Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Synthesis and characterization of epitope-imprinted polymers for purification of human hemoglobin†
Héctor Bagán a,
Tongchang Zhoua,
Nélida Leiva Eriksson
a,
Tongchang Zhoua,
Nélida Leiva Eriksson ab,
Leif Bülow
ab,
Leif Bülow a and
Lei Ye
a and
Lei Ye *a
*a
aDivision of Pure and Applied Biochemistry, Lund University, Box 124, 221 00 Lund, Sweden. E-mail: lei.ye@tbiokem.lth.se; Fax: +46 46 2224611; Tel: +46 46 2229560
bSchool of Biological Sciences, University of Essex, Wivenhoe Park, Colchester, CO4 3SQ, UK
First published on 29th August 2017
Abstract
One promising method to prepare protein-selective polymers is the epitope-imprinting approach, where surface-accessible peptides from a target protein are used as templates to create surface-exposed binding sites on molecularly imprinted polymers (MIPs). However, selection of a suitable peptide target is not always straightforward, and synthesis of peptide on a large scale can be costly. In this work, we developed a new approach that can be used to select peptide epitopes on protein surface to be used as templates to prepare protein-selective MIPs. In this case study, human hemoglobin (Hb) was immobilized on silica nanoparticles and then fragmented by tryptic digestion. The particle-supported peptides were then used as templates to synthesize the Hb-selective MIPs, which were obtained after removal of the silica support and the peptides. The MIPs were tested in equilibrium binding experiments to evaluate their protein separation performance. The new surface imprinted MIPs displayed high selectivity for Hb, and was able to separate different variants of Hb from protein mixtures and crude cell extracts.
Introduction
Molecular imprinting is a widely used technique for the preparation of polymers with selectivity for separation and purification of various substances. The technique uses functional monomers that can form supramolecular complexes with the target molecule, which subsequently is cross-linked to generate specific polymers. Removal of the template molecule leaves selective binding sites in the cross-linked polymers, in which the steric dimension and the distribution of functional groups are defined by the molecular template.1,2 Molecularly imprinted polymers (MIPs) are frequently used to achieve separation and detection of low molecular weight compounds such as drugs, metal ions, sugars, steroids and amino acid derivatives.3–6 However, application of MIPs to purify large bio-macromolecules (e.g. proteins) is still a challenging task. The large molecular size of proteins makes it difficult for them to diffuse in cross-linked polymers, leading to incomplete template removal and very slow rebinding kinetics. Another important issue, often associated with proteins as template, is their low stability. Proteins can easily be denatured at elevated temperature or by exposure to organic solvents, conditions that frequently are employed to synthesize MIPs.In spite of the many challenges, some successful protein-imprinted polymers (MIPs) have been reported in the literature using sol–gel process and free radical polymerization.7–9 In particular, the use of hydrophilic monomers combined with low crosslinking densities has been adopted to offer practical protein diffusion in soft MIP gels. However, soft MIP particles are not suitable to use on a large scale, as for instance to be packed in columns to afford high efficiency affinity separation.
In a recent study, we developed a new procedure of protein imprinting based on Pickering emulsion polymerization. In this approach the protein template, hemoglobin (Hb), was immobilized on silica nanoparticles to stabilize an oil (monomer)-in-water emulsion.10 During the Pickering emulsion polymerization the protein could act as template to create accessible binding sites on the surface of the obtained cross-linked polymer beads. The new MIP beads had a high mechanical rigidity and displayed high selectivity for Hb, and could be used to separate Hb from crude bacterial cultures. The imprinting process involved exposure of the immobilized Hb to nonpolar organic phase and polymerization at an elevated temperature, which likely caused partial denaturation of the protein template. Since the prepared MIP beads displayed selective Hb binding when tested in non-denaturing solvent, it became plausible that even unstructured peptides may leave sequence-specific binding sites on a surface-imprinted polymer. Based on this result, we anticipated that it should be possible to use surface-exposed peptides (epitopes) to synthesize MIPs for selective uptake of target protein through interaction with the peptide epitopes, a concept that has been described previously.11–13
With the epitope imprinting method, a short peptide, even without a defined tertiary structure, can be used as template to produce protein-selective polymers. The main challenge is then to identify the most addressable epitope on a target protein, and to synthesize sufficient amount of the epitope for the molecular imprinting reaction. For proteins with known structural information, epitope identification may be achieved or facilitated through bioinformatic analysis. Nevertheless, chemical synthesis of the peptide epitope still remains costly and is difficult to use for preparing large amounts of MIPs. For proteins that lack reliable structural information, it is difficult to identify appropriate peptides that can be used as epitope templates to prepare the protein-selective polymers through molecular imprinting.
