Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Heavy metal detoxification by recombinant ferritin from Apostichopus japonicus
Kaixue Sia,
Tinghong Minga,
Yanyan Lib,
Xiaoting Qiua,
Liping Chena,
Jun Zhoua,
Chenyang Lu*a,
Xiurong Su *a,
Ye Lia and
Ling-Zhi Cheonga
*a,
Ye Lia and
Ling-Zhi Cheonga
aSchool of Marine Science, Ningbo University, Ningbo, 315211, China. E-mail: suxiurong_public@163.com; luchenyang@nbu.edu.cn; Fax: +86 574 87608368; Tel: +86 574 87608368
bDepartment of Food Science, Cornell University, New York, NY, USA
First published on 30th August 2017
Abstract
Ferritin is a bionanomaterial that is widely applied in magnetic resonance imaging, drug delivery systems, biocompatible fluorescence, neutron-capture therapy and electrochemical markers. Ferritin also has great potential for use in environmental detection and heavy metal removal due to its hollow cage sequestration and maintenance of iron in a nontoxic and bio-available form. In this study, the heavy metal binding activity of ferritin from Apostichopus japonicus (AjFER) was elucidated using scanning electron microscopy (SEM). It was observed that ferritin aggregation morphology changed dramatically upon exposure to five metals. The reaction systems formed ferritin-Cd, ferritin-As, ferritin-Hg, ferritin-Pb, and ferritin-Cr aggregations. The aggregations of horse spleen ferritin (HSF) and AjFER are relatively unified rules. The enrichment capacity of ferritin was further analyzed using Inductively Coupled Plasma Mass spectrometry. The contents of Cd2+, Hg2+, Cr3+, Pb2+, and As3+ enriched by recombinant AjFER were higher than that for the standard HSF in ordinary groups. Moreover, the concentration levels of enriched heavy metal ions in groups treated with phosphate were higher than those in ordinary dialysis control AjFER groups. The conformation stability of AjFER binding to different heavy metal ions was determined using Circular Dichroism (CD) spectroscopy, which revealed that the second structure of Mn+-ferritin has relative stability. The affinity of ferritin for metal ions was determined using Isothermal Titration Calorimetry (ITC), and the order of binding constant was Pb-Fer > Hg-Fer > Cd-Fer > As-Fer > Cr-Fer. The ion channel was measured using site-directed mutagenesis, which revealed that heavy metal ions primarily enter the protein cage via a three-fold channel and that the ferroxidase center may be one of the key sites for metal ion diffusion into the interior of the protein shell.
1 Introduction
Heavy metal contamination has become one of the most serious environmental issues in recent years because of the ubiquity, difficult degradation, and easy accumulation of heavy metal pollutants, particularly in the marine environment, which has received global attention. Heavy metals can accumulate in the body and eventually lead to health issues.1 Heavy metals also threaten littoral and marine surroundings when industrial sewage and sediments are discharged into the ocean.2,3 In recent years, many techniques for heavy metal elimination have been researched, including chemical and biological methods. Chemical approaches include precipitation,4 ion-exchange,5 adsorption,6 membrane filtration,7 coagulation flocculation,8 flotation,9 and electrochemical10 methods. However, these approaches often have many limitations, such as high costs, secondary pollution, byproducts, and harsh reaction conditions. The bio-absorption of heavy metals from the environment is a relatively new approach that was shown to be very promising for the elimination of heavy metal ions. These bio-absorption materials include non-living,11 algal,12 and microbial13 biomasses. Despite these materials' inexpensive price and effective adsorption, unfortunately, these studies remain in experimental phases. Moreover, it is difficult to isolate biosorbents after adsorption.Apostichopus japonicus is a marine animal that belongs to Echinodermata, Holothuroidea, Aspidochirotida, Stichopodidae. A. japonicus is primarily detected on the coast of the Yellow Sea and the Bohai Sea in China, which is rich in sea grasses. The water depth in these areas is commonly 3 to 5 meters. Most young Apostichopus japonicus live in the intertidal zone or in eelgrass growing on fine sediment bottoms, which have a higher metal ion concentration. Artificially bred A. japonicus exist in the bottom of shrimp ponds, where the content of heavy metals is often higher than that in the ocean. Zhang et al.14 studied the effect of Cd2+ and Cu2+ treatment on ferritin expression, revealing that ferritin mRNA from Exopalaemon carinicauda is primarily expressed in the hepatopancreas and that its expression is significantly upregulated at different periods of time after the exposure of shrimp in Cu2+ and Cd2+ aqueous solution. Liu et al.15 also reported that ferritin expression is associated with heavy metal stimulation. Here, the ferritin gene from Apostichopus japonicus was recombined and overexpressed, and the functional characterization of recombinant ferritin was elucidated.
