly(rA) tail of mRNA would deliver 160–200 Gd3+ ions to a localized microenvironment which compares very favourably with dendrimeric contrast agents that have achieved a loading of 150–200 chelators using 6th generation PAMAM dendrimers.35 While there are 256 potential sites available in a G6 PAMAM dendrimer, a typical loading is closer to ∼170 Gd3+ ions per dendrimer. Other dendrimers up to G10 have been synthesised with a loading of 1860 Gd3+ ions.36 However, all these structures lack the specificity that is necessary for molecular MRI and incorporation of molecules such as polysaccharides,37–40 oligopeptides,41–44 proteins,44,45 antibodies46 and oligonucleotides47 is needed. In comparison, the (Gd(III)-DOTA)4–PNA probe not only consists of targeting oligomer but may deliver a high load of Gd3+ ions due to accumulation of probe on the repetitive sequence of the target. For actual application, another module would need to be added to the structure of the probe, that is, one that would facilitate cellular uptake and potentially nuclear localization. There are a variety of strategies already known to enhance cellular uptake of PNA48 such as the use of cationic analogues,49,50 use of cell penetrating peptides,19,51,52 or the inclusion of specific moiety that drives receptor mediated endocytosis,53 no name a few.
In summary, a poly(Gd(III)-DOTA)–PNA probe has been prepared conveniently via on-resin click chemistry utilizing a pre-metallated chelator. The resulting PNA conjugate recognized poly(rA) and putatively formed a stable triplex structure. The probe and its complex with poly(rA) was evaluated by NMRD studies. An increase in relaxivity was observed at ∼20 MHz for both the (Gd(III)-DOTA)4–PNA and the triplex, [(Gd(III)-DOTA)4–PNA]2:poly(rA) at 25 °C, as compared to a monomeric chelate. By binding to poly(rA), Gd3+ ions would be significantly loaded to a localized microenvironment, which may improve the enhancement of contrast in MR images.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Funding of this work was gratefully received (RHEH) from the Natural Sciences and Engineering Research Council of Canada.Notes and references
- T. F. Massoud and S. S. Gambhir, Genes Dev., 2003, 17, 545–580 CrossRef CAS PubMed.
- É. Tóth, L. Helm and A. E. Merbach, Top. Curr. Chem., 2002, 221, 123 CrossRef.
- K. B. Ghaghanda, M. Ravoori, D. Sabapathy, J. Bankson, V. Kundra and A. Annapraganda, PLoS One, 2009, 4, e7628 Search PubMed.
- L. S. Karfeld, S. R. Bull, N. E. Davis, T. J. Meade and A. E. Barron, Bioconjugate Chem., 2007, 18, 1697–1700 CrossRef CAS PubMed.
- S. Langereis, Q. G. de Lussanet, M. H. P. van Genderen, E. W. Meijer, R. G. H. Beets-Tan, A. W. Griffioen, J. M. A. van Engelshoven and W. H. Backes, NMR Biomed., 2006, 19, 133–141 CrossRef CAS PubMed.
- S. R. Bull, M. O. Guler, R. E. Bras, T. J. Meade and S. I. Stupp, Nano Lett., 2005, 5, 1–4 CrossRef CAS PubMed.
- R. Mishra, W. Su, R. Pohmann, J. Pfeuffer, M. G. Sauer and K. Ugurbil, Bioconjugate Chem., 2009, 20, 1860–1868 CrossRef CAS PubMed.
- J. D. Lewis, S. I. Gunderson and I. W. Mattaj, J. Cell Sci., Suppl., 1995, 19, 13–19 CrossRef CAS.
- A. Jacobson and S. W. Peltz, Annu. Rev. Biochem., 1996, 65, 693–739 CrossRef CAS PubMed.
- M. Wickens, P. Anderson and R. J. Jackson, Curr. Opin. Genet. Dev., 1997, 7, 220–232 CrossRef CAS PubMed.