In this work, we have explored a new method that can be used to simplify identification and preparation of peptide epitopes for molecular imprinting. Using human Hb as model, we demonstrate that suitable peptide epitopes can be selected/identified by enzymatic digestion of a target protein that is pre-immobilized on silica beads. Using the peptides-decorated silica beads as template, we have carried out free radical polymerization and obtained hemoglobin-selective polymer particles after removing the sacrificial silica. The epitope-imprinted polymers have been characterized with standard protein samples as well as with crude cell extracts to evaluate the molecular separation performance.
Experimental
Materials
Silica nanoparticles (10 nm), aminopropyltriethoxysilane, glutaraldehyde (25% in water), sodium borohydride, trypsin, sodium dithionite, potassium ferricyanide, 1,4-dithiothreitol (DTT), acetic acid, hydrochloric acid, sodium dodecyl sulfate (SDS), methacrylic acid (MAA), acrylamide (Am), ethylene glycol dimethacrylate (EGDMA), toluene, methanol, chloroform, acetonitrile and 2,2′-azobis(2-methylpropionitrile) (AIBN) from Sigma Aldrich were used without further purification. Polyacrylamide gels (4–20%, Mini-protean) from Bio-Rad were used for SDS-PAGE analysis. Adult human hemoglobin (HbA), bovine serum albumin (BSA), myoglobin from equine skeletal muscle (Mb) and haptoglobin (Hp) were all obtained from Sigma Aldrich. Recombinant fetal human hemoglobin (HbF) was produced in E. coli as described previously.14Instruments
Attenuated total reflection infrared spectra were recorded using a Nicolet iS5 FT-IR instrument (Thermo Scientific, Madison, WI, USA). UV-visible spectra were measured using a Beckman Coulter DU 800 spectrophotometer (Beckman Coulter, Inc.). SDS-PAGE was carried out using a Bio-Rad Mini-PROTEAN Tetra Cell (Bio-Rad, Hercules, CA, USA).Polymer synthesis
In a round bottom flask (50 mL), 240 μL of glutaraldehyde (25%) were dissolved in 15 mL of water. To this solution, 1.0 g of the previously modified silica nanoparticles suspended in 5 mL water was added slowly. The suspension was stirred at room temperature for 24 hours. After that, the particles were collected, rinsed with water, methanol, and finally dried in a depressurized desiccator.
To reduce the Schiff base, the dry particles were suspended in 20 mL of methanol and cooled in an ice-water bath. To this suspension, 100 mg of sodium borohydride was added in small portions. The suspension was stirred in ice-water bath for 1 hour, then at room temperature for 3 hours. The unreacted sodium borohydride was destroyed by adding 10 mL of dilute hydrochloric acid. Finally, the silica particles were rinsed with methanol, water, and dried in a depressurized desiccator.
Protein binding analysis
Polymer particles (5 mg) were suspended in 1 mL of phosphate buffer (20 mM) containing Hb (0.5 mg). The mixture was gently stirred in a cold room (4–6 °C) for 3 hours. After centrifugation, the supernatant was collected and measured by UV-Vis spectrometry to calculate the amount of unbound Hb. The absorption bands at 405 (ε405 = 179 mM−1 cm−1) and 419 nm (ε405 = 191 mM−1 cm−1) were used to quantify Hb that contained Fe(III) and Fe(II)–CO, respectively.17To study the influence of Hp on the binding of Hb, HbA (0.5 mg, 31.5 μM) and Hp (0.5 mg) were first mixed in 1 mL of pH 7.4 buffer and incubated at 4–6 °C for 30 min. To the protein solution, 5 mg of polymer particles were added. The mixture was stirred at 4–6 °C for 3 hours. After centrifugation, the supernatant was collected and analysed by UV-Vis spectrometry.
