Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Promiscuity of inhibitors of human protein kinases at varying data confidence levels and test frequencies
Dagmar Stumpfea,
Annachiara Tinivellab,
Giulio Rastelli b and
Jürgen Bajorath
b and
Jürgen Bajorath *a
*a
aDepartment of Life Science Informatics, B-IT, LIMES Program Unit Chemical Biology and Medicinal Chemistry, Rheinische Friedrich-Wilhelms-Universität, Dahlmannstr. 2, D-53113 Bonn, Germany. E-mail: bajorath@bit.uni-bonn.de; Fax: +49-228-2699-341; Tel: +49-228-2699-306
bDepartment of Life Sciences, University of Modena and Reggio Emilia, Via Campi 103, 41125, Modena, Italy
First published on 23rd August 2017
Abstract
More than 141![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 inhibitors of human kinases and their activity data were assembled to perform an in-depth analysis of inhibitor promiscuity (single- versus multi-kinase activity) at varying activity data confidence levels. For ∼20% of these inhibitors, it was also possible to consider test frequency and inactivity information. Only small subsets of highly promiscuous inhibitors were identified. Nearly 95% of more than 45
000 inhibitors of human kinases and their activity data were assembled to perform an in-depth analysis of inhibitor promiscuity (single- versus multi-kinase activity) at varying activity data confidence levels. For ∼20% of these inhibitors, it was also possible to consider test frequency and inactivity information. Only small subsets of highly promiscuous inhibitors were identified. Nearly 95% of more than 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 inhibitors with high-confidence data were only active against one or at most two kinases. At decreasing data confidence levels, more than 92
000 inhibitors with high-confidence data were only active against one or at most two kinases. At decreasing data confidence levels, more than 92![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 kinase inhibitors were on average active against two kinases. When taking all activity information without any restrictions into account, the mean promiscuity degree of kinase inhibitors was less than four and notably biased by small numbers of highly promiscuous inhibitors. Even under these conditions, more than 70% of all inhibitors were active against a single kinase. There was only small-scale progression of inhibitor promiscuity when data confidence criteria were iteratively removed during the analysis. Furthermore, the majority of inhibitors that were tested against 10 to 20 different kinases were only active against a single kinase. The results of our activity data-driven analysis indicate that promiscuity of kinase inhibitors cannot generally be assumed. Many inhibitors retain single-kinase activity at decreasing data confidence criteria or increasing test frequency. Hence, on the basis of currently available data, many kinase inhibitors are selective, which is an important aspect for drug development.
000 kinase inhibitors were on average active against two kinases. When taking all activity information without any restrictions into account, the mean promiscuity degree of kinase inhibitors was less than four and notably biased by small numbers of highly promiscuous inhibitors. Even under these conditions, more than 70% of all inhibitors were active against a single kinase. There was only small-scale progression of inhibitor promiscuity when data confidence criteria were iteratively removed during the analysis. Furthermore, the majority of inhibitors that were tested against 10 to 20 different kinases were only active against a single kinase. The results of our activity data-driven analysis indicate that promiscuity of kinase inhibitors cannot generally be assumed. Many inhibitors retain single-kinase activity at decreasing data confidence criteria or increasing test frequency. Hence, on the basis of currently available data, many kinase inhibitors are selective, which is an important aspect for drug development.
Introduction
Protein kinases are major drug targets1,2 and the promiscuity of classical ATP site-directed (type I) kinase inhibitors continues to be debated.3,4 In this context, promiscuity is rationalized as the ability of an inhibitor to interact with multiple kinases. Given that the ATP site is largely conserved across the kinome, it is often thought that these inhibitors might be active against many different kinases. The human kinome comprises 518 kinases,5 excluding complexes and isoforms. Some highly promiscuous kinase inhibitors were identified.6 Among these were compounds that have proven effective as anti-cancer agents for which promiscuity and ensuing polypharmacology often play a decisive role.3On the other hand, recent global analyses of high-confidence activity data for publicly available inhibitors of the human kinome have revealed that the majority of these inhibitors were only annotated with one or two kinase targets.6,7 Data incompleteness and the unavailability of test frequency information in the literature might at least in part explain these findings, but it is also conceivable that the promiscuity of many ATP site-directed kinase inhibitors is indeed lower than often thought.