Ferritin is composed of a protein shell with highly symmetrical subunits and of an iron core, which includes thousands of iron phosphate molecules.16–18 Ferritin is composed of 24 identical monomers, typically with high 432 symmetry. This geometry results in 6 four-fold, 8 three-fold, and 12 two-fold channels. These channels and their nature depend on the subunit type and source.19 In eukaryotes, the three-fold channel is a hydrophilic channel that connects the internal cavity with the outside environment. There are two three-fold negative amino acid rings situated on the D helix of the ion channel. Calculations of electrostatic surface potential revealed a negative electrostatic funnel that can attract many metal ions, such as Ca, Zn, Mn, Fe, and Mg.20 The unique shell of ferritin has also been employed as a protein cage to synthesize bionanomaterials.21,22 The ferritin shell can also be utilized as a template to prepare nanoparticle tags with distinct voltammetric signatures.23 Under specific in vitro conditions, metal compounds (FeS and CdS) and radioactive materials (uranium and nuclear energy) have been successfully assembled into the ferritin nano-space.22,24–27 In addition, the internal cavity of ferritin can absorb heavy metal ions, such as iron and small organic molecules.28,29 Heavy metal binding sites are widely distributed throughout the interior and exterior of ferritin, including the inner and outer surfaces of the protein cage, two- and three-phase iron phosphate core surfaces and the deep iron tunnel.30,31 Thus, ferritin can be used to store excess metal ions in the body and prevent metal ion poisoning.32,33 Using recombinant ferritin to eliminate heavy metal ions from water can decrease the cost of secondary contamination.34
Ferritin can be extracted and purified from animals or plants.35–37 However, this method is costly and not feasible for large-scale production. The present study aimed to isolate and characterize a new ferritin from the A. japonicus cDNA library constructed in our laboratory.38 Changes in terms of the morphology of the ferritin following heavy metals uptake were determined using scanning electron microscopy (SEM). The ability of isolated ferritin to take up heavy metals under in vitro conditions was compared with that of HSF using Inductively Coupled Plasma Mass Spectrometry (ICP-MS). We also evaluated the uptake efficiency of isolated ferritin following phosphate treatment. The secondary structures of refolded AjFER and standard HSF and Mn+-AjFER were determined using Circular Dichroism (CD) spectroscopy. The affinity of ferritin for metal ions was also characterized using Isothermal Titration Calorimetry (ITC). The main heavy metal ion channels were roughly confirmed by amino acid site-directed mutagenesis. The present study is expected to provide a foundation for developing a biological material for heavy metal ion removal.