To study the binding characteristics of different forms of Hb, the solution of HbA was first reduced with sodium dithionite and then saturated with CO.18 For the oxidation of HbF to the Met form, the protein was treated with potassium ferricyanide for two hours, followed by gel filtration to remove the excess potassium ferricyanide before the binding experiment.18
Optimization of elution conditions
To optimize the elution conditions, Hb was first allowed to bind to the polymer particles as described before. Next, the polymer particles were re-suspended in 1 mL of different elution solvents and gently stirred in a cold room (4–6 °C) for 30 min. After this step, the suspension was centrifuged and the supernatant was collected and analysed by UV-Vis spectrometry to determine the concentration of the eluted Hb. To test the functionality of the eluted Hb, the eluted protein was first reduced with sodium dithionite, then gently bubbled with CO or oxygen (O2) before it was analysed by UV-Vis spectrometry.Competitive protein binding assay
To study the influence of interfering proteins on Hb binding, 5 mg of polymer particles were mixed with 1 mL of pH 7.4 buffer containing 0.5 mg of Hb (31.5 μM) and 5 mg of the interfering protein (BSA, 76 μM; Mb, 315 μM). After 30 min incubation in a cold room, the samples were centrifuged, and the polymer particles were rinsed with 1 mL of pH 7.4 buffer. After this step, the polymer particles were incubated in 1 mL of pH 8 buffer containing 1 M NaCl for 30 min to elute the bound proteins. After centrifugation, the eluted proteins in the supernatants were analysed by SDS-PAGE.For the SDS-PAGE analysis, 50 μL of protein solution was mixed with 50 μL of the loading buffer. After 10 min of incubation at 100 °C, 10 μL of the solution was loaded to the gel. The electrophoresis was run for approximately 30 min at a voltage of 200 V in the pH 8.3 running buffer as specified by the supplier.
Separation of HbF from crude cell extracts
The crude extract from cell culture contained approximately 0.5 mg mL−1 of HbF (31.5 μM). The crude extract was first dialyzed against pH 7.4 phosphate buffer (20 mM). Next, 1 mL of the dialysed crude extract was mixed with 15 mg of polymer particles for 30 min in a cold room. Then, the polymer particles were rinsed with 1 mL of pH 7.4 buffer. After this step, the polymer particles were mixed with 100 μL of 1 M NaCl in pH 8.0 phosphate buffer (20 mM) for 30 min to elute the bound proteins. The different protein solutions collected were analysed by SDS-PAGE.Results and discussion
Identification and selection of epitope templates
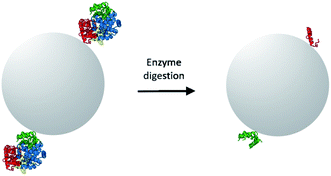
The objective of this study was to prepare MIPs selective for Hb by imprinting surface-exposed peptide sequences. As a starting point, we adopted the monomer composition used in our previous work where a Hb-selective MIP was synthesized by Pickering emulsion polymerization.10 In the previous work, the Pickering emulsion was stabilized by the protein-coated silica nanoparticles. The Hb immobilized on the silica surface acted as the molecular template to generate specific binding sites for Hb. One limitation of the previous work is that the hydrophilicity of the protein-coated nanoparticle stabilizer needs to be finely controlled in order to produce a thermodynamically stable, oil-in-water emulsion. While it is in principle possible to adjust the polarity of nanoparticle stabilizer by chemical modification or co-adsorption of additional molecules, the process of fine tuning the partition of the nanoparticles between the two immiscible phases is not always straightforward. To avoid the time-consuming step of optimizing Pickering emulsion, in the present work we selected to perform surface molecular imprinting in the interstices of nanoparticle-packed reactor. The surface exposed peptide sequences of Hb were immobilized on the nanoparticles to act as the template. Therefore, a pre-polymerization mixture