Open Access Article
Open Access ArticleLanthanide contraction and chelating effect on a new family of lanthanide complexes with tetrakis(O-isopropyl)methyle-nediphosphonate: synthesis, structures and terahertz time-domain spectroscopy†
Yan Maa,
Yong-Sheng Yanga,
Yu-Han Jianga,
Yue-Xue Lia,
Min Liub,
Zhong-Feng Lia,
Hong-Liang Hana,
Yu-Ping Yangc,
Xiu-Lan Xind and
Qiong-Hua Jin *a
*a
aDepartment of Chemistry, Capital Normal University, Beijing 100048, China. E-mail: jinqh@cnu.edu.cn
bThe College of Materials Science and Engineering, Beijing University of Technology, Beijing 100022, China
cSchool of Science, Minzu University of China, Beijing 100081, China
dSchool of Food and Chemical Engineering, Beijing Technology and Business University, Beijing 100048, China
First published on 25th August 2017
Abstract
Sixteen lanthanide–diphosphate complexes [LnL2(H2O)4Cl]Cl2 (Ln = La3+ for 1, Ln = Ce3+ for 2), [LnL2(H2O)4]Cl3·6H2O (Ln = Nd3+ for 3, Ln = Sm3+ for 4, Ln = Eu3+ for 5, Ln = Gd3+ for 6, Ln = Tb3+ for 7, Ln = Dy3+ for 8, Ln = Er3+ for 9), {[LaL2(H2O)4Cl]Cl2}2 (10), [PrL2(OH)2(H2O)Cl] [PrL2(H2O)4Cl]Cl2 (11), [LaL2(H2O)5Cl]Cl2·2H2O (12), [YbL2(H2O)3Cl]Cl2·H2O (13), [NdL2(H2O)3Cl]Cl2·3H2O (14), [HoL2(H2O)Cl2]Cl·H2O (15), [Sc2L2(H2O)4(OH)2]Cl4·4H2O·2CH3OH (16) (L = tetrakis(O-isopropyl)methylenediphosphonate) have been synthesized by the reaction of lanthanide chlorides and L ligand in the solvent of acetonitrile (with ethanol or DMF) at room temperature. The title complexes were characterized by IR, elemental analysis, single-crystal X-ray diffraction analysis, thermal analysis, 1HNMR and 31PNMR spectroscopy. Complexes 1–15 are mononuclear complexes and complex 16 is a binuclear cluster complex. Complex 12 is the intermediate of complex 1. Complexes 11–13 display a one-dimensional chain formed by hydrogen bonds and the structures of complexes 14, 16 can be simplified as two-dimensional mesh 63 topology formed by hydrogen bonds. In those lanthanide complexes, the lanthanide ion with an atomic number smaller than Pr element tends to form a nine-coordinated structure and that with the atomic number larger than Pr element tends to form an eight or lower number-coordinated structure. Pr element has two kinds of coordination situations, with coordination numbers of nine or eight. The terahertz (THz) time-domain spectra of these complexes were also studied.
Introduction
Supramolecular chemistry is the chemistry of the intermolecular bond, covering the structures and functions of the entities formed by association of two or more chemical species1,2 through weaker non-covalent bonding interactions such as hydrogen bonding, electrostatic interactions, electron-donor/electron-acceptor interactions and π–π stacking interactions.3,4 A considerable amount of the research activity on metal phosphonate chemistry has focused on the rational design and synthesis of multifunctional coordination polymers, mainly due to their interesting structures and potential application in the areas of catalysis, ion exchange, proton conductivity, intercalation chemistry, photochemistry, and materials chemistry.5,6 Many research groups have used phosphonate ligands to synthesize a large number of metal phosphonates that possess mainly extended structures including 1D-chained, 2D-layered and 3D-pillared structures. As a result of these studies, a wide range of 3d-metal phosphonate and 3d–4f-heterometallic phosphonate cages have been synthesized. In contrast to transition metal phosphonates, reported molecular assemblies of purely 4f-phosphonates are rare.7,8 According to the HSAB principle, lanthanide cations belong to hard acid, so it is easily coordinated by the hard base. For this reason, lanthanide cations are expected to interact strongly with phosphoryl (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) group in order to gain lanthanide–phosphate compounds.9 Compared with their monodentate analogues, the lanthanide complexes with chelating diphosphonate ligands have good stability. So they have been studied owing to their interesting coordination patterns, catalytic properties and the applications in certain specialized separation or recovery processes.10
O) group in order to gain lanthanide–phosphate compounds.9 Compared with their monodentate analogues, the lanthanide complexes with chelating diphosphonate ligands have good stability. So they have been studied owing to their interesting coordination patterns, catalytic properties and the applications in certain specialized separation or recovery processes.10
In rare earth coordination chemistry, the chelating effect and lanthanide contraction are very common laws. The chelating effect indicates that the reaction to produce the chelate is more advantageous in terms of energy than the reaction to produce the monodentated complex. Chelating effect is mainly caused by two factor, one is the entropy increase effect, the entropy in the reaction increase due to the need for less ligand molecules caused by chelating. The other is the role of enthalpy, the same multi-dentate ligand reduces the mutual repulsion of ligands in different sites, which will affect the coordination pattern of ligands and the stability of complexes.11–13 Due to the reason of lanthanide contraction the atomic radius and ionic radius of the lanthanide elements decrease with the increase of atomic number will affect coordination number of metal ions.14–16 Under the influence of these two factors, the coordination modes and properties of lanthanide complexes are very different.17–20 What is more, lanthanide ions have a larger radii and higher coordination numbers than transition metals and exhibit specific, line-shaped f–f emissions, covering a large spectroscopic range from the visible to the near-infrared (NIR). Lanthanide complexes have attracted attention due to their potential applications in electroluminescent devices, fluorescence materials, bioassays and luminescent probes.21–27
There are a few reports on the use of terahertz means to detect the properties of lanthanide complexes. In our previous work, we synthesized a number of lanthanide complexes with the bidentate ligand tetrakis(O-isopropyl)methylenediphosphonate, some of them were characterized by THz-TDS spectra.28–30 Terahertz spectrum covers the frequency region from 0.1 to 10 THz (3.3 cm−1 to 333 cm−1) which bridges the microwave and the far infrared. The THz spectrum contains a wealth of chemical information, such as intramolecular twist, crystal lattice vibration, hydrogen bond, van der Waals force and so on. Besides, many substances have fingerprints in range of the terahertz spectra band. Therefore, based on these fingerprints, it is possible to identify the components, to analyze the physicochemical properties and to study dynamic behaviour of the compound. Conclusively, terahertz spectroscopy is complementary to Fourier transform infrared spectroscopy and X-ray technology.31 Due to the relative infancy of THz spectroscopy, especially in the chemical sciences, work in this area is still limited and the literature is underdeveloped and disparate. This is also true with regard to THz metrology where further developments are required to standardise measurements and methodologies in order to allow THz spectroscopy to become a routine tool for the chemical sciences.32
In this paper, we will make a detailed discussion for a series of sixteen lanthanide–diphosphates complexes on the syntheses, crystal structures, 1HNMR and 31PNMR spectroscopy characteristics, thermal analysis and terahertz (THz) time-domain spectroscopy. In additions, these complexes afford an opportunity to study lanthanide contraction and chelating effect.
Experimental details
Materials and physical measures
All other materials were of analytical grade and used without further purification. All experiments were performed at ambient temperature. Elemental analyses (C, H, N) were performed on a Flash EA 1112 analyzer. IR spectra were recorded from KBr pellets on a Brucker EQUINOX 55 FT-spectrometer operating in the 4000–600 cm−1 region. NMR experiments were carried out on a VNMRS-600 spectrometer using CDCl3 as solvent. Emission spectrum was recorded on a Hitachi F-4500 Luminescence Spectrophotometer. The THz absorption spectrum was recorded on the THz time-domain device of Minzu University of China, based on photoconductive switches for generation and electro-optical crystal detection of the far-infrared light. The experimental apparatus for terahertz transmission measurements has been discussed in detail elsewhere.33 The preparation of the samples is by pressing the mixed powder of samples and polyethylene into a pellet. The detection of THz absorption spectra is carried out at N2 atmosphere to avoid the influence of water vapour. The thickness of the samples 1–16 and ligands is about 1 mm. The THz absorption spectra of the samples are obtained by the THz time-domain device and the effective spectrum range is 0.4–3.3 THz.Synthesis of complexes 1–2, 4–9, 11 and 12
A mixture of 0.2 mmol LnCl3·6H2O (Lafor 1, Ce for 2, Sm for 4, Eu for 5, Gd for 6, Tb for 7, Dy for 8, Er for 9, Pr for 11, La for 12), 1,10-phenanthroline (0.005 g), CH3CN (10 ml)and three drops of DMF was stirred for 3 hour before adding nine drops of L (0.4 mmol). Stirring was continued for 3 hours at ambient temperature to give a clear solution. The solution was filtered. Three days later, the clear solution became turbid. Several months later, lavender crystal of complex 4 and other colourless crystals of complexes not in quantitative yield were obtained. Single crystals suitable for X-ray diffraction were obtained by slow evaporation of a mixed solvent of CH3CN and DMF in the refrigerator.Synthesis of complexes 3, 10
A mixture of LnCl3·6H2O (0.2 mmol, 0.07172 g NdCl3·6H2O for 3, 0.2 mmol, 0.07427 g LaCl3·6H2O for 10), pyrazine (0.005 g), CH3CN (10 ml) was stirred for 3 hour before adding nine drops of L (0.4 mmol). Stirring was continued for 3 hours at ambient temperature to give a clear solution. The solution was filtered. Three days later, the clear solution became turbid. Several months later, lavender crystals of complex 3 not in quantitative yield were obtained. Single crystals suitable for X-ray diffraction were obtained by slow evaporation of a mixed solvent of CH3CN in the refrigerator.Synthesis of complexes 13, 15