2 Materials and methods
2.1 Expression and purification of A. japonicus ferritin
Ferritin cDNAs from A. japonicus were cloned in a previous study,38 a cDNA encoding full-length AjFER were identified and characterized by using approaches such as cDNA library preparation, rapid amplification of cDNA ends, fluorescence real-time quantitative polymerase chain reaction (PCR), and western blotting. AjFER cDNA was 1222 bp in length and comprised a 513-bp 5′-UTR containing a putative iron regulatory element, a 187-bp 3′-UTR, and a 522-bp complete open reading frame encoding a polypeptide with 173 amino acid residues. The conserved motifs of ferritin, including iron binding signature, H-specific ferroxidase center, and phosphorylation site, were present in the deduced amino acid sequence of AjFER. A PCR fragment encoding the mature polypeptide of AjFER was amplified using gene-specific primers Fer-R (5′-GGATCCATGCAGCCAAGCCAAGTC-3′) and Fer-F (5′-AAGCTTTTAATCCTCCTTCAGGTT-3′), both of which contained BamHI and HindIII sites at their 5′ ends. The PCR products were cloned into a pMD18-T vector (TakaRa), digested using restriction enzymes BamHI and HindIII (NEB), and subcloned into the BamHI/HindIII sites of pET-28a(+) expression vector (Novagen). The recombinant plasmid (pET-28a-AjFER) was transformed into Escherichia coli BL21 cells (Novagen). Positive clones were identified by kanamycin screening and sequencing. The recombinant plasmid (pET-28a-AjFER) was transformed into Escherichia coli BL21. A. japonicus ferritin was also expressed and purified.39 A positive clone was incubated overnight at 37 °C at 120 rpm, and after the culture reached an OD600 = 0.6 to 0.8, 1 mM isopropyl β-D-thiogalacto-pyranoside (IPTG) was added to induce AjFER expression for an additional 12 h. Ferritin was purified using a Ni-NTA affinity column. The expression and purified products were further incubated in GSH/GSSG buffer (50 mM Tris–HCl, 1 mM EDTA, 50 mM NaCl, 10% glycerol, 1% glycine, 2 mM reduced glutathione, and 0.2 mM oxide glutathione, pH 8.0) with a stepwise decrease of urea with the order of 6 M, 4 M and 2 M overnight at 4 °C to promote purified protein refolding and obtain AjFER with biological activity. The obtained activated AjFER was incubated in buffer solution (25 mmol L−1 Tris–HCl, 150 mmol L−1 NaCl, pH 8.0) with 0.5 U mg−1 thrombin at 25 °C for 2 h so as to remove the his-tag. Enzyme-digested products was separated by Sephacryl S-300 HR column equilibrated with 150 mmol L−1 NaCl at pH 8.0. The collected AjFER was examined by SDS-PAGE.2.2 Functional characterization of AjFER
2.3 SEM analysis
AjFER and HSF treated with five metal ions were evenly smeared onto the surface of pretreated mica sheets and dried for 4 days at room temperature (environmental humidity, 5%).39 The surface morphologies were observed using scanning electron microscopy (SEM) (S-3400N, Hitachi, Japan) with a 40-mm probe distance, 30° sample inclination, 21-mm working distance, and 100-s analysis time.2.4 ICP-MS analysis
A total of 15 ferritin groups (ten ordinary groups and five phosphate-stimulated groups) enriched with heavy metals were collected from dialysis and each group was subjected to five repeated trials. The concentration of ferritin sample was determined by using BCA kit (Sangon, Shanghai, China). For microwave digestion, 1 mL of each ferritin sample was accurately transferred directly to a polytetrafluoroethylene digestion vessel. The micro-wave digestion parameters were based on our previous work.39 Sample blanks were also prepared using the aforementioned full analytical procedure, except for sampling and analysis. The calibration standard solutions were prepared using a 10 mg L−1 multi-element standard solution (CLMS-2AN, Spex, CA, USA). All reagents were of analytical grade or better. For sample dilution and preparation of the standards, we used ultrapure water (MilliQ, Millipore). The data were then subjected to one-way analysis of variance (ANOVA). Differences were considered significant at P < 0.05. The molar ratio of metal ions vs. ferritin (Mn+/protein) can be calculated since the contents of metal ions binding to Ajfer per unit volume can be determined by ICP-MS, and the contents of AjFER per unit volume can be determined by BCA kit.2.5 Circular dichroism analysis