composed of functional monomer and crosslinker were used to fill up the interstices of the packed silica, where a radical polymerization was initiated to produce a crosslinked polymer. Removing the silica nanoparticles from the composite monolith would lead to porous, peptide-imprinted polymer particles.The normal procedure of epitope-imprinting requires rational selection and synthesis of peptide sequence before it can be used as a template for molecular imprinting. For protein targets that have known structural information, surface exposed peptide sequences can be selected based on the 3D structure of the target proteins. However, identification of accessible peptide sequences becomes difficult when the 3D structure of a target protein is not available. In this situation, it will be very useful if alternative method can lead to correct identification/selection of suitable peptide sequences as the template epitopes. The method that we introduced to assist identification/selection of template epitopes, using Hb as a model target protein, is shown in Scheme 1. In this method, the target protein (Hb) is first immobilized through its surface-exposed amino-groups on silica nanoparticles, which is then subjected to on-particle digestion using site-specific enzymes. Because the protein is immobilized through its surface-accessible peptide sequences, a complete enzyme digestion will leave only the surface-accessible peptides to remain covalently bound on the nanoparticle surface. In addition to assisting selection of the correct epitope template, this new method also avoids the expensive chemical synthesis of peptide templates. In this work we choose to use trypsin as the digestive enzyme because this is one of the mostly used proteases in biochemical investigations and it has well-defined specificity in peptide cleavage.
The aldehyde-functionalized silica nanoparticles (Si@CHO) were synthesized in two steps following standard sol–gel chemistry. To immobilize surface accessible peptide epitopes on the aldehyde-functionalized silica nanoparticles, Hb was first conjugated to Si@CHO particles through reductive alkylation reaction, followed by an on-particle tryptic digestion to give the epitope-modified Si@epi particles. In this way only the surface-accessible peptides on Hb were able to act as templates in the next imprinting reaction. To study the importance of using surface epitopes as templates, we also immobilized pre-digested Hb fragments on Si@CHO particles, and used the obtained Si@pep particles in the following imprinting reaction. In the latter case all the peptide fragments from the tryptic digestion were expected to act as the templates during the imprinting reaction.
For immobilization of Hb and the products of its tryptic digest, silica nanoparticles were first modified with aminopropyltriethoxysilane to introduce amino groups on the particle surface. Fig. 1 shows the FT-IR spectra of the silica nanoparticles before and after modification with aminopropyltriethoxysilane. After the chemical modification, the silanol signal at 3400 cm−1 (due to the O–H stretching) disappeared, and new absorption bands related to the amino group at 2932 and 2979 cm−1 for the NH2 stretching, 1482–1564 cm−1 for the NH2 bending and 1387 cm−1 for the C–O–C stretching emerged. After further modification with glutaraldehyde, the amino signals from the silica nanoparticles disappeared (Fig. 1, curve c), and the aldehyde-modified nanoparticles turned into a pale pink colour.
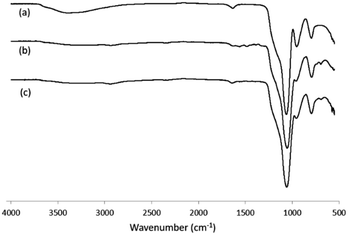 | ||
| Fig. 1 FT-IR spectra of (a) silica nanoparticles; (b) silica nanoparticles modified with amino groups; (c) silica nanoparticles modified with aldehyde groups (Si@CHO). | ||
For the two types of peptide-modified nanoparticles (Si@epi and Si@pep), their FT-IR spectra showed clearly the amide bond signals in the range of 1390–1650 cm−1 (Fig. 2), which confirmed the presence of the peptides on the surfaces of each particles.