A mixture of LnCl3·6H2O (YbCl3·6H2O 0.2 mmol, 0.07110 g for 13, HoCl3·6H2O 0.2 mmol, 0.07586 g for 15), CH3CN (10 ml) was stirred for 3 hour before adding nine drops of L (0.4 mmol). Stirring was continued for 3 hours at ambient temperature to give a clear solution. The solution was filtered. Three days later, the clear solution became turbid. Several months later, colourless crystals of complexes not in quantitative yield were obtained. Single crystals suitable for X-ray diffraction were obtained by slow evaporation of a mixed solvent of CH3CN in the refrigerator.Synthesis of complex 14
A mixture of NdCl3·6H2O (0.2 mmol, 0.07172 g), 1,10-phenanthroline (0.005 g), CH3CN (10 ml) was stirred for 3 hour before adding nine drops of L (0.4 mmol). Three days later, the clear solution became turbid. Several months later, lavender crystals of complex 14 not in quantitative yield were obtained. Single crystals suitable for X-ray diffraction were obtained by slow evaporation of a mixed solvent of CH3CN in the refrigerator.Synthesis of complex 16
A mixture of ScCl3·6H2O (0.2 mmol, 0.05186 g), CH3CN (5 ml) and CH3OH (5 ml) was stirred for 3 hour before adding nine drops of L (0.4 mmol). Stirring was continued for 3 hours at ambient temperature to give a clear solution. The solution was filtered. Three days later, the clear solution became turbid. Several months later, colourless crystals of complex 16 not in quantitative yield were obtained. Single crystals suitable for X-ray diffraction were obtained by slow evaporation of a mixed solvent of CH3CN and CH3OH in the refrigerator.X-ray crystallography
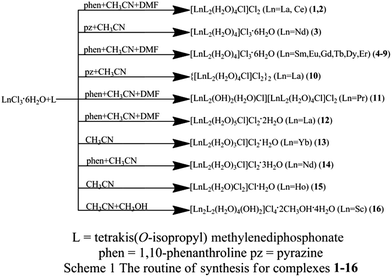
Single-crystal X-ray diffraction studies of complexes 1–16 are performed on a Bruker SMART diffractometer equipped with CCD area detector with a graphite monochromator situated in the incident beam for data collection. All data are corrected by semi-empirical method using SADABS program. The program SAINT is used for integration of the diffraction profiles. All structures are solved by direct methods using SHELXS program of the SHELXTL-97 package and refined with SHELXL.34,35 Metal atom centers were located from the E-maps and other non-hydrogen atoms are located in successive difference Fourier syntheses. The final refinements are performed by full matrix least-squares methods with anisotropic thermal parameters for non-hydrogen atoms on F2. All the hydrogen atoms are first found in difference electron density maps, and then placed in the calculated sites and included in the final refinement in the riding model approximation with displacement parameters derived from the parent atoms to which they are bonded. Further crystallographic data and experimental details for structural analyses of all the complexes are summarized in Table 1 and selected bond lengths and angles with their estimated standard deviations for products 1–16 in Table 2 respectively (Scheme 1).| a R = ∑(||F0| − |Fc||)/∑|F0|.b wR = [∑w(|F0|2 − |Fc|2)2/∑w(F02)]1/2. | ||||||||
|---|---|---|---|---|---|---|---|---|
| Complex | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| Formula | C26H66Cl3LaO16P4 | C26H68Cl3CeO16P4 | C26H78Cl3NdO22P4 | C26H80Cl3SmO22P4 | C26H80Cl3EuO22P4 | C26H80Cl3GdO22P4 | C26H80Cl3TbO22P4 | C26H80Cl3DyO22P4 |
| Mr | 1003.93 | 1007.15 | 1117.35 | 1125.48 | 1127.09 | 1132.38 | 1134.05 | 1137.63 |
| Temperature (K) | 298(2) | 93(2) | 298(2) | 298(2) | 298(2) | 298(2) | 298(2) | 298(2) |
| Crystal system | Triclinic | Triclinic | Triclinic | Triclinic | Triclinic | Triclinic | Triclinic | Triclinic |
| Space group | P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
| Crystal size (mm) | 0.50 × 0.40 × 0.20 | 0.43 × 0.33 × 0.27 | 0.48 × 0.35 × 0.08 | 0.41 × 0.38 × 0.26 | 0.45 × 0.37 × 0.26 | 0.45 × 0.43 × 0.35 | 0.42 × 0.39 × 0.24 | 0.38 × 0.35 × 0.23 |
| a (Å) | 12.2667(11) | 11.978(3) | 10.3830(10) | 10.3690(10) | 10.4108(12) | 10.3390(11) | 10.3470(7) | 10.3287(9) |
| b (Å) | 13.8644(13) | 13.572(3) | 15.2969(14) | 15.2349(14) | 15.3989(16) | 15.1721(15) | 15.1905(13) | 15.1740(18) |
| c (Å) | 16.750(2) | 16.458(4) | 18.6231(18) | 18.6571(16) | 18.8774(19) | 18.6529(17) | 18.6403(18) | 18.563(2) |
| α (°) | 99.0390(10) | 97.679(2) | 85.654(2) | 85.598(2) | 84.974(2) | 85.223(2) | 85.087(2) | 85.156(2) |
| β (°) | 108.566(2) | 109.5330(10) | 82.4010(10) | 82.4860(10) | 81.8110(10) | 82.5970(10) | 82.3520(10) | 82.2570(10) |
| γ (°) | 110.314(2) | 109.940(2) | 76.2460(10) | 76.3060(10) | 76.2280(10) | 76.4460(10) | 76.5000(10) | 76.1480(10) |
| V (Å3) | 2414.9(4) | 2275.7(9) | 2844.8(5) | 2835.7(4) | 2904.8(5) | 2816.5(5) | 2819.0(4) | 2794.9(5) |
| Z | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Dcalc (Mg m−3) | 1.381 | 1.470 | 1.304 | 1.318 | 1.289 | 1.335 | 1.336 | 1.352 |
| θ range (°) | 25.01 | 27.48 | 25.02 | 25.02 | 25.02 | 25.02 | 25.02 | 25.02 |
| F(000) | 1036 | 1042 | 1162 | 1170 | 1172 | 1174 | 1176 | 1178 |
| Data/restraint/parameters | 8371/0/467 | 9996/0/496 | 9887/0/551 | 9865/0/521 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 041/0/522 041/0/522 |
9691/0/521 | 9792/0/521 | 9703/0/521 |
| Reflections collected | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 576 576 |
18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 681 681 |
14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 781 781 |
14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 859 859 |
14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 719 719 |
14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 326 326 |
14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 605 605 |
14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 493 493 |
| Independent reflections | 8371 | 9996 | 9887 | 9865 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 041 041 |
9691 | 9792 | 9703 |
| Goodness-of-fit on F2 | 1.070 | 0.999 | 1.040 | 1.116 | 1.066 | 1.021 | 1.013 | 1.062 |
| Rint | 0.0331 | 0.0360 | 0.0411 | 0.0225 | 0.0752 | 0.0590 | 0.0265 | 0.0262 |
| R1[I > 2σ(I)]a | 0.0504 | 0.0415 | 0.0628 | 0.0431 | 0.0707 | 0.0944 | 0.0384 | 0.0415 |
| wR2[I > 2σ(I)]b | 0.1273 | 0.0833 | 0.1596 | 0.1182 | 0.1853 | 0.2455 | 0.0901 | 0.0935 |
| R1 (all data) | 0.0708 | 0.0474 | 0.0997 | 0.0610 | 0.0893 | 0.1339 | 0.0545 | 0.0651 |
| wR2 (all data) | 0.1478 | 0.0867 | 0.1882 | 0.1307 | 0.1997 | 0.2657 | 0.0970 | 0.1077 |
| Residuals (e Å−3) | 0.916, −1.824 | 1.191, −1.589 | 1.938, −1.241 | 1.550, −0.760 | 2.007, −2.433 | 5.265, −3.173 | 0.900, −1.008 | 0.964, −0.596 |
| Complex | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 |
| Formula | C26H80Cl3ErO22P4 | C26H68Cl3LaO16P4 | C52H132Cl4Pr2O31P8 | C26H74Cl3LaO19P4 | C26H68Cl3YbO16P4 | C26H72Cl3NdO18P4 | C26H64Cl3HoO14P4 | C28H86Cl4Sc2O24P4 |
| Mr | 1142.39 | 1005.94 | 1924.96 | 1059.99 | 1040.07 | 1047.31 | 995.93 | 1162.57 |
| Temperature (K) | 298(2) | 298(2) | 298(2) | 298(2) | 298(2) | 298(2) | 298(2) | 298(2) |
| Crystal system | Triclinic | Triclinic | Triclinic | Triclinic | Monoclinic | Orthorhombic | Triclinic | Triclinic |
| Space group | P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
P2(1)/n | P2(1)2(1)2(1) | P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
| Crystal size (mm) | 0.47 × 0.32 × 0.30 | 0.44 × 0.40 × 0.27 | 0.40 × 0.34 × 0.31 | 0.45 × 0.39 × 0.36 | 0.40 × 0.35 × 0.23 | 0.43 × 0.40 × 0.31 | 0.48 × 0.45 × 0.21 | 0.33 × 0.27 × 0.25 |
| a (Å) | 10.4150(10) | 13.8000(12) | 12.3732(14) | 12.1400(11) | 15.2938(16) | 10.6178(11) | 12.3651(11) | 10.2541(11) |
| b (Å) | 15.6001(16) | 16.7321(14) | 15.0540(18) | 14.9259(13) | 16.7518(19) | 20.6875(19) | 13.0974(13) | 12.3626(14) |
| c (Å) | 18.1899(19) | 22.430(2) | 29.867(3) | 16.2591(15) | 20.3380(17) | 23.129(3) | 16.4608(15) | 12.4492(16) |
| α (°) | 89.380(2) | 107.433(2) | 97.4780(10) | 113.440(2) | 90 | 90 | 69.5060(10) | 77.1300(10) |
| β (°) | 75.5300(10) | 97.1030(10) | 94.5400(10) | 103.8080(10) | 111.9320(10) | 90 | 68.1450(10) | 81.321(2) |
| γ (°) | 74.2320(10) | 98.8000(10) | 108.099(2) | 91.4690(10) | 90 | 90 | 77.605(2) | 67.0670(10) |
| V (Å3) | 2748.8(5) | 4803.6(7) | 5200.5(10) | 2600.8(4) | 4833.5(8) | 5080.4(9) | 2306.7(4) | 1413.3(3) |
| Z | 2 | 4 | 2 | 2 | 4 | 4 | 2 | 1 |
| Dcalc (Mg m−3) | 1.380 | 1.391 | 1.229 | 1.354 | 1.429 | 1.369 | 1.434 | 1.366 |
| θ range (°) | 25.02 | 25.02 | 25.02 | 25.02 | 25.02 | 25.02 | 25.02 | 25.02 |
| F(000) | 1182 | 2080 | 1996 | 1100 | 2132 | 2172 | 1020 | 616 |
| Data/restraint/parameters | 9604/0/581 | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 704/0/933 704/0/933 |
18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 018/0/1037 018/0/1037 |
9044/0/494 | 8528/0/467 | 8956/0/485 | 7950/0/449 | 4890/0/289 |
| Reflections collected | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 054 054 |
24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 293 293 |
26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 047 047 |
13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 536 536 |
24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 048 048 |
25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 667 667 |
11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 509 509 |
7178 |
| Independent reflections | 9604 | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 704 704 |
18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 018 018 |
9044 | 8528 | 8956 | 7950 | 4890 |
| Goodness-of-fit on F2 | 1.048 | 1.016 | 1.018 | 0.998 | 1.137 | 1.043 | 1.034 | 0.998 |
| Rint | 0.0826 | 0.0334 | 0.0311 | 0.0320 | 0.0665 | 0.0436 | 0.0445 | 0.0356 |
| R1[I > 2σ(I)]a | 0.0803 | 0.0397 | 0.0629 | 0.0468 | 0.0495 | 0.0346 | 0.0613 | 0.0711 |
| wR2[I > 2σ(I)]b | 0.1956 | 0.0886 | 0.1575 | 0.1080 | 0.1067 | 0.072 | 0.1440 | 0.2115 |
| R1 (all data) | 0.1071 | 0.1131 | 0.1102 | 0.0834 | 0.1136 | 0.0473 | 0.1153 | 0.1155 |