Data-driven promiscuity assessment benefits from concentrating on high-confidence activity data. This requires careful data curation, but provides the best possible basis for arriving at sound promiscuity estimates. Although such estimates are intrinsically conservative, they are least influenced by experimental heterogeneity. For compound data mining, this is an important aspect to consider.6
Of course, further differentiated data mining strategies might be considered. For example, given that the majority of kinase inhibitors were not found to be promiscuous on the basis of high-confidence activity data, one might assess the issue of data sparseness by softening confidence criteria and taking increasing amounts of activity data into account. This would make it possible to evaluate an anticipated progression of inhibitor promiscuity as activity data increases and determine its magnitude. Thus, monitoring promiscuity progression was a primary goal of our current analysis.
In addition, our study was further motivated by the remarkable increase in the number of kinase inhibitors that are becoming publicly available.8 For example, our survey of inhibitors of the human kinome in 20156 was based upon nearly 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 kinase inhibitors for which high-confidence activity data were available in ChEMBL,9 the major public repository of compounds from the medicinal chemistry literature. These inhibitors were active against a total of 266 human kinases.6 However, early in 2017, more than 45
000 kinase inhibitors for which high-confidence activity data were available in ChEMBL,9 the major public repository of compounds from the medicinal chemistry literature. These inhibitors were active against a total of 266 human kinases.6 However, early in 2017, more than 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 kinase inhibitors with high-confidence activity data were available in ChEMBL.8 These inhibitors were active against 286 human kinases. Hence, within merely two years, the number of public kinase inhibitors more than doubled, although kinome coverage only slightly increased. Notably, 70% of the qualifying inhibitors available in 2015 were only annotated with a single kinase.6 For the much larger number of inhibitors available in early 2017, this proportion further increased to 76%.8
000 kinase inhibitors with high-confidence activity data were available in ChEMBL.8 These inhibitors were active against 286 human kinases. Hence, within merely two years, the number of public kinase inhibitors more than doubled, although kinome coverage only slightly increased. Notably, 70% of the qualifying inhibitors available in 2015 were only annotated with a single kinase.6 For the much larger number of inhibitors available in early 2017, this proportion further increased to 76%.8
Moreover, we have also addressed the issue of promiscuity versus test frequency by including compounds from PubChem BioAssays11 in our analysis. Extensively tested screening compounds with activity in kinase assays were identified and their target-based assay frequency and promiscuity were determined. Thus, in addition to studying promiscuity at varying data confidence levels for large numbers of kinase inhibitors, it was also possible to compare inhibitor promiscuity in the presence and absence of test frequency information. The results of our analysis are reported in the following.
Methods and materials
Human kinase targets
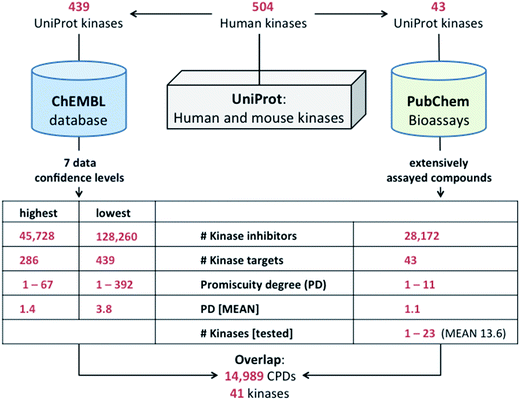
Human kinases were extracted from UniProt10 that organizes both human and mouse kinases and contains 504 human entries. UniProt IDs for human kinases were linked to ChEMBL9 and PubChem11 target IDs to consistently associate compounds with kinases.Kinase inhibitors
Classical (type I) kinase inhibitors are ATP site-directed, compete with ATP, and bind to the “DFG-in” conformation of the activation loop in the active site.12 Although other classes (type II–IV) of kinase inhibitors exist,12,13 which partly bind to regions distant from the ATP site and are allosteric in nature (type III–IV),12–14 the vast majority of publicly available kinase inhibitors, estimated to be more than 98%,6 are type I compounds. Thus, for large-scale activity data mining, other types of inhibitors can currently be neglected.Compounds active against at least one of all available human kinases were selected from ChEMBL release 22 and a subset of screening compounds from PubChem BioAssays.11 This subset of PubChem consisted of 437![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 257 compounds that were tested in both primary assays (percentage of inhibition from a single dose) and confirmatory assays (dose-response assays yielding IC50 values).16 From this subset of extensively assayed compounds, with a mean and median of 411 and 437 assays per compound, respectively,15 kinase inhibitors were selected. Kinase inhibitors from ChEMBL were analyzed under varying activity data confidence criteria, as specified below. From ChEMBL, inactivity records from assays or test frequency data cannot be obtained. However, for PubChem inhibitors, the number of kinases against which they were tested (also referred to as test frequency) was determined and taken into account in assessing their promiscuity. Fig. 1 summarizes the target and compound selection process.