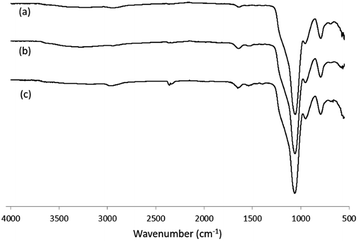 | ||
| Fig. 2 FT-IR spectra of (a) Si@CHO nanoparticles; (b) Si@pep nanoparticles; (c) Si@epi nanoparticles. | ||
Effects of the template peptides on Hb binding
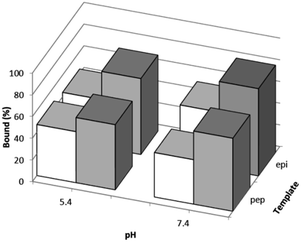
The effects of the template peptides used during the imprinting reaction on Hb binding was studied using equilibrium binding analysis. Hb (0.5 mg mL−1) was incubated with 5 mg of the two different types of imprinted polymers, prepared using Si@epi and Si@pep as the particle-supported templates, before the unbound fraction was collected and quantified by UV-Vis spectroscopy. As shown in Fig. 3, the two types of MIP particles displayed higher Hb binding than the NIP particles, and the highest specific Hb binding was observed at pH 7.4 for the MIP particles prepared using the Si@epi nanoparticles. Based on these results, it can be concluded that the use of the surface epitopes as templates led to MIP particles with higher selectivity for Hb. Therefore, in the remaining experiments, we focused on investigation of the epitope-imprinted polymer particles.The influence of more basic pH values on Hb binding to the epitope-imprinted polymer was further investigated. As shown in Fig. 4, Hb binding to the epitope-imprinted polymer decreased when the pH was changed from 7.4 to 9.0, suggesting that the bound Hb can be eluted from the MIP by increasing the pH of the elution buffer.
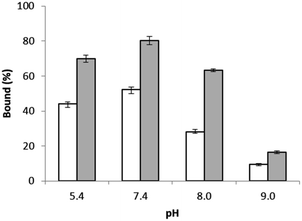 | ||
| Fig. 4 Hb binding to the epitope-imprinted polymer (solid bars) and the non-imprinted polymer particles (empty bars) at different pH values. | ||
Instead of eluting Hb using high pH buffer (pH = 9.0), we also investigated the use of NaCl in more neutral buffer to recover the bound Hb from the MIP particles. Table 1 shows the effect of addition of NaCl on the Hb recovery from the epitope-imprinted polymer particles at two different pH values. By increasing the salt concentration and using a slightly basic pH buffer, up to 72% Hb bound to the MIP particles could be eluted in one single step.
| pH | NaCl conc. (M) | Hb eluted (%) | |
|---|---|---|---|
| NIP | MIP | ||
| 7.4 | 0.2 | 56 | 51 |
| 7.4 | 1 | 71 | 62 |
| 8.0 | 0.2 | 60 | 62 |
| 8.0 | 1 | 72 | 72 |
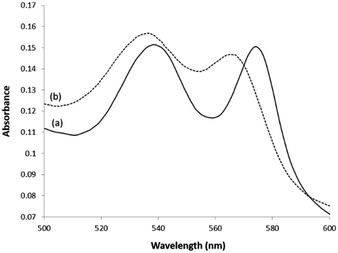
To test the activity of the Hb eluted by 1 M NaCl in pH 8.0 buffer, the eluted Hb was first reduced with sodium dithionite, then saturated with CO or O2 separately before being characterized with UV-Vis spectroscopy. As shown in Fig. 5, the UV-Vis absorbance spectra of the Hb in the CO- and O2-binding status are in agreement with a functionally active Hb.19 Therefore, in the remaining experiments we selected to use pH 8.0 buffer containing 1 M NaCl as the elution buffer to recover Hb from the epitope-imprinted polymer particles.
In the next step, the capability of the epitope-imprinted polymer to bind adult Hb (HbA) and fetal Hb (HbF) in different forms was investigated. The recombinant HbF tested in this work was expressed in E. coli and was purified by standard ion exchange chromatography prior to use. The HbA and HbF proteins at different oxidation states, and in the CO-bound form were incubated with the polymer particles to evaluate their binding under the different conditions.