| wR2 (all data) | 0.2187 | 0.1402 | 0.1763 | 0.1257 | 0.1477 | 0.0794 | 0.1834 | 0.2474 |
| Residuals (e Å−3) | 3.051, −1.547 | 0.549, −0.997 | 2.281, −1.278 | 1.306, −0.761 | 1.408, −0.715 | 0.478, −0.476 | 1.800, −1.641 | 0.638, −0.587 |
| Complex | Ln | Coordination number | Ln–O (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) O) |
O–Ln–O (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) O) |
Ln–Cl | ||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | La | 9 | 2.513(4) | 2.542(4) | 2.555(4) | 2.571(4) | 70.8(13) | 71.8 (13) | 2.916(16) |
| 2 | Ce | 9 | 2.470(2) | 2.500(2) | 2.514(2) | 2.552(2) | 73.2(7) | 71.8(7) | 2.903(10) |
| 3 | Nd | 8 | 2.422(5) | 2.435(5) | 2.465(5) | 2.470(6) | 73.1 (18) | 73.3(18) | — |
| 4 | Sm | 8 | 2.412(4) | 2.444(4) | 2.404(4) | 2.449(4) | 73.8(13) | 73.4(13) | — |
| 5 | Eu | 8 | 2.412(5) | 2.418(5) | 2.453(5) | 2.466(5) | 73.5(17) | 74.0(17) | — |
| 6 | Gd | 8 | 2.369(8) | 2.441(8) | 2.419(8) | 2.389(8) | 73.6(3) | 74.0(3) | — |
| 7 | Tb | 8 | 2.368(3) | 2.379(3) | 2.408(3) | 2.413(3) | 74.3(10) | 73.8(9) | — |
| 8 | Dy | 8 | 2.363(3) | 2.390(4) | 2.402(3) | 2.347(4) | 74.2(12) | 74.2(12) | — |
| 9 | Er | 8 | 2.321(7) | 2.343(6) | 2.347(7) | 2.384(7) | 74.3(2) | 73.5(2) | — |
| 10 | La | 9,9 | 2.539(11) | 2.553(10) | 2.564(11) | 2.474(11) | 72.7(4) | 70.7(4) | 2.911(5) |
| 2.538(10) | 2.566(11) | 2.556(10) | 2.524(11) | 71.4(4) | 71.6(4) | 2.910(4) | |||
| 11 | Pr | 8,9 | 2.444(4) | 2.460(4) | 2.495(4) | 2.521(4) | 71.0(14) | 70.6(14) | 2.902(18) |
| 2.441(6) | 2.450(6) | 2.461(6) | 2.430(5) | 72.2(2) | 73.2(19) | 2.848(3) | |||
| 12 | La | 9 | 2.503(3) | 2.534(3) | 2.574(3) | 73.6(11) | 2.916(14) | ||
| 13 | Yb | 7 | 2.240(6) | 2.287(6) | 2.319(6) | 75.3(2) | 2.556(2) | ||
| 14 | Nd | 8 | 2.402(3) | 2.435(3) | 2.477(3) | 2.477(3) | 72.1(11) | 71.5(12) | 2.818(13) |
| 15 | Ho | 7 | 2.333(6) | 2.353(6) | 2.357(6) | 2.362(6) | 73.8(2) | 73.7(2) | 2.591(3) |
| 2.611(3) | |||||||||
| 16 | Sc | 6,6 | 2.088(4) | 2.078(4) | Sc–Sc 3.164(19) | O(L1)–Sc–O(L2) 175.1(15) | |||
Results and discussion
Synthesis of the complexes
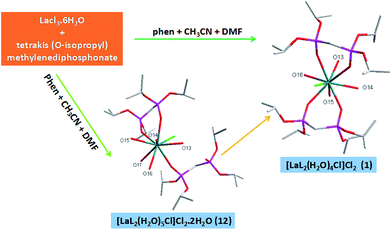
Sixteen complexes 1–16 were synthesized by the reactions of lanthanide metal(III) chloride with tetrakis(O-isopropyl)methylenediphosphonate (L) under the presence of a small amount of 1,10-phenanthroline (phen) or pyrazine (pz) at room temperature in acetonitrile or in a mixture of ethanol and DMF solution. This shows that the choice of the solvent has influence on the experimental product, just as reported in the literature.36 It is worth noting that a small amount of 1,10-phenanthroline (phen) or pyrazine (pz) is added into the reaction. According to HSAB theory, compared with nitrogen atoms of 1,10-phenanthroline or pyrazine, rare earth metals tend to be coordinated with oxygen of tetrakis(O-isopropyl)methylenediphosphonate or crystal water. So we can guess that phen or pz play can facilitate the synthesis of unsolvated products with good crystallization quality.It is very interesting that the crystals of complex 12 was found in the process of synthesizing complex 1. Therefore, we believe that complex 12 is an intermediate product of complex 1. The complex 12 was colourless and the complex 1 was yellow. During the evaporation of the solvent, the colour change from the colourless to yellow was observed in the beaker. From the perspective of the structure of the complexes, the L ligand of monodentate coordination in 12 is converted to one of bidentate coordination in 1, which makes the structure of 1 more stable. It is well-known that the thermodynamic stability of the complex is expressed by the stability constant, and the factors that affect the stability constant are the enthalpy change and the entropy change of the coordination reaction. Chelating effect is essentially an entropy increase effect.23,24 The conversion of complex 12 to complex 1 can be understood as L-ligand substitution with H2O molecules acting on the metal center. This substitution reaction increases the number of molecules in the reaction system and can be regarded as an entropy increase process (Scheme 2). The experimental results show that the presence of chelating effect will enhance the stability of the complex.
Crystal structure
![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) . Here, we only discussed complex 1 deeply to demonstrate the common structural features. In complex 1, the average of distance of La–O (P
. Here, we only discussed complex 1 deeply to demonstrate the common structural features. In complex 1, the average of distance of La–O (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) bonds is 2.545 Å as usual, the average of distance of La–O (P
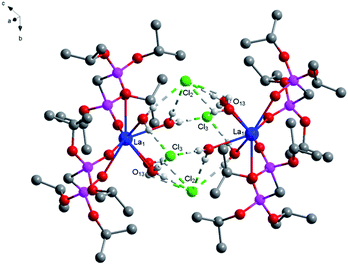
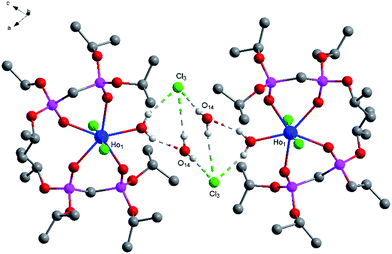
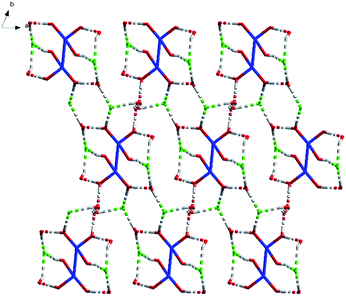
O) bonds is 2.545 Å as usual, the average of distance of La–O (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) bonds being observed between the reported complexes NaLa[(PO3H)2CH–C6H4–CH(PO3H)2]·4H2O37 and La[(EtO)2P(O)CH2P(O)(EtO)2]2(NO3)3 (ref. 38) varies between 2.558 Å and 2.518 Å. The average of (P)O–La–O(P) bond angle is 71.3° and it is smaller than those reported the compounds [La(Ph2P(O)CH2P(O)Ph2)4](OTf)3·1.5H2O (72.7°)39 [H3N(CH2)4NH3]La[hedpH][hedpH2] (hedp = 1-hydroxyethylidenediphosphonate) (74.0°)40 and La[(EtO)2P(O)CH2P(O)(EtO)2]2(NO3)3 (74.0°)38. The smaller (P)O–La–O(P) bond angle is a great extent on the different substituent group of phosphorus. Meanwhile, coordination water and free chlorine atoms also form hydrogen bonds with distances of 2.278 Å for [O(13)–H(13F)⋯Cl(2)], 2.657 Å for [O(14)–H(14C)⋯Cl(2)], 2.673 Å for [O(14)–H(14A)⋯Cl(2)], 2.677 Å for [O(15)–H(15C)⋯Cl(2)], 2.361 Å for [O(16)–H(16F)⋯Cl(2)], 2.296 Å for [O(13)–H(13G)⋯Cl(3)], 2.312 Å for [O(15)–H(15D)⋯Cl(3)], 2.283 Å for [O(16)–H(16G)⋯Cl(3)] and 2.374 Å for [O(14)–H(14C)⋯O(13)]. These strong hydrogen bonds enhance the stability of the two adjacent units and affect the crystal packing of the molecules (Fig. 2).
O) bonds being observed between the reported complexes NaLa[(PO3H)2CH–C6H4–CH(PO3H)2]·4H2O37 and La[(EtO)2P(O)CH2P(O)(EtO)2]2(NO3)3 (ref. 38) varies between 2.558 Å and 2.518 Å. The average of (P)O–La–O(P) bond angle is 71.3° and it is smaller than those reported the compounds [La(Ph2P(O)CH2P(O)Ph2)4](OTf)3·1.5H2O (72.7°)39 [H3N(CH2)4NH3]La[hedpH][hedpH2] (hedp = 1-hydroxyethylidenediphosphonate) (74.0°)40 and La[(EtO)2P(O)CH2P(O)(EtO)2]2(NO3)3 (74.0°)38. The smaller (P)O–La–O(P) bond angle is a great extent on the different substituent group of phosphorus. Meanwhile, coordination water and free chlorine atoms also form hydrogen bonds with distances of 2.278 Å for [O(13)–H(13F)⋯Cl(2)], 2.657 Å for [O(14)–H(14C)⋯Cl(2)], 2.673 Å for [O(14)–H(14A)⋯Cl(2)], 2.677 Å for [O(15)–H(15C)⋯Cl(2)], 2.361 Å for [O(16)–H(16F)⋯Cl(2)], 2.296 Å for [O(13)–H(13G)⋯Cl(3)], 2.312 Å for [O(15)–H(15D)⋯Cl(3)], 2.283 Å for [O(16)–H(16G)⋯Cl(3)] and 2.374 Å for [O(14)–H(14C)⋯O(13)]. These strong hydrogen bonds enhance the stability of the two adjacent units and affect the crystal packing of the molecules (Fig. 2).
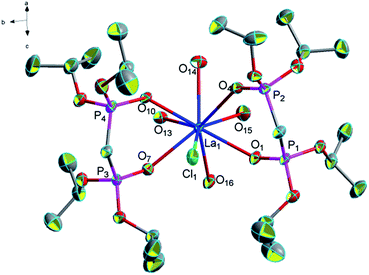 | ||
| Fig. 1 Molecular structure of complex 1. Hydrogen atoms and solvent molecule were omitted for clarity. | ||
![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) , each unit contains one Nd(III) atom, two bidentate L ligands, four coordinated water molecules, six lattice water molecules, and three free chlorine atoms. Nd(III) atoms are eight-coordinated by four phosphonate oxygen atoms (O1, O4, O7, O10) from two bidentate L ligands and four oxygen atom (O13, O14, O15, O16) from four coordinated water molecules (Fig. 3). In complex 3, the average distance of Nd–O (P
, each unit contains one Nd(III) atom, two bidentate L ligands, four coordinated water molecules, six lattice water molecules, and three free chlorine atoms. Nd(III) atoms are eight-coordinated by four phosphonate oxygen atoms (O1, O4, O7, O10) from two bidentate L ligands and four oxygen atom (O13, O14, O15, O16) from four coordinated water molecules (Fig. 3). In complex 3, the average distance of Nd–O (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) bonds (2.466 Å) is shorter than the average distance of Nd–O (P
O) bonds (2.466 Å) is shorter than the average distance of Nd–O (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) bonds (2.474 Å) in complex [NH4][Nd(hedp)(H2O)]·H2O (hedpH4 = 1-hydroxy ethylidenediphosphonic acid)6 and longer than the average distance of Nd–O (P
O) bonds (2.474 Å) in complex [NH4][Nd(hedp)(H2O)]·H2O (hedpH4 = 1-hydroxy ethylidenediphosphonic acid)6 and longer than the average distance of Nd–O (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) bonds (2.432 Å) in complex Nd[(PO3H)2CH–C6H4–CH(PO3H)(PO3H2)]·4H2O.33 The average (P)O–Nd–O(P) bond angle (73.2°) is quite similar to those reported complexes Nd[(PO3H)2CH–C6H4–CH(PO3H)(PO3H2)]·4H2O (73.2°)41 and [Nd((iPrO)2P(O)CH2–P(O)(OiPr)2)2(NO3)3]·(CH3CN)2 (73.6°).20 Moreover, O–H⋯Cl hydrogen bonds formed by coordinated water molecules with free chlorine ions 2.380 Å for [O(13)–H(13I)⋯Cl(1)], 2.397 Å for [O(13)–H(13J)⋯Cl(1)], 2.470 Å for [O(20)–H(20F)⋯Cl(1)], 2.309 Å for [O(22)–H(22F)⋯Cl(1)], 2.206 Å for [O(16)–H(16F)⋯Cl(2)], 2.392 Å for [O(18)–H(18C)⋯Cl(2)], 2.247 Å for [O(19)–H(19F)⋯Cl(2)], 2.238 Å for [O(16)–H(16G)⋯Cl(3)], 2.343 Å for [O(17)–H(17G)⋯Cl(3)] and 2.237 Å for [O(18)–H(18D)⋯Cl(3)] and the O–H⋯O hydrogen bonds formed by coordinated water molecules with free water molecules 1.954 Å for [O(15)–H(15C)⋯O(17)], 2.299 Å for [O(20)–H(20G)⋯O(17)], 1.954 Å for [O(15)–H(15D)⋯O(18)], 1.890 Å for [O(14)–H(14C)⋯O(19)], 1.905 Å for [O(17)–H(17F)⋯O(19)], 1.945 Å for [O(21)–H(21D)⋯O(20)], 1.961 Å for [O(22)–H(22G)⋯O(20)], 1.881 Å for [O(19)–H(19G)⋯O(21)], 1.922 Å for [O(14)–H(14D)⋯O(22)], 1.942 Å for [O(21)–H(21C)⋯O(22)]. The complex displays a one-dimensional chain formed by these twenty hydrogen bonds and these non-covalent bond interactions make this complex stable (Fig. 4).