CD was performed using a JASCO J-815 CD spectrophotometer in the far UV range of 260–190 nm at room temperature using a 0.1 cm quartz cuvette. The obtained results were analyzed using the CDPro software, and the proportions of α-helices, β-sheets, β-turns and unordered conformations in the protein were obtained.2.6 Isothermal titration calorimetry analysis
High-sensitivity ITC experiments were conducted at 25 °C using a MicroCal ITC200 (Malvern Instruments). The Mn+ solution was prepared in a buffer containing 25 mM MES, pH 5.5, and 150 mM NaCl. A large number of pre-experiments of titration of metal ions to ferritin was performed so as to obtain the suitable titration concentration, since it is a complex binding reaction of metal ions binding to ferritin. The final concentrations of Cd2+, Cr3+, As3+, Pb2+, and Hg2+ were 16.7 mM, 2.5 mM, 2.5 mM, 556 μM, and 800 μM, respectively. AjFER was used at concentrations of 2.5, 0.5, 5, 0.25, and 0.5 μM, respectively. Mn+ was titrated into the protein solution in a series of 19 injections of 2 μL (the first injection was a dummy injection of 0.4 μL), at 2 min apart, with a stirring speed of 750 rpm. All data analysis has been performed using Origin Pro version 8.0.402.7 Preparation of mutants
In a previous study, the ferroxidase center sites and ion channels were predicted using the SMART procedure.38 The results indicated that the ferroxidase center was composed of Glu-25, Tyr-32, Glu-59, Glu-60, His-63, Glu-105, and Gln-139, and the ion channels was composed of His-116, Asp-129, and Glu-131. The mutant ferroxidase center (S1) (E25A, Y32A, E59A, E60A, H63A, E105A, and Q139A) and ion channels (S2) (H116A, D129A, and E131A) were prepared using gene synthesis.3 Results
3.1 Recombinant expression of AjFER
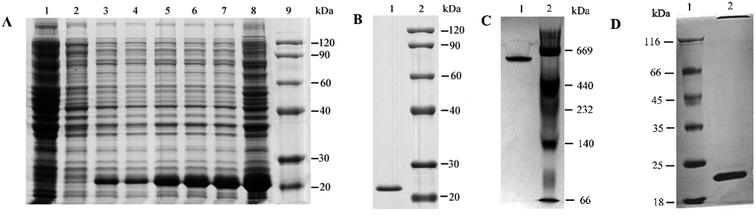
The lysate was analyzed using SDS-PAGE. Obvious protein bands with a molecular weight of 24 kDa were detected after IPTG induction (Fig. 1A and B) and could be purified to homogeneity using HiTrap Chelating Columns. The result was consistent with the theoretical molecular mass of a subunit. Moreover, the content of overexpressed recombinant ferritin increased with time. The expression of recombinant ferritin peaked at 5 h after adding IPTG to the culture. A single protein band was observed after purification using Ni-NTA. Native-PAGE was used to characterize the multi-subunit refolding and recombination (Fig. 1C). A protein band with a molecular weight of 576 kDa was observed in lane 1, indicating that 24 monomers were packed together. As treated with thrombin, the His-tage was removed from obtained overexpressed ferritin. A protein band with a molecular weight of 20 kDa was observed in lane 2, indicating that His-tage were removed from ferritin (Fig. 1D).3.2 Morphological characterization of recombinant ferritin following different treatments
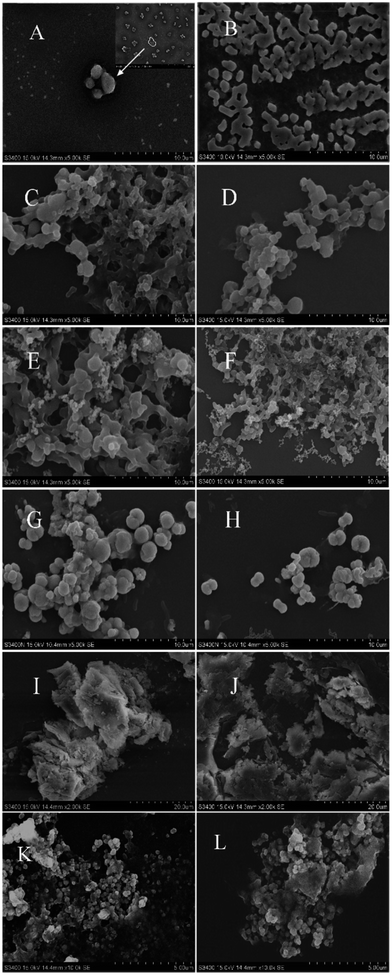