257 compounds that were tested in both primary assays (percentage of inhibition from a single dose) and confirmatory assays (dose-response assays yielding IC50 values).16 From this subset of extensively assayed compounds, with a mean and median of 411 and 437 assays per compound, respectively,15 kinase inhibitors were selected. Kinase inhibitors from ChEMBL were analyzed under varying activity data confidence criteria, as specified below. From ChEMBL, inactivity records from assays or test frequency data cannot be obtained. However, for PubChem inhibitors, the number of kinases against which they were tested (also referred to as test frequency) was determined and taken into account in assessing their promiscuity. Fig. 1 summarizes the target and compound selection process.
 | ||
| Fig. 1 Kinases and inhibitors. Human kinases were mapped and inhibitors extracted from ChEMBL and PubChem. On the left, kinase inhibitor data obtained from ChEMBL are summarized for varying selection criteria (according to Fig. 2). On the right, extensively tested inhibitors of kinases with ChEMBL target IDs available in PubChem are reported. | ||
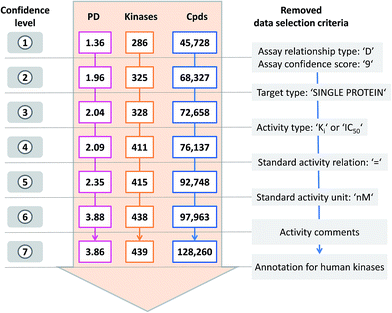
Data confidence criteria
From ChEMBL, inhibitors can be selected following a hierarchy of seven database-specific criteria9 to gradually increase (or decrease) the degree of activity data confidence,16 as illustrated in Fig. 2. For our analysis, we first extracted kinase inhibitors on the basis of high activity data confidence, corresponding to confidence level 1 in Fig. 2. Then, selection criteria were iteratively removed (from the top to bottom in Fig. 2) to gradually transition from high- to low-confidence data, producing a total of seven different confidence levels (1–7). The following criteria were applied:(i) Direct interaction assays with highest confidence: assay relationship type ‘D’, assay confidence sore ‘9’;
(ii) Specific targets: target type ‘SINGLE PROTEIN’;
(iii) Defined activity measurements: activity type ‘Ki’ or ‘IC50’;
(iv) Specified activity values: standard relation ‘=’;
(v) Standard activity unit: ‘nM’;
(vi) Activity comments: removal of compounds designated as inconclusive, not active, inactive, not evaluated/determined.
(vii) Kinase organism annotation: ‘Homo sapiens’.
Accordingly, kinase inhibitors at the highest confidence level 1 were required to meet all seven selection criteria, yielding the smallest set. By contrast, for kinase inhibitors at the lowest confidence level 7, all available activity data were taken into account, without any confidence measures, hence producing the largest set of inhibitors. The increase in the number of inhibitors between confidence level 1 and 7 was not dependent on the order in which selection criteria were applied. Each confidence level defines activity criteria for ChEMBL compounds. We note that Ki and IC50 values were not separately considered here to support increasing promiscuity levels for inhibitors.
In PubChem, similar data confidence criteria cannot be applied. However, in addition to focusing on extensively assayed inhibitors, the requirement of qualifying PubChem compounds to be tested in both primary and confirmatory assays also represented a data confidence criterion. For example, under these conditions, low-confidence kinase profiling data from single experiments incorporated into PubChem did not qualify for the analysis. For kinase inhibitors from PubChem, activity annotations from primary and confirmatory assays against different kinase targets were combined to yield upper level promiscuity estimates.