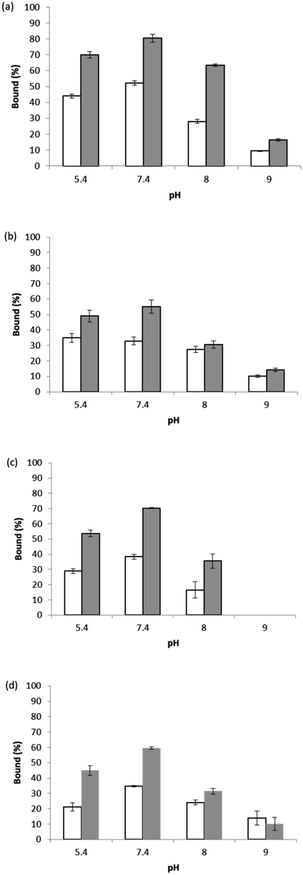
Fig. 6 shows the binding of the two Hb proteins in two different forms measured at four different pH values. In general, the specific binding of HbF is lower than the specific binding of HbA, and the specific binding of Hb in its Fe(III) form is slightly higher than in its Fe(II) form saturated with CO. The difference between HbA and HbF binding can be explained by the fact that unlike HbA, the HbF is a α2γ2 tetramer. The amino acid sequence in the γ-chain is not identical to the β-chain of HbA that was used to prepare the epitope templates. In addition, in the recombinant HbF, the two α- and two γ-subunits contain an additional N-terminal Met residue. Despite these variations, specific binding to the epitope-imprinted particles for HbA and HbF was observed.
The capability of the epitope-imprinted polymer to bind both HbA and HbF can be explained by that the two proteins have the same α-subunits. In the epitope-imprinting process, HbA was immobilized on aldehyde-modified silica nanoparticles using the same chemical conjugation as previously used by Hu et al. to link poly(ethylene glycol) (PEG) to Hb.20 Based on the results of Hu et al., there are six types of surface-exposed amino groups in HbA that are able to react with terminal aldehyde on large PEG molecules, with a decreasing order of reactivity as: Val-1(α) = Val-1(β) ≫ Lys-8(β) > Lys-11(α) > Lys-40(α) > Lys-56(α). Compared to the lysine residues, the four N-terminal valine residues were significantly more reactive to conjugate to PEG via its terminal aldehyde. Most likely, the HbA immobilized on the Si@CHO nanoparticles prepared in our present work was coupled to the silica nanoparticles via the N-terminal valine residues in the α- and the β-chains. Based on the amino acid sequence of the two polypeptide chains in HbA and considering that trypsin cleaves peptide bond next to lysine residue, it is likely that the predominant epitopes presented on the silica nanoparticles, after tryptic digestion, are derived from the two N-terminal sequences in the α- and the β-chains:
VLSPADK (α-chain)
VHLTPEEK (β-chain)
Although the presence of other peptide sequences on the Si@epi nanoparticles due to Hb immobilization through the lysine residues can not be excluded, these sequences may have only a low abundance and therefore contribute much less to the overall imprinting effect. On the bases of specific binding observed for HbF, it is likely that the N-terminal sequence in the α-chain of HbA played a major role in defining the specific binding sites.