O) bonds (2.432 Å) in complex Nd[(PO3H)2CH–C6H4–CH(PO3H)(PO3H2)]·4H2O.33 The average (P)O–Nd–O(P) bond angle (73.2°) is quite similar to those reported complexes Nd[(PO3H)2CH–C6H4–CH(PO3H)(PO3H2)]·4H2O (73.2°)41 and [Nd((iPrO)2P(O)CH2–P(O)(OiPr)2)2(NO3)3]·(CH3CN)2 (73.6°).20 Moreover, O–H⋯Cl hydrogen bonds formed by coordinated water molecules with free chlorine ions 2.380 Å for [O(13)–H(13I)⋯Cl(1)], 2.397 Å for [O(13)–H(13J)⋯Cl(1)], 2.470 Å for [O(20)–H(20F)⋯Cl(1)], 2.309 Å for [O(22)–H(22F)⋯Cl(1)], 2.206 Å for [O(16)–H(16F)⋯Cl(2)], 2.392 Å for [O(18)–H(18C)⋯Cl(2)], 2.247 Å for [O(19)–H(19F)⋯Cl(2)], 2.238 Å for [O(16)–H(16G)⋯Cl(3)], 2.343 Å for [O(17)–H(17G)⋯Cl(3)] and 2.237 Å for [O(18)–H(18D)⋯Cl(3)] and the O–H⋯O hydrogen bonds formed by coordinated water molecules with free water molecules 1.954 Å for [O(15)–H(15C)⋯O(17)], 2.299 Å for [O(20)–H(20G)⋯O(17)], 1.954 Å for [O(15)–H(15D)⋯O(18)], 1.890 Å for [O(14)–H(14C)⋯O(19)], 1.905 Å for [O(17)–H(17F)⋯O(19)], 1.945 Å for [O(21)–H(21D)⋯O(20)], 1.961 Å for [O(22)–H(22G)⋯O(20)], 1.881 Å for [O(19)–H(19G)⋯O(21)], 1.922 Å for [O(14)–H(14D)⋯O(22)], 1.942 Å for [O(21)–H(21C)⋯O(22)]. The complex displays a one-dimensional chain formed by these twenty hydrogen bonds and these non-covalent bond interactions make this complex stable (Fig. 4).
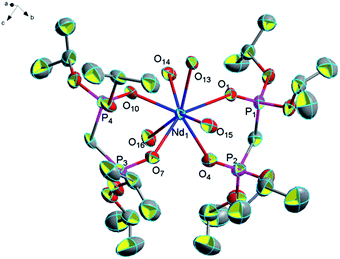 | ||
| Fig. 3 Molecular structure of complex 3. Hydrogen atoms and solvent molecule were omitted for clarity. | ||
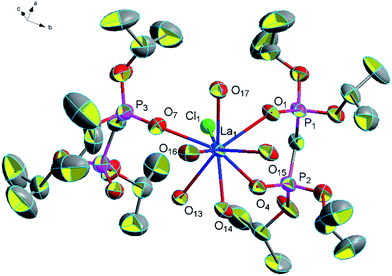
![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) . Here, we only discuss complex 11 to research structural features. In complex 11, one of the central atoms Pr is eight-coordinated by four oxygen atoms from two L ligands, one oxygen atom from a coordinated water molecule, two oxygen atoms from two hydroxyls and a coordinated chlorine atom; while the other central atom Pr is nine-coordinated by four oxygen atoms from two L ligands, four oxygen atoms from four coordinated water molecules and a coordinated chlorine atom (Fig. 5). The average distance of Pr–O (P
. Here, we only discuss complex 11 to research structural features. In complex 11, one of the central atoms Pr is eight-coordinated by four oxygen atoms from two L ligands, one oxygen atom from a coordinated water molecule, two oxygen atoms from two hydroxyls and a coordinated chlorine atom; while the other central atom Pr is nine-coordinated by four oxygen atoms from two L ligands, four oxygen atoms from four coordinated water molecules and a coordinated chlorine atom (Fig. 5). The average distance of Pr–O (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) bonds is 2.463 Å and the average (P)O–Pr–O(P) bond angle is about 71.8°. The average Pr–O (P
O) bonds is 2.463 Å and the average (P)O–Pr–O(P) bond angle is about 71.8°. The average Pr–O (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) bond distance being observed between the reported complexes [Pr(NO3)2(Ph2P(O)CH2P(O)Ph2)2] [Pr(NO3)4(Ph2P(O)CH2P(O)Ph2)2]41 and [NH4][Pr(hedp)(H2O)]·H2O (hedpH4 = 1-hydroxy ethylidenediphosphonic acid)12 varies between 2.429 Å and 2.474 Å. The average (P)O–Pr–O(P) bond angle (71.8°) in the title structure is smaller than the (P)O–Pr–O(P) bond angle in the complexes [Pr((iPrO)2P(O)CH2–P(O)(OiPr)2)2(NO3)3]·(CH3CN)2 (73.3°),20 [Pr(NO3)2(Ph2P(O)CH2P(O)Ph2)2][Pr(NO3)4(Ph2P(O)CH2P(O)Ph2)2] (74.0°)42 and [NH4][Pr(hedp)(H2O)]·H2O (76.1°) (hedpH4 = 1-hydroxy ethylidenediphosphonic acid).6 In addition, it is also having a number of hydrogen bonds (2.361 Å for [O(13)–H(13F)⋯Cl(3)], 2.339 Å for [O(15)–H(15C)⋯Cl(3)], 2.336 Å for [O(16)–H(16F)⋯Cl(3)], 2.314 Å for [O(16)–H(16G)⋯Cl(3)], 2.257 Å for [O(13)–H(13G)⋯Cl(4)], 2.638 Å for [O(14)–H(14E)⋯Cl(4)], 2.258 Å for [O(14)–H(14F)⋯Cl(4)] and 2.274 Å for [(O15)–H(15D)⋯Cl(4)]) which formed a one-dimensional chain of complex 11 (Fig. 6).
O) bond distance being observed between the reported complexes [Pr(NO3)2(Ph2P(O)CH2P(O)Ph2)2] [Pr(NO3)4(Ph2P(O)CH2P(O)Ph2)2]41 and [NH4][Pr(hedp)(H2O)]·H2O (hedpH4 = 1-hydroxy ethylidenediphosphonic acid)12 varies between 2.429 Å and 2.474 Å. The average (P)O–Pr–O(P) bond angle (71.8°) in the title structure is smaller than the (P)O–Pr–O(P) bond angle in the complexes [Pr((iPrO)2P(O)CH2–P(O)(OiPr)2)2(NO3)3]·(CH3CN)2 (73.3°),20 [Pr(NO3)2(Ph2P(O)CH2P(O)Ph2)2][Pr(NO3)4(Ph2P(O)CH2P(O)Ph2)2] (74.0°)42 and [NH4][Pr(hedp)(H2O)]·H2O (76.1°) (hedpH4 = 1-hydroxy ethylidenediphosphonic acid).6 In addition, it is also having a number of hydrogen bonds (2.361 Å for [O(13)–H(13F)⋯Cl(3)], 2.339 Å for [O(15)–H(15C)⋯Cl(3)], 2.336 Å for [O(16)–H(16F)⋯Cl(3)], 2.314 Å for [O(16)–H(16G)⋯Cl(3)], 2.257 Å for [O(13)–H(13G)⋯Cl(4)], 2.638 Å for [O(14)–H(14E)⋯Cl(4)], 2.258 Å for [O(14)–H(14F)⋯Cl(4)] and 2.274 Å for [(O15)–H(15D)⋯Cl(4)]) which formed a one-dimensional chain of complex 11 (Fig. 6).
 | ||
| Fig. 5 Molecular structure of complex 11. Hydrogen atoms and solvent molecule were omitted for clarity. | ||
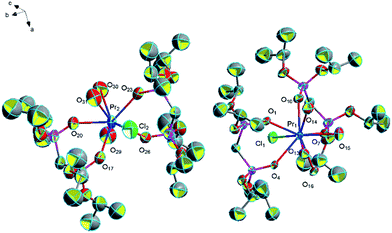
![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) (Fig. 7). We believe that complex 12 is an intermediate product of complex 1. The average distance of La–O (P
(Fig. 7). We believe that complex 12 is an intermediate product of complex 1. The average distance of La–O (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) bonds of complex 12 is 2.537 Å, and it is longer than that of complex 1 (2.450 Å). The (P)O–La–O(P) bond angle is 73.6°, and it is larger than that of complex 1 (71.3°). This phenomenon is due to the different coordination modes of complex 1 and complex 12. The steric hindrance of two bidentate ligands in complex 1 is larger than that of a monodentate L ligand and a bidentate L ligand in complex 12, so complex 12 has a longer La–O (P
O) bonds of complex 12 is 2.537 Å, and it is longer than that of complex 1 (2.450 Å). The (P)O–La–O(P) bond angle is 73.6°, and it is larger than that of complex 1 (71.3°). This phenomenon is due to the different coordination modes of complex 1 and complex 12. The steric hindrance of two bidentate ligands in complex 1 is larger than that of a monodentate L ligand and a bidentate L ligand in complex 12, so complex 12 has a longer La–O (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) bond and a larger (P)O–La–O(P) bond angle. A part of hydrogen bonds can be seen in complex 12 between free water molecules, coordinated water molecules and P
O) bond and a larger (P)O–La–O(P) bond angle. A part of hydrogen bonds can be seen in complex 12 between free water molecules, coordinated water molecules and P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of L ligands (1.876 Å for [O(13)–H(13F)⋯O(10)], 1.984 Å for [O(19)–H(19F)⋯O(10)], 1.928 Å for [O(19)–H(19G)⋯O(14)], 2.018 Å for [O(13)–H(13G)⋯O(18)], 2.109 Å for [O(14)–H(14E)⋯O(18)], 1.969 Å for [O(18)–H(18F)⋯O(19)]), which is much shorter than the sum of the van der Waals radii (3.22 Å). These hydrogen bonds connect the two adjacent units strongly and it is helpful for forming a one-dimensional chain. The other part of hydrogen bonds is formed by free water molecules, coordinated water molecules and two free chlorine atoms (2.323 Å for [O(15)–H(15C)⋯Cl(2)], 2.369 Å for [O(16)–H(16G)⋯Cl(2)], 2.348 Å for [O(17)–H(17F)⋯Cl(2)], 2.303 Å for [O(18)–H(18C)⋯Cl(2)], 2.303 Å for [O(14)–H(14F)⋯Cl(3)], 2.240 Å for [O(15)–H(15D)⋯Cl(3)], 2.498 Å for [O(16)–H(16F)⋯Cl(3)], 2.264 Å for [O(17)–H(17G)⋯Cl(3)]). These hydrogen bonds act as a linker linked the dual-core units and it is necessary for forming a one-dimensional chain (Fig. 8).