Promiscuity degree
For our analysis, the promiscuity degree (PD) of an inhibitor was defined as the number of human kinases it was active against.Data mining and analysis
All calculations were performed using in-house scripts and KNIME17 protocols with the aid of the OpenEye18 chemistry toolkit.Results and discussion
Analysis scheme
Our comprehensive analysis of kinase inhibitors and their activity data in ChEMBL and PubChem is summarized in Fig. 1. We first mapped UniProt IDs of human kinases to ChEMBL and PubChem. A total of 439 kinase entries were detected in ChEMBL. In addition, the extensively assayed subset of PubChem included compounds tested against 43 kinases. For all human kinases in ChEMBL and PubChem, inhibitors were systematically identified and assigned to UniProt IDs. Then their activity records were analyzed.ChEMBL
We first analyzed data from ChEMBL. Compounds and activity data in ChEMBL primarily originate from the medicinal chemistry literature and are manually curated. Given the source of the data, no records of test frequency or inactivity are provided in this database.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 260 inhibitors for 439 kinases. For 45
260 inhibitors for 439 kinases. For 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 728 of these inhibitors, which were active against 286 human kinases, high-confidence activity data were also available, corresponding to confidence level 1 in Fig. 2. Hence, an unprecedentedly large number of inhibitors was analyzed for nearly 300 (level 1) and more than 400 (level 7) human kinases.
728 of these inhibitors, which were active against 286 human kinases, high-confidence activity data were also available, corresponding to confidence level 1 in Fig. 2. Hence, an unprecedentedly large number of inhibitors was analyzed for nearly 300 (level 1) and more than 400 (level 7) human kinases.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 748 inhibitors. An increase in the mean PD from 2.4 to 3.9 was only observed when standard activity units were no longer required and all types of measurements were considered including, for example, percentage of inhibition or residual activity. Thus, as long as at least the standard activity unit (nM) was reported in activity records, the mean promiscuity of human kinase inhibitors from ChEMBL was low, even if no additional data confidence criteria were applied. At highest measurement confidence, corresponding to confidence level 3, the mean PD value was 2.0 and decreased to 1.4 when highest assay confidence was also required (proceeding from level 3 to 1). For all three mean promiscuity values of 1.4 (level 1), 2.0 (3), and 3.9 (7), the corresponding median values were 1.0, hence indicating that small numbers of highly promiscuous inhibitors were mostly responsible for the PD increase, especially from 2.0 to 3.9.
748 inhibitors. An increase in the mean PD from 2.4 to 3.9 was only observed when standard activity units were no longer required and all types of measurements were considered including, for example, percentage of inhibition or residual activity. Thus, as long as at least the standard activity unit (nM) was reported in activity records, the mean promiscuity of human kinase inhibitors from ChEMBL was low, even if no additional data confidence criteria were applied. At highest measurement confidence, corresponding to confidence level 3, the mean PD value was 2.0 and decreased to 1.4 when highest assay confidence was also required (proceeding from level 3 to 1). For all three mean promiscuity values of 1.4 (level 1), 2.0 (3), and 3.9 (7), the corresponding median values were 1.0, hence indicating that small numbers of highly promiscuous inhibitors were mostly responsible for the PD increase, especially from 2.0 to 3.9.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 260 inhibitors were active against at least five and at least 10 kinases, respectively. At decreasing levels of data confidence, only small subsets of highly promiscuous inhibitors were detected. Thus, the increase in mean PD values from 2.4 to 3.9 observed at level 5 and 7 was largely due to small numbers of highly promiscuous inhibitors (which may also include activity artifacts), as indicated by the difference between increasing mean and constant median PD values discussed above.
260 inhibitors were active against at least five and at least 10 kinases, respectively. At decreasing levels of data confidence, only small subsets of highly promiscuous inhibitors were detected. Thus, the increase in mean PD values from 2.4 to 3.9 observed at level 5 and 7 was largely due to small numbers of highly promiscuous inhibitors (which may also include activity artifacts), as indicated by the difference between increasing mean and constant median PD values discussed above.PubChem
Given that no records of assay frequency or inactivity are available in ChEMBL, we extended the analysis to the PubChem BioAssay database.11 Inactivity or assay frequency data for screening compounds are not provided in PubChem records either, but can be obtained by determining in which assays individual compounds have been tested and found to be active. We analyzed 437![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 257 PubChem compounds that were extensively tested in both primary and confirmatory assays, identified inhibitors of human kinases, and determined their target-based test frequency.