Selectivity of protein binding to the epitope-imprinted polymer particles
To evaluate the protein selectivity of the epitope-imprinted polymer, the uptake of HbA and its mixture with haptoglobin (Hp) by the imprinted polymer were compared with that of bovine serum albumin (BSA) and myoglobin (Mb). When the Hb-Hp complex was tested, the two proteins were pre-incubated to allow formation of their complex before the polymer particles were added. Since Hp is known to bind Hb with a very high affinity,21 the uptake of Hb by the imprinted polymer is expected to decrease when Hp is introduced. This expected effect of Hp is clearly seen in Fig. 7. The inhibition of Hb binding by Hp is understandable, as complexation with Hp makes the N-terminal of the α-chain in HbA become inaccessible.21 The lack of differential binding between the MIP and NIP particles for BSA and Mb is due to that these proteins do not have surface epitopes similar to the peptide templates used during the imprinting reaction.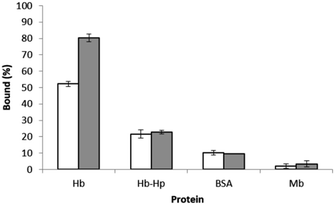 | ||
| Fig. 7 Binding of HbA in the absence and presence of Hp, BSA and Mb to 5 mg of the polymer particles in 1 mL of phosphate buffer at pH 7.4. The concentration of each proteins was 0.5 mg mL−1. | ||
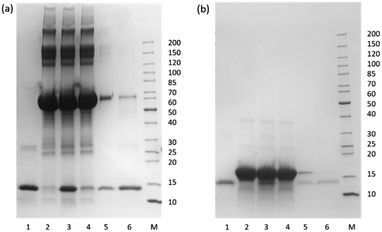
To test the specific Hb binding in the presence of a large excess of interfering proteins, HbA was mixed with BSA or Mb before the protein mixture was treated with the epitope-imprinted polymer particles. BSA and Mb were selected as representative interfering proteins due to their similarity to Hb in terms of molecular weight and tertiary structure, respectively. In the competitive binding experiment, 1 mL of protein solution containing Hb and the interfering protein were first incubated with 5 mg of the MIP particles. The polymer particles were then washed with the binding buffer, before the bound proteins were eluted with 1 mL of pH 8.0 buffer containing 1 M NaCl. After each step the protein solutions collected were analysed by SDS-PAGE. In the case of using Mb as the interfering protein, to avoid the problem of merging protein bands from the Hb subunits and Mb, all the protein solutions collected were diluted three times before the samples were analysed by SDS-PAGE.
From the SDS-PAGE results shown in Fig. 8, it is clear that the MIP particles could selectively bind Hb in the presence of a large excess of BSA and Mb. Also, after a simple washing, the adsorbed protein could be eluted from the MIP particles in a single elution step to give the purified Hb. The capability of the MIP particles to separate and purify Hb from the closely related Mb is particularly interesting, as both the primary and tertiary structures of these proteins have a high similarity, and the clearly demonstrated protein selectivity of the MIP particles is encouraging for demanding protein separations in complex biological samples.
Separation of recombinant HbF from crude cell culture
The higher O2 affinity and stability of fetal hemoglobin are attractive for developing hemoglobin based oxygen carriers (HBOCs) for therapeutic purposes.14 One possible approach to producing large quantity of recombinant HbF is to express the protein in bacteria.22 To test if the epitope-imprinted polymer can be used to affect protein separation in early stage of downstream purification, we used crude extract of E. coli cells expressing HbF as a model. The E. coli cells used for the HbF production were first disintegrated by ultrasound sonication. After removing the particulates, the supernatant was dialyzed against pH 7.4 buffer, and then saturated with CO to keep the HbF in its Fe(II) form before the sample was treated with the imprinted polymer particles. Most of the proteins in the crude cell extract did not bind to the imprinted polymer particles, and was removed during the loading and washing steps. In the protein solution that was finally eluted from the MIP particles, the concentration of HbF became significantly higher than in all the other fractions (Fig. S1, in ESI†). Although the effect of protein purification offered by this single step treatment is limited, the preliminary result is encouraging for further optimizing and scaling up the MIP synthesis in the next stage of development. We expect that more effective purification of recombinant HbF can be realized by using optimized MIPs in affinity column chromatography setting.Conclusions
In this work we have developed a new approach for synthesizing epitope-imprinted polymers for selective protein separation. Using tryptic digestion of target protein immobilized on silica nanoparticles, it was demonstrated that the selected peptide epitopes could act as molecular templates to generate protein-specific binding sites on the surface of crosslinked polymer particles. The proposed method for epitope identification should be particularly useful when the structure of the target protein is not known. In the case of hemoglobin separation, our synthetic approach provided MIP particles that displayed clearly selective binding for HbA and recombinant HbF. The possibility of using sterilisable MIP to enable protein purification is encouraging for larger scale bioseparations, in particular for purification of therapeutic proteins in the biomedical and biotechnology areas.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Swedish Foundation for Strategic Research project “Production of Blood Substitutes” (contract RBP 14-0055).Notes and references
- L. Ye, Adv. Biochem. Eng./Biotechnol., 2015, 150, 1–24 CrossRef CAS PubMed.