O of L ligands (1.876 Å for [O(13)–H(13F)⋯O(10)], 1.984 Å for [O(19)–H(19F)⋯O(10)], 1.928 Å for [O(19)–H(19G)⋯O(14)], 2.018 Å for [O(13)–H(13G)⋯O(18)], 2.109 Å for [O(14)–H(14E)⋯O(18)], 1.969 Å for [O(18)–H(18F)⋯O(19)]), which is much shorter than the sum of the van der Waals radii (3.22 Å). These hydrogen bonds connect the two adjacent units strongly and it is helpful for forming a one-dimensional chain. The other part of hydrogen bonds is formed by free water molecules, coordinated water molecules and two free chlorine atoms (2.323 Å for [O(15)–H(15C)⋯Cl(2)], 2.369 Å for [O(16)–H(16G)⋯Cl(2)], 2.348 Å for [O(17)–H(17F)⋯Cl(2)], 2.303 Å for [O(18)–H(18C)⋯Cl(2)], 2.303 Å for [O(14)–H(14F)⋯Cl(3)], 2.240 Å for [O(15)–H(15D)⋯Cl(3)], 2.498 Å for [O(16)–H(16F)⋯Cl(3)], 2.264 Å for [O(17)–H(17G)⋯Cl(3)]). These hydrogen bonds act as a linker linked the dual-core units and it is necessary for forming a one-dimensional chain (Fig. 8).
 | ||
| Fig. 7 Molecular structure of complex 12. Hydrogen atoms and solvent molecule were omitted for clarity. | ||
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) bonds is 2.282 Å and the average (P)O–Yb–O(P) bond angle is about 75.3°. We also found the complex formed by Yb(III) and diphosphate ligands, in which Yb(III) is also coordinated by a monodentatediphosphate ligand and a bidentate diphosphate ligand ([Yb(NO3)(Ph2P(O)CH2P(O)Ph2)3]·(NO3)2).42 Complex 13 is also a one-dimensional chain formed by a lot of hydrogen bonds. Among them, two groups of these three hydrogen bonds (2.170 Å for [O(15)–H(15C)⋯Cl(2)], 2.541 Å for [O(16)–H(16G)⋯Cl(2)] and 1.873 Å for [O(15)–H(15D)⋯O(16)]) formed a six-membered ring and two groups of these two hydrogen bonds (2.208 Å for [O(13)–H(13F)⋯Cl(3)] and 2.349 Å for [O(13)–H(13G)⋯Cl(3)]) formed a four-membered ring. These two kinds of rings and the other three hydrogen bonds (2.220 Å for [O(14)–H(14E)⋯Cl(2)], 2.288 Å for [O(14)–H(14G)⋯Cl(3)] and 1.984 Å for [O(16)–H(16D)⋯O(10)]) made the structure more stable collectively (Fig. 10).
O) bonds is 2.282 Å and the average (P)O–Yb–O(P) bond angle is about 75.3°. We also found the complex formed by Yb(III) and diphosphate ligands, in which Yb(III) is also coordinated by a monodentatediphosphate ligand and a bidentate diphosphate ligand ([Yb(NO3)(Ph2P(O)CH2P(O)Ph2)3]·(NO3)2).42 Complex 13 is also a one-dimensional chain formed by a lot of hydrogen bonds. Among them, two groups of these three hydrogen bonds (2.170 Å for [O(15)–H(15C)⋯Cl(2)], 2.541 Å for [O(16)–H(16G)⋯Cl(2)] and 1.873 Å for [O(15)–H(15D)⋯O(16)]) formed a six-membered ring and two groups of these two hydrogen bonds (2.208 Å for [O(13)–H(13F)⋯Cl(3)] and 2.349 Å for [O(13)–H(13G)⋯Cl(3)]) formed a four-membered ring. These two kinds of rings and the other three hydrogen bonds (2.220 Å for [O(14)–H(14E)⋯Cl(2)], 2.288 Å for [O(14)–H(14G)⋯Cl(3)] and 1.984 Å for [O(16)–H(16D)⋯O(10)]) made the structure more stable collectively (Fig. 10).
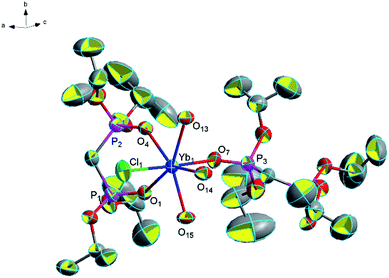 | ||
| Fig. 9 Molecular structure of complex 13. Hydrogen atoms and solvent molecule were omitted for clarity. | ||
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) bonds of complex 14 (2.448 Å) is similar to that of complex 3 (2.446 Å) and the (P)O–Nd–O(P) bond angle (71.8°) is smaller than that of complex 3 (73.2°). This phenomenon is due to the different coordination modes of complex 3 and complex 14. The steric hindrance of three coordinated water molecules and a coordinated chlorine atom in complex 14 is larger than that of four coordinated water molecules in complex 3, so complex 14 has a smaller (P)O–Nd–O(P) bond angle. In addition, it is also having a number of hydrogen bonds (2.444 Å for [O(16)–H(16G)⋯Cl(1)], 2.376 Å for [O(17)–H(17G)⋯Cl(1)], 2.276 Å for [O(13)–H(13F)⋯Cl(2)], 2.262 Å for [O(14)–H(14F)⋯Cl(2)], 2.295 Å for [O(18)–H(18C)⋯Cl(2)], 2.308 Å for [O(13)–H(13G)⋯Cl(3)], 2.250 Å for [O(15)–H(15C)⋯Cl(3)], 2.395 Å for [O(16)–H(16E)⋯Cl(3)], 2.328 Å for [O(17)–H(17F)⋯Cl(3)], 2.328 Å for [O(18)–H(18D)⋯O(16)], 1.917 Å for [O(14)–H(14E)⋯O(17)] and 1.842 Å for [O(15)–H(15D)⋯O(18)]) which formed a two-dimensional mesh of complex 14 (Fig. 12).
O) bonds of complex 14 (2.448 Å) is similar to that of complex 3 (2.446 Å) and the (P)O–Nd–O(P) bond angle (71.8°) is smaller than that of complex 3 (73.2°). This phenomenon is due to the different coordination modes of complex 3 and complex 14. The steric hindrance of three coordinated water molecules and a coordinated chlorine atom in complex 14 is larger than that of four coordinated water molecules in complex 3, so complex 14 has a smaller (P)O–Nd–O(P) bond angle. In addition, it is also having a number of hydrogen bonds (2.444 Å for [O(16)–H(16G)⋯Cl(1)], 2.376 Å for [O(17)–H(17G)⋯Cl(1)], 2.276 Å for [O(13)–H(13F)⋯Cl(2)], 2.262 Å for [O(14)–H(14F)⋯Cl(2)], 2.295 Å for [O(18)–H(18C)⋯Cl(2)], 2.308 Å for [O(13)–H(13G)⋯Cl(3)], 2.250 Å for [O(15)–H(15C)⋯Cl(3)], 2.395 Å for [O(16)–H(16E)⋯Cl(3)], 2.328 Å for [O(17)–H(17F)⋯Cl(3)], 2.328 Å for [O(18)–H(18D)⋯O(16)], 1.917 Å for [O(14)–H(14E)⋯O(17)] and 1.842 Å for [O(15)–H(15D)⋯O(18)]) which formed a two-dimensional mesh of complex 14 (Fig. 12).
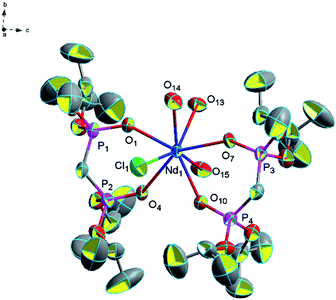 | ||
| Fig. 11 Molecular structure of complex 14. Hydrogen atoms and solvent molecule were omitted for clarity. | ||
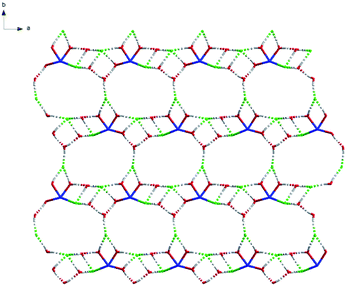
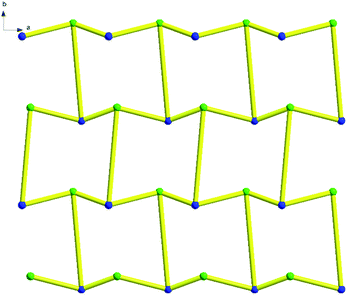
Topologically, each Nd(III) atom and chlorine atom (Cl) is considered as a 3-connected quadrangle node and the complex can be simplified as two-dimensional mesh 63 topology formed by Nd(III) atom and chlorine atom (Cl) (Fig. 13 and S9†). Type of this net consists of six connected nodes shared by three hexagonal units. The hexagonal unit forms a six-membered ring, comprising three Nd(III) atoms and three chlorine atom. This two-dimensional mesh 63 topology makes complex structure more stable.
![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) (Fig. 14). The average distance of Ho–O (P
(Fig. 14). The average distance of Ho–O (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) bonds is 2.351 Å and it is longer than the average distance of Ho–O (P
O) bonds is 2.351 Å and it is longer than the average distance of Ho–O (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) bonds (2.341 Å) in complex [Ho(NO3)2(Ph2P(O)CH2P(O)Ph2)2H2O]·NO3. The average (P)O–Ho–O(P) bond angle is about 73.8° and it is smaller than the average (P)O–Ho–O(P) bond angle (76.7°) in complex [Ho(NO3)2(Ph2P(O)CH2P(O)Ph2)2H2O]·NO3.42 There are four hydrogen bonds in complex 15 (1.969 Å for [O(13)–H(13F)⋯O(14)], 2.258 Å for [O(13)–H(13G)⋯Cl(3)], 2.792 Å for [O(14)–H(14E)⋯Cl(3)] and 2.299 Å for [O(14)–H(14F)⋯Cl(3)]) and these hydrogen bonds connect the two adjacent units strongly and it is helpful for the stability of this structure (Fig. 15).
O) bonds (2.341 Å) in complex [Ho(NO3)2(Ph2P(O)CH2P(O)Ph2)2H2O]·NO3. The average (P)O–Ho–O(P) bond angle is about 73.8° and it is smaller than the average (P)O–Ho–O(P) bond angle (76.7°) in complex [Ho(NO3)2(Ph2P(O)CH2P(O)Ph2)2H2O]·NO3.42 There are four hydrogen bonds in complex 15 (1.969 Å for [O(13)–H(13F)⋯O(14)], 2.258 Å for [O(13)–H(13G)⋯Cl(3)], 2.792 Å for [O(14)–H(14E)⋯Cl(3)] and 2.299 Å for [O(14)–H(14F)⋯Cl(3)]) and these hydrogen bonds connect the two adjacent units strongly and it is helpful for the stability of this structure (Fig. 15).
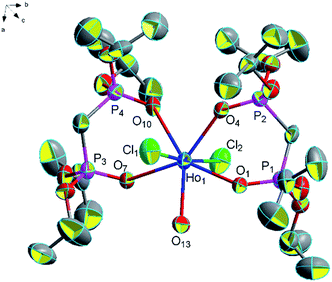 | ||
| Fig. 14 Molecular structure of complex 15. Hydrogen atoms and solvent molecule were omitted for clarity. | ||
![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) (Fig. 16). The average distance of Sc–O (P
(Fig. 16). The average distance of Sc–O (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) bonds is 2.083 Å and it is similar to the average distance of Sc–O (P
O) bonds is 2.083 Å and it is similar to the average distance of Sc–O (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) bonds (2.090 Å) in complex [Sc(Ph2P(O)CH2P(O)Ph2)3](OTf)3.39 What is more, there are a lot of hydrogen bonds in complex 16 (2.576 Å for [O(7)–H(7D)⋯Cl(1)], 2.300 Å for [O(10)–H(10G)⋯Cl(1)], 2.630 Å for [O(12)–H(12F)⋯Cl(1)], 2.104 Å for [O(8)–H(8E)⋯Cl(2)], 2.251 Å for [O(10)–H(10F)⋯Cl(2)], 2.309 Å for [O(11)–H(11C)⋯Cl(2)], 1.738 Å for [O(8)–H(8D)⋯O(10)], 1.822 Å for [O(9)–H(9F)⋯O(11)], 2.330 Å for [O(11)–H(11D)⋯O(11)] and 1.668 Å for [O(9)–H(9G)⋯O(12)]) and these hydrogen bonds form a two-dimensional mesh.