257 PubChem compounds that were extensively tested in both primary and confirmatory assays, identified inhibitors of human kinases, and determined their target-based test frequency.
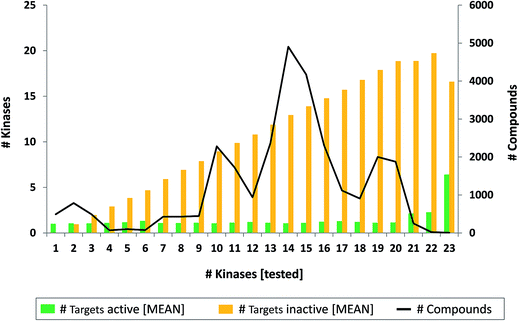
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 172 kinase inhibitors with activity against 43 different kinases. These inhibitors from PubChem were tested against one to 23 human kinases, with a mean of 13.6 kinases per compound. Notably, 14
172 kinase inhibitors with activity against 43 different kinases. These inhibitors from PubChem were tested against one to 23 human kinases, with a mean of 13.6 kinases per compound. Notably, 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 989 of the inhibitors detected in PubChem, with activity against 41 different kinases, were also found in ChEMBL (owing to the fact that ChEMBL also incorporates data from the PubChem BioAssay collection).
989 of the inhibitors detected in PubChem, with activity against 41 different kinases, were also found in ChEMBL (owing to the fact that ChEMBL also incorporates data from the PubChem BioAssay collection).Exemplary inhibitors
Fig. 4 shows different sets of structurally analogous kinase inhibitors from ChEMBL (top, middle) and PubChem (bottom) having varying promiscuity degrees. In the first set (top), the promiscuous compound on the left had a PD value of 5 at confidence level 1 and a PD of 7 at level 7. Therefore, this compound was promiscuous at highest data confidence and displayed a moderate increase in promiscuity at the lowest confidence level. By contrast, its two structural analogs were only active against a single kinase at the highest and lowest confidence level. Furthermore, all three inhibitors in the second set (middle) were active against a single kinase at confidence level 1. Proceeding to confidence level 7, the PD value of only the inhibitor on the left increased from 1 to 3, whereas it remained constant for the other two analogs. Hence, these compounds were examples for selective inhibitors under varying data confidence. Moreover, the inhibitors of the third series from PubChem (bottom) were tested (from the left to the right) against 13, 10, and 18 different kinases. The promiscuous inhibitor on the left was found to be active against three of 13 kinases, while the other two analogs were only active against a single kinase. Thus, there were also selective within their experimental boundaries. These examples illustrate the prevalence of single-kinase inhibitors and also show that closely related analogs might have varying promiscuity degrees. In addition to data sparseness, assay variance and different assay formats might also influence promiscuity degrees, but it is evident that intrinsic promiscuity is far from being the rule among the kinase inhibitors investigated here.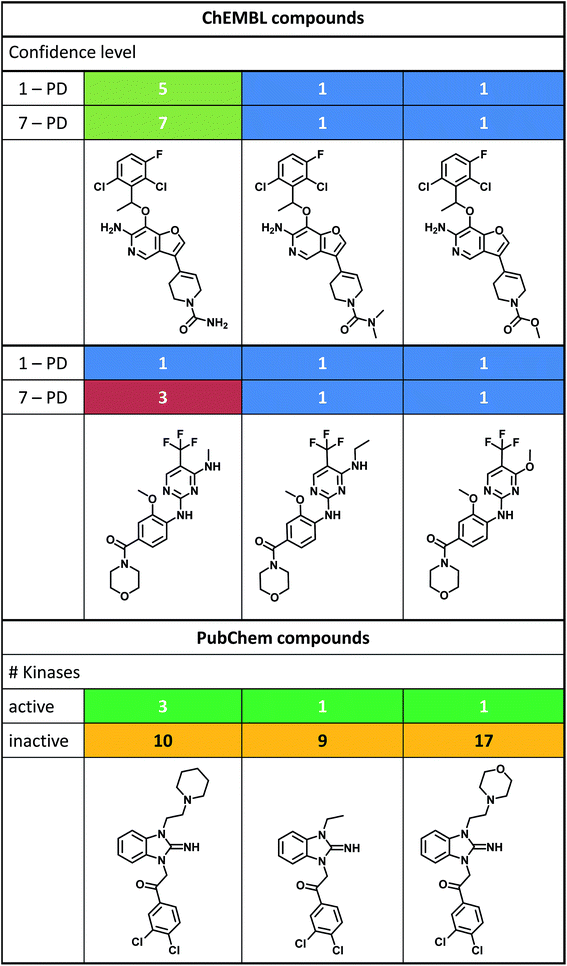 | ||
| Fig. 4 Kinase inhibitors with varying promiscuity degrees. Shown are three different sets of structurally analogous promiscuous or non-promiscuous kinase inhibitors. Inhibitors in the top and middle panel originated from ChEMBL. For these inhibitors, high-confidence activity data were available and their PD values are reported for confidence level 1 and 7. Cells containing different PD values are color-coded. Inhibitors in the bottom panel originated from PubChem and the number of kinases they were active or inactive against is reported (color-coded according to Fig. 3). | ||
Conclusions