- L. Chen, X. Wang, W. Lu, X. Wu and J. Li, Chem. Soc. Rev., 2016, 45, 2137–2211 RSC.
- T. Zhou, L. Jørgensen, M. A. Mattebjerg, I. S. Chronakis and L. Ye, RSC Adv., 2014, 4, 30292–30299 RSC.
- Y.-G. Zhao, X.-H. Chen, S.-D. Pan, H. Zhu, H.-Y. Shen and M.-C. Jin, J. Mater. Chem. A, 2013, 1, 11648 CAS.
- Y. Ma, G. Pan, Y. Zhang, X. Guo and H. Zhang, Angew. Chem., Int. Ed., 2013, 52, 1511–1514 CrossRef CAS PubMed.
- P. Parmpi and P. Kofinas, Biomaterials, 2004, 25, 1969–1973 CrossRef CAS PubMed.
- A. Nematollahzadeh, W. Sun, C. S. Aureliano, D. Lütkemeyer, J. Stute, M. J. Abdekhodaie, A. Shojaei and B. Sellergren, Angew. Chem., Int. Ed., 2011, 50, 495–498 CrossRef CAS PubMed.
- S. Yu, A.-Q. Luo, D. Biswal, J. Z. Hilt and D. Puleo, Talanta, 2010, 83, 156–161 CrossRef CAS PubMed.
- X. Pang, G. Cheng, R. Li, S. Lu and Y. Zhang, Anal. Chim. Acta, 2005, 550, 13–17 CrossRef CAS.
- T. Zhou, K. Zhang, T. Kamra, L. Bülow and L. Ye, J. Mater. Chem. B, 2015, 3, 1254–1260 RSC.
- A. Rachkov and N. Minoura, Biochim. Biophys. Acta, 2001, 1544, 255–266 CrossRef CAS.
- H. Nishino, C.-S. Huang and K. J. Shea, Angew. Chem., Int. Ed., 2006, 45, 2392–2396 CrossRef CAS PubMed.
- M. Kempe and K. Mosbach, J. Chromatogr. A, 1995, 691, 317–323 CrossRef CAS PubMed.
- K. Ratanasopa, T. Cedervall and L. Bülow, Adv. Exp. Med. Biol., 2016, 876, 445–453 CrossRef CAS PubMed.
- P. K. Jal, S. Patel and B. K. Mishra, Talanta, 2004, 62, 1005–1028 CrossRef CAS PubMed.
- H. K. Hustoft, H. Malerod, S. R. Wilson, L. Reubsaet, E. Lundanes and T. Greibrokk, A Critical Review of Trypsin Digestion for LC-MS Based Proteomics, ed. H.-C. Leung, InTech, Integrative Proteomics, 2012, DOI:10.5772/29326.
- F. Meng and A. I. Alayash, Anal. Biochem., 2017, 521, 11–19 CrossRef CAS PubMed.
- J. B. Conant and N. D. Scott, J. Biol. Chem., 1926, 69, 575–587 CAS.
- W. G. Zulstra and A. Buursma, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 1987, 88, 251–255 CrossRef.
- T. Hu, M. Prabhakaran, S. Acharya and B. N. Manjula, Biochem. J., 2005, 392, 555–564 CrossRef CAS PubMed.
- C. B. F. Andersen, M. Torvund-Jensen, M. J. Nielsen, C. L. P. de Oliveira, H.-P. Hersleth, N. H. Andersen, J. S. Pedersen, G. R. Andersen and S. K. Moestrup, Nature, 2012, 489, 456–460 CrossRef CAS PubMed.
- H. W. Kim and A. G. Greenburg, Artif. Cells, Blood Substitutes, Biotechnol., 2006, 34, 537–550 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Additional SDS-PAGE results of protein samples. See DOI: 10.1039/c7ra07674f |
| This journal is © The Royal Society of Chemistry 2017 |