O) bonds (2.090 Å) in complex [Sc(Ph2P(O)CH2P(O)Ph2)3](OTf)3.39 What is more, there are a lot of hydrogen bonds in complex 16 (2.576 Å for [O(7)–H(7D)⋯Cl(1)], 2.300 Å for [O(10)–H(10G)⋯Cl(1)], 2.630 Å for [O(12)–H(12F)⋯Cl(1)], 2.104 Å for [O(8)–H(8E)⋯Cl(2)], 2.251 Å for [O(10)–H(10F)⋯Cl(2)], 2.309 Å for [O(11)–H(11C)⋯Cl(2)], 1.738 Å for [O(8)–H(8D)⋯O(10)], 1.822 Å for [O(9)–H(9F)⋯O(11)], 2.330 Å for [O(11)–H(11D)⋯O(11)] and 1.668 Å for [O(9)–H(9G)⋯O(12)]) and these hydrogen bonds form a two-dimensional mesh.
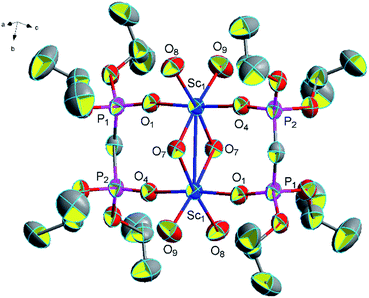 | ||
| Fig. 16 Molecular structure of complex 16. Hydrogen atoms and solvent molecule were omitted for clarity. | ||
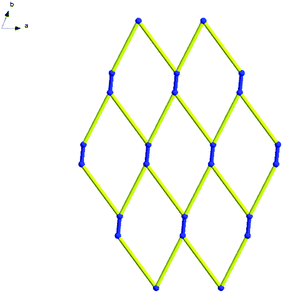
Topologically, all the Sc(III) are viewed as a 3-connected quadrangle node. In this way, hydrogen bonds link the Sc–Sc units to form a two-dimensional mesh 63 topology. The type of this net is similar to that of complex 14 and consists of six connected nodes shared by three hexagonal units. The hexagonal unit constitutes a six-membered ring, comprising six Sc(III) atoms (Fig. 17). However, every line of complex 14 in the topological structure is simplified by hydrogen bonds and the blue lines of complex 16 are the real metallic bonds. Comparing the two complexes, the topological structure of complex 14 is a distorted two-dimensional mesh 63 topology and the net of complex 16 is a classic one (Fig. 18 and S10†).
Chelating effect and lanthanide contraction
In this work, chelating effect and lanthanide contraction affect the coordination modes of the ligand tetrakis(O-isopropyl)methylenediphosphonate, the coordination numbers of the Ln(III) ions, and the average Ln(III)–Oligand bond lengths of lanthanide complexes.The single crystal diffraction data shows that the two L ligands have the same bidentate coordination mode in the complexes 1–11, 14–16, but two ligands in complexes 12 and 13 adopt two different modes – monodentate mode and bidentate mode. The complex 12 is the intermediate of complex 1, in the process of reaction the monodentate ligand L in 12 is spontaneously converted into bidentate coordination mode. All these show that the ligand is more prone to take bidentate chelation mode when forming the complex, which agrees with the result found in literature25 that the values of ΔEbidentate is lower than ΔEmonodentate when bis-phosphinoxides coordinate with rare earth ions. Because of the lanthanide contraction, the radius of the Yb3+ in 13 is smaller than the other rare earth ions (La3+, Ce3+, Pr3+, Nd3+, Sm3+, Eu3+, Gd3+, Tb3+, Dy3+, Ho3+, Er3+), which leads to a large steric hindrance hence results in the existence of monodentate coordination in 13. In summary, we believe that it is chelating effect and lanthanide contraction which decide the coordination mode and crystal structure of complexes.
In addition, there are two interesting phenomena associated with lanthanide contraction in this work. One is that we selected the similar reaction route and ligands, but we obtained products with different coordination number. Complexes 1, 2, 10, 12 all have a coordination number of nine with central atom La or Ce. Complexes 3–9 and 14 all have a coordination number of eight with central atom Nd, Sm, Eu, Gd, Tb, Dy or Er. However, complex 11 with central atom Pr has two kinds of coordination number, nine or eight. The lanthanide with atomic ordinal number smaller than Pr element tends to form nine-coordinated structure and the metal with atomic number larger than Pr element tends to form eight or lower number-coordinated structure. Pr element has two kinds of coordination numbers-nine or eight. In complexes 13 and 15, the coordination number is 7, whose central atoms are Yb and Ho. Complex 16 is a binuclear cluster compound with the central atom Sc and the coordination number is six. This phenomenon is considered to be due to the different radii of the central atom.
In the complexes 1–8, 15, 9 and 13, the normalized average distances from Ln to the oxygen of ligand decrease as the atomic number of the lanthanide increases [2.545 Å for La–O (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 2.509 Å for Ce–O (P
O), 2.509 Å for Ce–O (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 2.446 Å for Nd–O (P
O), 2.446 Å for Nd–O (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 2.427 Å for Sm–O (P
O), 2.427 Å for Sm–O (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 2.437 Å for Eu–O (P
O), 2.437 Å for Eu–O (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 2.405 Å for Gd–O (P
O), 2.405 Å for Gd–O (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 2.392 Å for Tb–O (P
O), 2.392 Å for Tb–O (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 2.376 Å for Dy–O (P
O), 2.376 Å for Dy–O (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 2.351 Å for Ho–O (P
O), 2.351 Å for Ho–O (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 2.349 Å for Er–O (P
O), 2.349 Å for Er–O (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) and 2.282 Å for Yb–O (P
O) and 2.282 Å for Yb–O (P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)]. This phenomenon can be attributed to “lanthanide contraction”.43,44 Furthermore, the (P)O–Ln–O(P) bond angles change almost increasingly as the atomic number of the lanthanide increases (71.3° for (P)O–La–O(P), 72.5° for (P)O–Ce–O(P), 73.2° for (P)O–Nd–O(P), 73.6° for (P)O–Sm–O(P), 73.7° for (P)O–Eu–O(P), 73.8° for (P)O–Gd–O(P), 74.1° for (P)O–Tb–O(P), 74.2° for (P)O–Dy–O(P), 73.8° for (P)O–Ho–O(P), 73.9° for (P)O–Er–O(P) and 75.3° for (P)O–Yb–O(P)).
O)]. This phenomenon can be attributed to “lanthanide contraction”.43,44 Furthermore, the (P)O–Ln–O(P) bond angles change almost increasingly as the atomic number of the lanthanide increases (71.3° for (P)O–La–O(P), 72.5° for (P)O–Ce–O(P), 73.2° for (P)O–Nd–O(P), 73.6° for (P)O–Sm–O(P), 73.7° for (P)O–Eu–O(P), 73.8° for (P)O–Gd–O(P), 74.1° for (P)O–Tb–O(P), 74.2° for (P)O–Dy–O(P), 73.8° for (P)O–Ho–O(P), 73.9° for (P)O–Er–O(P) and 75.3° for (P)O–Yb–O(P)).
NMR spectra of complexes
The 1H NMR and 31P NMR spectra of complexes 1–16 were measured at room temperature in CDCl3 solution. In 1H NMR spectra of complexes 1–16, the resonance signals at range 1.08–1.89 ppm (1.24–1.52 ppm for 1, 1.09–1.68 ppm for 2, 1.19–1.54 ppm for 3, 1.12–1.35 ppm for 4, 1.25–1.89 ppm for 5, 1.39–1.55 ppm for 6, 1.16–1.41 ppm for 7, 1.20–1.40 ppm for 8, 1.28–1.38 ppm for 9, 1.24–1.47 ppm for 10, 1.15–1.25 ppm for 11, 1.35–1.40 ppm for 12, 1.08–1.18 ppm for 13, 1.23–1.55 ppm for 14, 1.38–1.48 ppm for 15 and 1.27–1.35 ppm for 16) are assigned to the methyl groups of L ligand in the solution. The resonance signals at range 2.07–2.69 ppm (2.53 ppm for 1, 2.64 ppm for 2, 2.69 ppm for 4, 2.45 ppm for 5, 2.13 ppm for 7, 2.51 ppm for 10, 2.07 ppm for 11, 2.53 ppm for 12 and 2.36 ppm for 16) is assigned to the methylene of L ligand in the solution. And the resonance signals at range 4.08–5.97 ppm (4.94 ppm for 1, 5.50 ppm for 2, 5.28 ppm for 3, 4.86 ppm for 4, 4.32 ppm for 5, 4.85 ppm for 6, 4.08 ppm for 7, 4.15 ppm for 8, 4.62 ppm for 9, 4.91 ppm for 10, 5.97 ppm for 11, 4.99 ppm for 12, 4.12 ppm for 13, 5.27 ppm for 14, 4.85 ppm for 15 and 4.76 ppm for 16) are assigned to the methane of L ligand in the solution. The resonance signals at 7.25–7.26 ppm are assigned to the CDCl3 solution.In 31P NMR spectra of complexes 1–11 and 14–16, all phosphorus atoms in each molecule are chemically equivalent because only a single resonance signal is found (18.24 ppm for 1, 18.45 ppm for 2, 18.43 ppm for 3, 17.90 ppm for 4, 15.77 ppm for 5, 16.70 ppm for 6, 16.68 ppm for 7, 16.32 ppm for 8, 17.07 ppm for 9, 18.11 ppm for 10, 18.72 ppm for 11, 19.64 ppm for 14, 17.44 ppm for 15 and 17.38 ppm 16). The similarity of resonance signals in solution of complexes 1–11 and 14–16 shows that the chemistry environment for the phosphorus atom from L in complexes 1–11 and 14–16 is similar. In 31P NMR spectra of complex 12 and 13, two resonance signals are found (21.49 ppm, 39.02 ppm for 12 and 16.88 ppm, 40.08 ppm for 13). In the 31P NMR spectra, the type of P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O exhibits two characteristic signals indicating that there are two kinds of coordinated situation of L ligands (a monodentate L ligand and a bidentate L ligand) of complex 12 and 13 (Table 3).
O exhibits two characteristic signals indicating that there are two kinds of coordinated situation of L ligands (a monodentate L ligand and a bidentate L ligand) of complex 12 and 13 (Table 3).
| 1H NMR (ppm) | 31P NMR (ppm) | |||||
|---|---|---|---|---|---|---|
| Complex 1 | 1.24–1.52 | 2.53 | 4.94 | 7.26 | 18.24 | |
| Complex 2 | 1.09–1.68 | 2.64 | 5.50 | 7.26 | 18.45 | |
| Complex 3 | 1.19–1.54 | 5.28 | 7.26 | 18.43 | ||
| Complex 4 | 1.12–1.35 | 2.69 | 4.86 | 7.26 | 17.90 | |
| Complex 5 | 1.25–1.89 | 2.45 | 4.32 | 7.26 | 15.77 | |
| Complex 6 | 1.39–1.55 | 4.85 | 7.26 | 16.70 | ||
| Complex 7 | 1.16–1.41 | 2.13 | 4.08 | 7.26 | 16.68 | |
| Complex 8 | 1.20–1.40 | 4.15 | 7.26 | 16.32 | ||
| Complex 9 | 1.28–1.38 | 4.62 | 7.26 | 17.07 | ||
| Complex 10 | 1.24–1.47 | 2.51 | 4.91 | 7.26 | 18.11 | |
| Complex 11 | 1.15–1.25 | 2.07 | 5.97 | 7.26 | 18.72 | |
| Complex 12 | 1.35–1.40 | 2.53 | 4.99 | 7.25 | 21.49 | 39.02 |
| Complex 13 | 1.08–1.18 | 4.12 | 7.26 | 16.88 | 40.08 | |
| Complex 14 | 1.23–1.55 | 5.27 | 7.26 | 19.64 | ||
| Complex 15 | 1.38–1.48 | 4.85 | 7.25 | 17.44 | ||
| Complex 16 | 1.27–1.35 | 2.36 | 4.76 | 7.26 | 17.38 | |
Thermogravimetric analysis of complexes
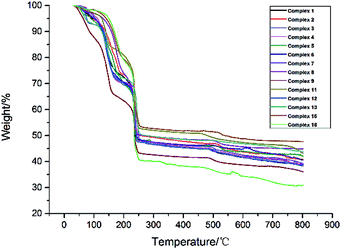
TG experiments of the samples of complexes 1–9, 11–13, 15–16 were carried out from room temperature to 800 °C (Fig. 19). The TGA curves for complex 1 and complex 2 display an initial weight loss of 18.36% (calcd 17.82%) for 1 and 18.68% (calcd 17.72%) for 2 at the temperature range of 114–168 °C and 120–184 °C, suggesting the loss of one coordinated chlorine atom, two uncoordinated chlorine atoms and four coordinated water molecules. Then, in the temperature range from 168 °C for 1 (184 °C for 2) to 242 °C, this part of weight loss is the decomposition of L ligands. At the temperature of 242 °C the weight percentage of complex 1 and complex 2 are 48.48% and 50.55%, respectively and then they are slowly decomposed.The TGA curves for complexes 3–9 display initial weight losses of 25.64% (calcd 25.28%) for 3, 25.46% (calcd 26.28%) for 4, 25.42% (calcd 25.68%) for 5, 25.30% (calcd 25.01%) for 6, 25.26% (calcd 24.37%) for 7, 25.18% (calcd 24.46%) for 8, and 25.07% (calcd 24.22%) for 9 between 100–111 °C and 210–214 °C (105–213 °C for 3, 103–213 °C for 4, 100–211 °C for 5, 106–214 °C for 6, 108–213 °C for 7, 111–212 °C for 8 and 111–210 °C for 9), suggesting the loss of three coordinated chlorine atoms, four coordinated water molecules and six free water molecules. The weight loss of the temperature range from 210–214 °C to 250 °C is the decomposition of L ligands and the rest of weight percentage were 47.46% for 3, 47.98% for 4, 48.44% for 5, 48.63% for 6, 48.32% for 7, 50.25% for 8 and 43.08% for 9. At the temperature higher than 250 °C, complexes 3–9 are slowly decomposed.