In this study, we have – for the first time – analyzed the promiscuity of human kinase inhibitors at varying data confidence levels and taking test frequency information into account. The vast majority of publicly available kinase inhibitors are directed against the ATP binding site. Since this site is largely conserved across the kinome, these types of inhibitors are often thought to be promiscuous, which has also become a paradigm for the use of kinase inhibitors in cancer treatment.Over the past two years, the number of kinase inhibitors for which high-confidence activity data are available has more than doubled. Combining ChEMBL and PubChem as compound data sources, more than 141![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 kinase inhibitors with at least low-confidence activity data were obtained. For about 20% of these inhibitors, it was possible to determine target-based test frequency and the proportion of targets the compounds were active against. Thus, there was an excellent basis for re-visiting the issue of kinase inhibitor promiscuity versus selectivity by large-scale compound and activity data analysis, which has motivated our investigation.
000 kinase inhibitors with at least low-confidence activity data were obtained. For about 20% of these inhibitors, it was possible to determine target-based test frequency and the proportion of targets the compounds were active against. Thus, there was an excellent basis for re-visiting the issue of kinase inhibitor promiscuity versus selectivity by large-scale compound and activity data analysis, which has motivated our investigation.
Through systematic compound data mining only small subsets of highly promiscuous kinase inhibitors were identified. By contrast, the majority of inhibitors from medicinal chemistry and screening sources were only active against a single kinase. For more than 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 inhibitors with available high-confidence activity data, a mean PD of 1.4 was obtained. At decreasing data confidence levels, mean PD values of more than 92
000 inhibitors with available high-confidence activity data, a mean PD of 1.4 was obtained. At decreasing data confidence levels, mean PD values of more than 92![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 kinase inhibitors remained low at around 2. Even in the absence of data confidence criteria, taking all activity information without restrictions into account, mean PD values were smaller than 4, but these values were biased by small numbers of highly promiscuous inhibitors (as shown by comparison of mean and median PD values). These findings were consistent with previous analyses focusing exclusively on high-confidence activity data. Accordingly, including increasing amounts of low-confidence activity data in promiscuity analysis did not lead to substantial increases in promiscuity degrees. Similarly, many kinase inhibitors that were tested in screening assays against 10 to 20 different kinase were only active against a single kinase and the mean PD value of all kinase inhibitors from screening sources was also close to 1.
000 kinase inhibitors remained low at around 2. Even in the absence of data confidence criteria, taking all activity information without restrictions into account, mean PD values were smaller than 4, but these values were biased by small numbers of highly promiscuous inhibitors (as shown by comparison of mean and median PD values). These findings were consistent with previous analyses focusing exclusively on high-confidence activity data. Accordingly, including increasing amounts of low-confidence activity data in promiscuity analysis did not lead to substantial increases in promiscuity degrees. Similarly, many kinase inhibitors that were tested in screening assays against 10 to 20 different kinase were only active against a single kinase and the mean PD value of all kinase inhibitors from screening sources was also close to 1.