- H. Fuke and M. Ohno, Nucleic Acids Res., 2008, 36, 1037–1049 CrossRef CAS PubMed.
- A. Scorilas, Crit. Rev. Clin. Lab. Sci., 2002, 39, 193–224 CrossRef CAS PubMed.
- S. L. Topalian, S. Kaneko, M. I. Gonzales, G. L. Bond, Y. Ward and J. L. Manley, Mol. Cell. Biol., 2001, 21, 5614–5623 CrossRef CAS PubMed.
- S. L. Topalian, M. I. Gonzales, Y. Ward, X. Wang and R. F. Wang, Cancer Res., 2002, 62, 5505–5509 CAS.
- F. Xing, G. Song, J. Ren, J. B. Chaires and X. Qu, FEBS Lett., 2005, 579, 5035–5039 CrossRef CAS PubMed.
- P. Giri and G. Suresh Kumar, Mol. BioSyst., 2010, 6, 81–88 RSC.
- P. Giri and G. S. Kumar, Arch. Biochem. Biophys., 2008, 474, 183–192 CrossRef CAS PubMed.
- N. V. Amirkhanov, I. Dimitrov, A. W. Opitz, K. Zhang, J. P. Lackey, C. A. Cardi, S. Lai, N. J. Wagner, M. L. Thakur and E. Wickstrom, Biopolymers, 2008, 89, 1061–1076 CrossRef CAS PubMed.
- W. Su, R. Mishra, J. Pfeuffer, K.-H. Wiesmuller, K. Ugurbil and J. Engelmann, Contrast Media Mol. Imaging, 2007, 2, 42–49 CrossRef CAS PubMed.
- M. R. Lewis, F. Jia, F. Gallazzi, Y. Wang, J. Zhang, N. Shenoy, S. Z. Lever and M. Hannink, Bioconjugate Chem., 2002, 13, 1176–1180 CrossRef CAS PubMed.
- X. Sun, H. Fang, X. Li, R. Rossin, M. J. Welch and J. S. Taylor, Bioconjugate Chem., 2005, 16, 294–305 CrossRef CAS PubMed.
- K. Westerlund, H. Honarvar, V. Tolmachev and A. Eriksson Karlström, Bioconjugate Chem., 2015, 26, 1724–1736 CrossRef CAS PubMed.
- X. Wang and R. H. E. Hudson, ChemBioChem, 2015, 16, 2156–2161 CrossRef CAS PubMed.
- V. Castro, H. Rodríguez and F. Albericio, ACS Comb. Sci., 2016, 18, 1–14 CrossRef CAS PubMed.
- A. H. St Amant, C. Engbers and R. H. E. Hudson, Artif. DNA PNA XNA, 2013, 4, 4–10 CrossRef PubMed.
- N. V. Amirkhanov and E. Wickstrom, Nucleosides, Nucleotides Nucleic Acids, 2005, 24, 423–426 CAS.
- A. Gourishankar and K. N. Ganesh, Artif. DNA PNA XNA, 2012, 3, 5–13 CrossRef PubMed.
- O. Almarsson and T. C. Bruice, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 9542–9546 CrossRef CAS.
- S. K. Kim, P. E. Nielsen, M. Egholm, O. Buchardt, R. H. Berg and B. N. Vf, J. Am. Chem. Soc., 1993, 115, 6477–6481 CrossRef CAS.
- R. F. H. Viguier and A. N. Hulme, J. Am. Chem. Soc., 2006, 128, 11370–11371 CrossRef CAS PubMed.
- D. E. Prasuhn, R. M. Yeh, A. Obenaus, M. Manchester and M. G. Finn, Chem. Commun., 2007, 1269–1271 RSC.
- F. Wojciechowski, A. Groß, I. T. Holder, L. Knörr, M. Drescher and J. S. Hartig, Chem. Commun., 2015, 13850–13853 RSC.
- S. Aime, P. L. Anelli, M. Botta, F. Fedeli, M. Grandi, P. Paoli and F. Uggeri, Inorg. Chem., 1992, 31, 2421–2428 Search PubMed.