The TGA curves for complex 11 displays an initial weight loss of 12.21% (calcd 10.12%) between 114 °C and 155 °C, suggesting the loss of two coordinated chlorine atoms, two free chlorine atoms, five coordinated water molecules and two coordinated hydroxide radicals. The weight loss of the temperature range from 155 °C to 252 °C is the decomposition of L ligands and the rest of weight percentage is 52.68%. At the temperature higher than 252 °C, complex 11 is slowly decomposed.
The TGA curves for complex 12 and complex 13 display initial weight loss of 21.00% (calcd 21.93%) for 12 and 18.68% (calcd 17.16%) for 13 at the temperature range of 111–182 °C and 116–181 °C, suggesting the loss of one coordinated chlorine atom, two free chlorine atoms, coordinated water molecules (five for complex 12 and three for complex 13) and free water molecules (two for complex 12 and one for complex 13). Then, in the temperature range from 111 °C for 12 (116 °C for 13) to 247 °C for 12 (255 °C for 13), this part of weight loss is the decomposition of L ligands. At the temperature of 247 °C and 255 °C the weight percentage of complex 12 and complex 13 are 47.48% and 50.55%, respectively and then they are slowly decomposed.
The TGA curves for complex 15 displays an initial weight loss of 15.60% (calcd 14.31%) between 131 °C and 189 °C, suggesting the loss of two coordinated chlorine atoms, one free chlorine atom, one coordinated water molecule and one free water molecule. The weight loss in the temperature range from 189 °C to 253 °C is the decomposition of L ligands and the rest of weight percentage is 53.46%. At the temperature higher than 253 °C, complex 15 is slowly decomposed.
The TGA curves for complex 16 displays an initial weight loss of 5.42% (calcd 5.51%) between room temperature and 150 °C, suggesting the loss of two free methanol molecules. The weight loss in the temperature range from 150 °C to 200 °C is four coordinated water molecules, four free water molecules and two coordinated hydroxide radicals. Then at the temperature range from 200 °C to 255 °C, this part of weight loss is the decomposition of L ligands and the rest of weight percentage is 40.37%. At the temperature higher than 255 °C, complex 16 is slowly decomposed.
According to the results of thermogravimetric analysis, we can clearly find that complex 1 begins to decompose at 114 °C and complex 12 begins to decompose at 111 °C. This phenomenon also demonstrates that the stability of complex 1 is superior to that of complex 12, namely the structure formed by two ligands adopting chelating coordination modes is more stable than the structure formed by two ligands adopting mono-dentate and chelating coordination modes.
Terahertz (THz) time-domain spectroscopy of the complexes
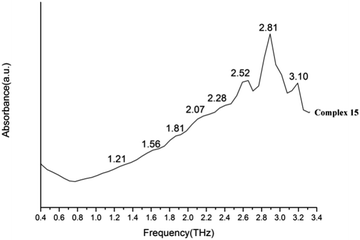
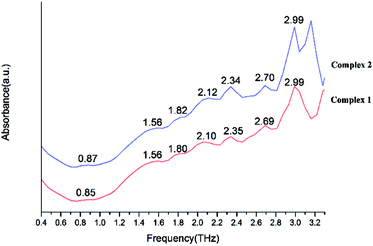
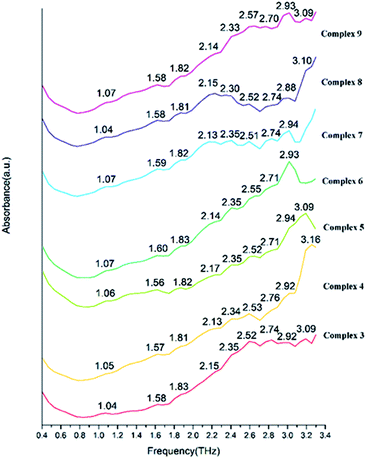
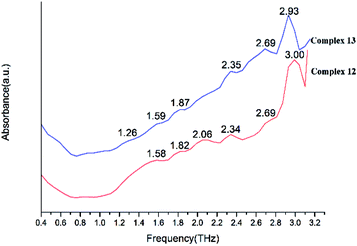
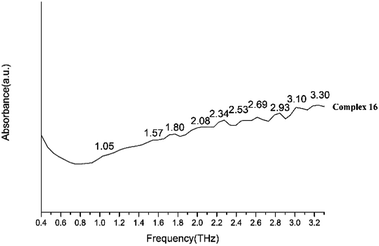
We present absorptivity and dispersion spectra of three different retinal isomers in the far-infrared region between 10 and 100 cm−1 (0.3–3.0 THz) measured by terahertz time-domain spectroscopy at 298 K. The bands in the THz region may be related to various modes. Hydrogen bonds, rotation of dipole, vdW (van der Waals' force), low-frequency vibrations of lattices and other modes may be related to the bands in the THz region.45 The observed signal terahertz pulses after passing through the sample material is usually interpreted as a possible reduction of the material by absorption and scattering.The room temperature terahertz (THz) absorption spectra of complexes 1–9, 11–13, 15, 16 and relative ligand (tetrakis(O-isopropyl)methylenediphosphonate) are measured (Table 4). The found peaks for each compound are as following: L (1.05, 1.22, 1.41, 1.58, 1.76 and 1.93 THz), complex 1 (0.85, 1.56, 1.80, 2.10, 2.35, 2.69 and 2.99 THz), complex 2 (0.87, 1.56, 1.80, 2.12, 2.34, 2.70 and 2.99 THz), complex 3 (1.04, 1.58, 1.83, 2.15, 2.35, 2.52, 2.74, 2.92 and 3.09 THz), complex 4 (1.05, 1.57, 1.81, 2.13, 2.34, 2.53, 2.76, 2.92 and 3.16 THz), complex 5 (1.06, 1.56, 1.82, 2.17, 2.35, 2.52, 2.71, 2.94 and 3.09 THz), complex 6 (1.07, 1.60, 1.83, 2.14, 2.35, 2.55, 2.71 and 2.93 THz), complex 7 (1.07, 1.59, 1.82, 2.13, 2.35, 2.51, 2.74 and 2.94 THz), complex 8 (1.04, 1.58, 1.81, 2.15, 2.30, 2.52, 2.74, 2.88 and 3.10 THz), complex 9 (1.07, 1.58, 1.82, 2.14, 2.33, 2.57, 2.70, 2.93 and 3.09 THz), complex 11 (1.07, 1.58, 1.82, 2.12, 2.34, 2.52, 2.69, 2.87 and 3.04 THz), complex 12 (1.58, 1.82, 2.06, 2.34, 2.69, and 3.00 THz), complex 13 (1.26, 1.59, 1.87, 2.35, 2.69 and 2.93 THz), complex 15 (1.21, 1.56, 1.81, 2.07, 2.28, 2.52, 2.81 and 3.10 THz in Fig. 20) and complex 16 (1.05, 1.57, 1.80, 2.08, 2.34, 2.53, 2.69, 2.93, 3.10 and 3.30 THz).
| Complex 1 | 0.85 | 1.56 | 1.80 | 2.10 | 2.35 | 2.69 | 2.99 | ||||
| Complex 2 | 0.87 | 1.56 | 1.80 | 2.12 | 2.34 | 2.70 | 2.99 | ||||
| Complex 3 | 1.04 | 1.58 | 1.83 | 2.15 | 2.35 | 2.52 | 2.74 | 2.92 | 3.09 | ||
| Complex 4 | 1.05 | 1.57 | 1.81 | 2.13 | 2.34 | 2.53 | 2.76 | 2.92 | 3.16 | ||
| Complex 5 | 1.06 | 1.56 | 1.82 | 2.17 | 2.35 | 2.52 | 2.71 | 2.94 | 3.09 | ||
| Complex 6 | 1.07 | 1.60 | 1.83 | 2.14 | 2.35 | 2.55 | 2.71 | 2.93 | |||
| Complex 7 | 1.07 | 1.59 | 1.82 | 2.14 | 2.35 | 2.51 | 2.74 | 2.94 | |||
| Complex 8 | 1.04 | 1.58 | 1.81 | 2.15 | 2.30 | 2.52 | 2.74 | 2.88 | 3.10 | ||
| Complex 9 | 1.07 | 1.58 | 1.82 | 2.14 | 2.33 | 2.57 | 2.70 | 2.93 | 3.09 | ||
| Complex 11 | 1.07 | 1.58 | 1.82 | 2.12 | 2.34 | 2.52 | 2.69 | 2.87 | 3.04 | ||
| Complex 12 | 1.58 | 1.82 | 2.06 | 2.34 | 2.69 | 3.00 | |||||
| Complex 13 | 1.26 | 1.59 | 1.87 | 2.35 | 2.69 | 2.93 | |||||
| Complex 15 | 1.21 | 1.56 | 1.81 | 2.07 | 2.28 | 2.52 | 2.81 | 3.10 | |||
| Complex 16 | 1.05 | 1.57 | 1.80 | 2.08 | 2.34 | 2.53 | 2.69 | 2.93 | 3.10 | 3.30 | |
| Ligand | 1.051.221.411.581.761.93 | ||||||||||
By comparing the THz absorption spectra of the products with those of the reactants, we can note that after reaction, the peaks of ligand disappeared or moved. The new peaks of complexes have a tendency to blue shifted compared with that of ligand (Fig. S14†) (1.41, 1.76, 1.93 THz for ligand and 1.56, 1.80, 2.06 THz for complexes). By comparing the THz absorption spectra of complexes 1–9 (Fig. 21 and 22), they all have the peaks of 1.58, 1.83, 2.15, 2.35, 2.74 and 2.92 THz and it is due to the similar structure with one rare earth metal atom and two ligands. However, there also have some different peaks for these complexes (0.87 THz for complexes 1–2 and 1.04, 2.52 THz of complexes 3–9). From the perspective of structural analysis, complexes 1–2 have one more chlorine atom and the extra chlorine atom changes the hydrogen bonding interaction of the complex. In complexes 12–13 (Fig. 23), the central atom coordinated by a monodentate diphosphate ligand and a bidentate diphosphate ligand. In the spectrum of complexes 12–13, we found that the peaks of 2.12 and 2.52 THz were missing comparing with complexes 1–9. This phenomenon indicates that these terahertz absorption peaks were assigned to two bidentate diphosphate ligands coordinating to the central atom. By comparing the THz absorption spectra of complex 16 with the other complexes, the spectrum of complex 16 (Fig. 24) almost have all terahertz absorption peaks of the other complexes and even have the peaks of 3.30 THz. This result is due to the special structure of complex 16, and which is a cluster compound and have some metallic bonds.