Taken together, the results of our large-scale analysis show that promiscuity of ATP site-directed inhibitors of human kinases cannot be generalized. Rather, a differentiated view is required and potential selectivity of kinases inhibitors needs to be taken into consideration. Clearly, on the basis of currently available screening and activity data, the majority of kinase inhibitors are only active against one or at most two kinases at varying data confidence levels. These findings also have important implications for kinase inhibitor development. In many instances, it should be possible to chemically advance ATP site directed inhibitors and render them selective.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The use of OpenEye's toolkits was made possible by their free academic licensing program. D.S. is supported by Sonderforschungsbereich 704 of the Deutsche Forschungsgemeinschaft.References
- P. Cohen, Nat. Rev. Drug Discovery, 2002, 1, 309–315 CrossRef CAS PubMed.
- Kinase Drug Discovery, ed. R. A. Ward and F. W. Goldberg, RSC, Cambridge, U.K., 2011 Search PubMed.
- Z. A. Knight, H. Lin and K. M. Shokat, Nat. Rev. Cancer, 2010, 10, 130–137 CrossRef CAS PubMed.
- A. Levitzki, Annu. Rev. Pharmacol. Toxicol., 2013, 53, 161–185 CrossRef CAS PubMed.
- G. Manning, D. B. Whyte, R. Martinez, T. Hunter and S. Sudarsanam, Science, 2002, 298, 1912–1934 CrossRef CAS PubMed.
- Y. Hu, N. Furtmann and J. Bajorath, J. Med. Chem., 2015, 58, 30–40 CrossRef CAS PubMed.
- Y. Hu, R. Kunimoto and J. Bajorath, Chem. Biol. Drug Des., 2017, 89, 834–845 CAS.
- D. Dimova and J. Bajorath, Molecules, 2017, 22, E730 CrossRef PubMed.
- A. P. Bento, A. Gaulton, A. Hersey, L. J. Bellis, J. Chambers, M. Davies, F. A. Krüger, Y. Light, L. Mak, S. McGlinchey, M. Nowotka, G. Papadatos, R. Santos and J. P. Overington, Nucleic Acids Res., 2014, 42, D1083–D1090 CrossRef CAS PubMed.
- UniProt Consortium, Reorganizing the Protein Space at the Universal Protein Resource (UniProt), Nucleic Acids Res., 2012, 40, D142–D148 CrossRef PubMed.
- Y. Wang, S. H. Bryant, T. Cheng, J. Wang, A. Gindulyte, B. A. Shoemaker, P. A. Thiessen, S. He and J. Zhang, Nucleic Acids Res., 2017, 45, D955–D963 CrossRef PubMed.
- L. K. Gavrin and E. Saiah, Med. Chem. Commun., 2013, 4, 41–51 RSC.
- S. Laufer and J. Bajorath, J. Med. Chem., 2014, 57, 2167–2168 CrossRef CAS PubMed.
- S. Müller, A. Chaikuad, N. S. Gray and S. Knapp, Nat. Chem. Biol., 2015, 11, 818–821 CrossRef PubMed.
- S. Jasial, Y. Hu and J. Bajorath, PLoS One, 2016, 11, e0153873 Search PubMed.
- Y. Hu and J. Bajorath, J. Chem. Inf. Model., 2014, 54, 3056–3066 CrossRef CAS PubMed.
- M. R. Berthold, N. Cebron, F. Dill, T. R. Gabriel, T. Kötter, T. Meinl, P. Ohl, C. Sieb, K. Thiel and B. Wiswedel, KNIME: The Konstanz Information Miner, in Studies in Classification, Data Analysis, and Knowledge Organization, ed. C. Preisach, H. Burkhardt, L. Schmidt-Thieme and R. Decker, Springer, Berlin, Germany, 2008, pp. 319−326 Search PubMed.
- OEChem TK, OpenEye Scientific Software, Inc., Santa Fe, NM, U.S., 2012 Search PubMed.
| This journal is © The Royal Society of Chemistry 2017 |