- S. Aime, M. Botta and E. Terreno, Adv. Inorg. Chem., 2005, 57, 173–232 CrossRef CAS.
- E. C. Wiener, M. W. Brechbiel, H. Brothers, R. L. Magin, O. A. Gansow, D. A. Tomalia and P. C. Lauterbur, Magn. Reson. Med., 1994, 31, 1–8 CrossRef CAS PubMed.
- L. H. Bryant, M. W. Brechbiel, C. Wu, J. W. M. Bulte, V. Herynek and J. A. Frank, J. Magn. Reson. Imag., 1999, 9, 348–352 CrossRef PubMed.
- E. K. Woller, E. D. Walter, J. R. Morgan, D. J. Singel and M. J. Cloninger, J. Am. Chem. Soc., 2003, 125, 8820–8826 CrossRef CAS PubMed.
- D. Zanini and S. Pamam, J. Org. Chem., 1998, 63, 3486–3491 CrossRef CAS.
- M. L. Wolfenden and M. J. Cloninger, Bioconjugate Chem., 2006, 17, 958–966 CrossRef CAS PubMed.
- D. Zanini and R. Roy, J. Am. Chem. Soc., 1997, 119, 2088–2095 CrossRef CAS.
- K. Sadler and J. P. Tam, Rev. Mol. Biotechnol., 2002, 90, 195–229 CrossRef CAS PubMed.
- L. Crespo, G. Sanclimens, M. Pons, E. Giralt, M. Royo and F. Albericio, Chem. Rev., 2005, 105, 1663–1681 CrossRef CAS PubMed.
- D. T. S. Rijkers, G. W. van Esse, R. Merkx, A. J. Brouwer, H. J. F. Jacobs, R. J. Pieters and R. M. J. Liskamp, Chem. Commun., 2005, 4581–4583 RSC.
- I. van Baal, H. Malda, S. A. Synowsky, J. L. J. van Dongen, T. M. Hackeng, M. Merkx and E. W. Meijer, Angew. Chem., Int. Ed. Engl., 2005, 44, 5052–5057 CrossRef CAS PubMed.
- R. Kluger and J. Zhang, J. Am. Chem. Soc., 2003, 125, 6070–6071 CrossRef CAS PubMed.
- C. Wu, M. W. Brechbiel, R. W. Kozak and O. A. Gansow, Bioorg. Med. Chem. Lett., 1994, 4, 449–454 CrossRef CAS.
- Y. Choi, A. Mecke, B. G. Orr, M. M. Banaszak Holl and J. R. Baker, Nano Lett., 2004, 4, 391–397 CrossRef CAS.
- U. Koppelhus and P. E. Nielsen, Adv. Drug Delivery Rev., 2003, 55, 267–280 CrossRef CAS PubMed.
- P. Zhou, M. Wang, L. Du, G. W. Fisher, A. Waggoner and D. H. Ly, J. Am. Chem. Soc., 2003, 125, 6878–6879 CrossRef CAS PubMed.
- R. Mitra and K. N. Ganesh, J. Org. Chem., 2012, 77, 5696–5704 CrossRef CAS PubMed.
- F. Gallazzi, Y. Wang, F. Jia, N. Shenoy, L. A. Landon, M. Hannink, S. Z. Lever and M. R. Lewis, Bioconjugate Chem., 2003, 14, 1083–1095 CrossRef CAS PubMed.
- K. Kaihatsu, K. E. Huffman and D. R. Corey, Biochemistry, 2004, 43, 14340–14347 CrossRef CAS PubMed.
- M. Równicki, M. Wojciechowska, A. J. Wierzba, J. Czarnecki, D. Bartosik, D. Gryko and J. Trylska, Sci. Rep., 2017, 7, 7644 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details and additional experiments. See DOI: 10.1039/c7ra09040d |
| This journal is © The Royal Society of Chemistry 2017 |