These results indicate that although the central atom and the ligand are the same, but different coordination situations make the terahertz absorption peaks have significant differences. The coordination number and ligand species of complexes can also affect the terahertz spectral absorption peaks appear or disappear. Therefore, THz spectroscopy may be a novel means to characterize the situation with the coordination of the ligand, and even more characterization methods in the field of coordination chemistry will have better prospects.22
Conclusion
In summary, a series of sixteen lanthanide–diphosphates complexes (L = tetrakis(O-isopropyl)methylenediphosphonate)have been synthesized in the solvent of acetonitrile, ethanol or methanol at room temperature and characterized by IR, elemental analyses, single-crystal X-ray diffraction analysis, thermal analysis, 1HNMR and 31P NMR spectroscopy. Complexes 11–13 displays a one-dimensional chain formed by hydrogen bonds and complexes 14 and 16 can be simplified as two-dimensional mesh 63 topology formed by hydrogen bonds. It is worth mentioning that complex 16 is a cluster compound and the metallic bond between adjacent Sc(III) atoms is 3.164 Å. In these complexes, the ligand adopts monodentate mode or chelating mode. As the atomic number increases, the coordination number of the Ln(III) ions change from nine to seven, the average Ln(III)–Oligand bond lengths of lanthanide complexes decrease. From the above facts we can see that the chelating effect and lanthanide contraction can affect the coordination modes of the ligand tetrakis(O-isopropyl)methylenediphosphonate, the coordination number of the Ln(III) ions, and the average Ln(III)–Oligand bond lengths of lanthanide complexes. With the study and exploration of terahertz (THz) time-domain spectroscopy, we reach the following conclusions: the new peaks of complexes have a tendency of blue shifted compared with that of the ligand. Although the central atom and the ligand are basically the same, different coordination situations make the terahertz absorption peaks significantly different. The coordination number and ligand species of complexes can also affect the existence of the terahertz spectral absorption peaks. So, THz spectroscopy may be a sensitive method to distinguish and determine small differences between the lanthanide–organic complexes.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work has been supported by the National Natural Science Foundation of China (Grant No. 21171119, 21376008, 81573822, 11574408 and 11204191), the National High Technology Research and Development Program 863 of China (Grant No. 2012AA063201), and the Scientific Research Base Development Program of the Beijing Municipal Commission of Education, Beijing Natural Science Foundation (Grant No. 2172017).Notes and references
- J. P. Byrne, J. A. Kitchen and T. Gunnlaugsson, Chem. Soc. Rev., 2014, 43, 5302 RSC.
- C. Lescop, Acc. Chem. Res., 2017, 50, 885 CrossRef CAS PubMed.
- X.-J. Zheng and L.-P. Jin, Polyhedron, 2003, 22, 2617 CrossRef CAS.
- P. H. Dinolfo and J. T. Hupp, Chem. Mater., 2001, 13, 3113 CrossRef CAS.
- R. Fu, S. Hu and X. Wu, Cryst. Growth Des., 2016, 16, 5074 CAS.
- D.-P. Dong, L. Liu, Z.-G. Sun, C.-Q. Jiao, Z.-M. Liu, C. Li, Y.-Y. Zhu, K. Chen and C.-L. Wang, Cryst. Growth Des., 2011, 11, 5346 CAS.
- K. H. Zangana, E. M. Pineda and R. E. Winpenny, Dalton Trans., 2014, 43, 17101 RSC.
- T. H. Zhou, F. Y. Yi, P. X. Li and J. G. Mao, Inorg. Chem., 2010, 49, 905 CrossRef CAS PubMed.
- (a) M. Visseaux, A. Dormond and D. Baudry, Bull. Soc. Chim. Fr., 1993, 130, 173 CAS; (b) V. A. Petrov, W. J. Marshall and V. V. Grushin, Chem. Commun., 2002, 520 RSC; (c) B. Matthew, L. William, P. Tajesh, C. P. Michael and W. Michael, Polyhedron, 2001, 20, 2055 CrossRef; (d) F. John, W. G. P. Andrew and R. R. David, Polyhedron, 2002, 21, 287 CrossRef.
- (a) S. Zdenek, N. Marek and P. Jiri, Inorg. Chem., 2005, 44, 2074 CrossRef PubMed; (b) S. Zdenek, N. Marek, P. Jiri and Z. Zbynek, Polyhedron, 2006, 25, 809 CrossRef; (c) S. Zdenek, P. Vaclav, P. Jiri and N. Marek, Polyhedron, 2008, 27, 283 CrossRef; (d) I. E. Nifant'ev, A. N. Tavtorkin, A. V. Shlyahtin, S. A. Korchagina, I. F. Gavrilenko, N. N. Glebova and A. V. Churakov, Dalton Trans., 2013, 42, 1223 RSC.
- S. Lis and G. Pawlicki, J. Lumin., 2010, 130, 832 CrossRef CAS.
- D. Reddy and D. Jaganyi, Transition Met. Chem., 2006, 31, 792 CrossRef CAS.
- B. Coupez, C. Boehme and G. Wipff, Phys. Chem. Chem. Phys., 2002, 4, 5716 RSC.
- X. Zou, M. Li, P. Yan, J. Zhang, G. Hou and G. Li, Dalton Trans., 2013, 42, 9482 RSC.
- S. Sairenji, S. Akine and T. Nabeshima, Dalton Trans., 2016, 45, 14902 RSC.
- Q. Cheng, W.-Y. Huang, Q.-H. Huang, Y.-J. Xiong, J.-F. Fang, Y. Li, F.-F. Zhu and S.-T. Yue, J. Coord. Chem., 2015, 68, 1980 CrossRef CAS.
- S. Tang, X. Liu and J. Zhou, J. Cluster Sci., 2017, 28, 1 CrossRef.
- W. Xu, Y. Zhou, D. Huang, W. Xiong, M. Su, K. Wang, S. Han and M. Hong, Cryst. Growth Des., 2013, 13, 5420 CAS.
- R. Ilmi and K. Iftikhar, J. Photochem. Photobiol., A, 2016, 325, 68 CrossRef CAS.
- R. J. Ellis, D. M. Brigham, L. Delmau, A. S. Ivanov, N. J. Williams, M. N. Vo, B. Reinhart, B. A. Moyer and V. S. Bryantsev, Inorg. Chem., 2017, 56, 1152 CrossRef CAS PubMed.
- F. Zinna, M. Pasini, F. Galeotti, C. Botta, L. Di Bari and U. Giovanella, Adv. Funct. Mater., 2017, 27, 1603719 CrossRef.
- J. Xu, Y. Zhang, H. Chen, W. Liu and Y. Tang, Dalton Trans., 2014, 43, 7903 RSC.
- M. D. Ward, Coord. Chem. Rev., 2007, 251, 1663 CrossRef CAS.
- S. K. Ghosh, J.-P. Zhang and S. Kitagawa, Angew. Chem., Int. Ed., 2007, 119, 8111 CrossRef.
- M. L. Cable, J. P. Kirby, K. Sorasaenee, H. B. Gray and A. Ponce, J. Am. Chem. Soc., 2007, 129, 1474 CrossRef CAS PubMed.
- L. Pan, K. M. Adams, H. E. Hernandez, X. Wang, C. Zheng, Y. Hattori and K. Kaneko, J. Am. Chem. Soc., 2003, 125, 3062 CrossRef CAS PubMed.
- T. M. Reineke, M. Eddaoudi, M. Fehr, D. Kelley and O. M. Yaghi, J. Am. Chem. Soc., 1999, 121, 1651 CrossRef CAS.
- Q. Jin, L. Ricard and F. Nief, Polyhedron, 2005, 24, 549 CrossRef CAS.
- S. Xu, M. Liu, H.-L. Han, Z.-F. Li, Q.-H. Jin, J. Hou, W. Su, Y.-Y. Chen and J.-Y. Yao, Polyhedron, 2015, 85, 69 CrossRef CAS.
- Y.-S. Yang, M. Liu, Y.-P. Yang, Q.-H. Jin, Z.-F. Li, X.-N. Xue, Z.-J. Zhang and W.-J. Zheng, Polyhedron, 2015, 93, 66 CrossRef CAS.
- (a) S. R. Keiding, J. Phys. Chem. A, 1997, 101, 5250–5254 CrossRef CAS; (b) L. Yang, H. Sun, S. Weng, K. Zhao, L. Zhang, G. Zhao, Y. Wang, Y. Xu, X. Lu, C. Zhang, J. Wu and J. Chen, Spectrochim. Acta, Part A, 2008, 69, 160–166 CrossRef PubMed; (c) L. Zhang, H. Zhong, C. Deng, C. Zhang and Y. Zhao, Appl. Phys. Lett., 2009, 94, 211106 CrossRef; (d) L. Tian, Q. Zhou, B. Jin, K. Zhao, S. Zhao, Y. Shi and C. Zhang, Sci. China, Ser. G: Phys., Mech. Astron., 2009, 39, 1938–1943 CrossRef; (e) P. Shumyatsky and R. R. Alfano, SPIE Rev., 2011, 16, 1–9 Search PubMed.
- A. I. McIntosh, B. Yang, S. M. Goldup, M. Watkinson and R. S. Donnan, Chem. Soc. Rev., 2012, 41, 2072 RSC.
- H. B. Liu, X. C. Zhang, Y. Hu, X. H. Wang, L. T. Guo and C. L. Zhang, Acta Phys. Sin. (in chinese), 2005, 54, 4124 Search PubMed.
- G. M. Sheldrick, SHELXS-97 and SHELXL-97, Software for Crystal Structure Analysis, Siemens Analytical X-ray Instruments Inc., Wisconsin, Madison, USA, 1997 Search PubMed.
- G. M. Sheldrick, SHELXTL NT Version 5.1, Program for Solution and Refinement of Crystal Structures, University of Göttingen, Germany, 1997 Search PubMed.
- L.-N. Cui, K.-Y. Hu, Q.-H. Jin, Z.-F. Li, J.-Q. Wu and C.-L. Zhang, Polyhedron, 2011, 30, 2253 CrossRef CAS.
- M. Plabst, L. B. McCusker and T. Bein, J. Am. Chem. Soc., 2009, 131, 18112 CrossRef CAS PubMed.
- A. M. J. Lees, R. A. Kresinski and A. W. G. Platt, New J. Chem., 2004, 28, 1457 RSC.
- J. Fawcett and A. W. G. Platt, Polyhedron, 2003, 22, 967 CrossRef CAS.
- F.-Y. Liu, L. Roces, R. A. Sa Ferreira, S. García-Granda, J. R. García, L. D. Carlos and J. Rocha, J. Mater. Chem., 2007, 17, 3696 RSC.
- M. Plabst and T. Bein, Inorg. Chem., 2009, 48, 4331 CrossRef CAS PubMed.
- A. M. J. Lees and A. W. G. Platt, Inorg. Chem., 2003, 42, 4673 CrossRef CAS PubMed.
- E. A. Quadrelli, Inorg. Chem., 2002, 41, 167 CrossRef CAS PubMed.
- M. Nishio, S. Inami, M. Katayama, K. Ozutsumi and Y. Hayashi, Inorg. Chem., 2012, 51, 784 CrossRef CAS PubMed.
- L. Yang, G. Zhao, W. Li, Y. Liu, X. Shi, X. Jia, K. Zhao, X. Lu, Y. Xu, D. Xie, J. Wu and J. Chen, Spectrochim. Acta, Part A, 2009, 73, 884 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: CCDC 707117 (complex 1), 708865 (complex 2), 868853 (complex 3), 848975 (complex 4), 849206 (complex 5), 848976 (complex 6), 849207 (complex 7), 849208 (complex 8), 868724 (complex 9), 891950 (complex 10), 848977 (complex 11), 868725 (complex 12), 868365 (complex 13), 1561233 (complex 14), 868364 (complex 15) and 868854 (complex 16). For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c7ra07888a |
| This journal is © The Royal Society of Chemistry 2017 |