As shown in Fig. 2A, the aggregation of recombinant AjFER in the negative group exhibited divergent spherical shapes with cage-like morphologies. The HSF aggregated close together, forming a lattice (Fig. 2B). Each HSF aggregate in the negative group was similar, with sizes ranging from 1 to 2 μm. In contrast to HSF, the AjFER aggregates were approximately 5 μm in size. After treatment with different heavy metals, the surface morphology of the HSF and AjFER aggregates changed dramatically. Different metals resulted in different surface morphologies. In the Hg2+ treatment groups (Fig. 2C and D), the aggregation monomers of HSF and AjFER adhered together like a honeycomb. The edge shapes of the aggregation were fuzzy. The two kinds of ferritin aggregations exhibited adhesion in a honeycomb shape after absorbing Cr3+ (Fig. 2E and F). The edge of the aggregation was not obvious. Aggregations with larger monomer sizes gathered into a honeycomb with holes containing an extensive amount of small ferritin aggregations. In the Pb2+ treatment groups (Fig. 2G and H), the ferritin aggregates were ellipsoid with non-uniform sizes. Some of the aggregate adhered together, also with a single aggregate. In the As3+ treatment groups (Fig. 2I and J), the aggregates formed an irregular block layer, which adhered to the surface of irregular similar layers. Compared to HSF, the coverage of recombinant AjFER aggregations was more intense in the five metal treatment groups. In addition, the aggregations were larger in the Pb2+ treatment groups than in groups treated with other metals. In the Cd2+ treatment groups (Fig. 2K and L), the aggregates of HSF and AjFER were present in the form of small and uniform sphere piles.3.3 Quantitative analysis to determine heavy metal enrichment capacity
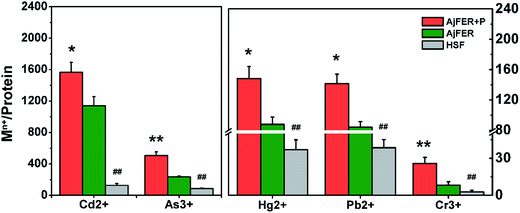
ICP-MS analysis was used to further confirm the content of heavy metal elements enriched by AjFER. As shown in Fig. 3, in the AjFER groups, the content enrichment order from high to low was Cd2+ > As3+ > Hg2+ > Pb2+ > Cr3+. In the HSF group, the enrichment order from high to low was Cd2+ > As3+ > Hg2+ > Cr3+ > Pb2+. The enrichment of the five heavy metals in recombinant AjFER was higher than that in standard HSF (P < 0.01) in the ordinary group. The concentration of heavy metals in the ordinary dialysis control groups was lower than that in the phosphate-stimulated groups (P < 0.05). The contents of Cd, As, Hg, Pb and Cr in ordinary groups are 1139.5, 234.2, 87.8, 84 and 8.2, respectively. While the enrichment of metal ions increased obviously in phosphate-stimulated group and the contents are 1566.7, 507.5, 148.1, 141.3 and 25.8, respectively. The PBS may contribute to form the metal ion core that attract more metal ion entering into the inner of protein cage.3.4 Secondary structure determination
To further describe the conformational changes of ferritin in the presence of heavy metal ions, CD spectra were examined. As shown in Table 1, the contents of the secondary structure in the native AjFER group were similar to those of the HSF group, indicating that recombined AjFER has a similar secondary structure. Cd-AjFER, Cr-AjFER, As-AjFER, Pb-AjFER, and Hg-AjFER were also prepared and the contents of the secondary structure show that the main variations were observed among α-helices, β-turns, and random coils. The number of α-helices in the treatment group decreased, whereas the number of random coils generally increased compared with the native ferritin group, indicating a change in conformation, possibly because the α-helices transformed into random coils. Among the treatment groups, there were also differences in the secondary structure, indicating that metal species have different influences on structures. In particular, the conformation of Pb-AjFER changed the most, for which the β-sheet increased by 13.1% and the random coils decreased to 19.3% from 28.3%, suggesting that Pb2+ has a larger effect on AjFER.| Categories | Secondary structure contents (%) | ||||||
|---|---|---|---|---|---|---|---|
| HSF | AjFER | AjFER-Cd | AjFER-Cr | AjFER-As | AjFER-Pb | AjFER-Hg | |
| α-Helix | 25.9 | 34.5 | 31.6 | 32 | 27.4 | 32.7 | 28 |
| β-Sheet | 0 | 0 | 0 | 0 | 0 | 13.1 | 0 |
| β-Turn | 32.5 | 37.2 | 37.3 | 38.1 | 33.3 | 34.9 | 33.4 |
| Random coils | 41.5 | 28.3 | 31.1 | 30 | 39.3 | 19.3 | 38.6 |
3.5 ITC of Mn+ binding to AjFER
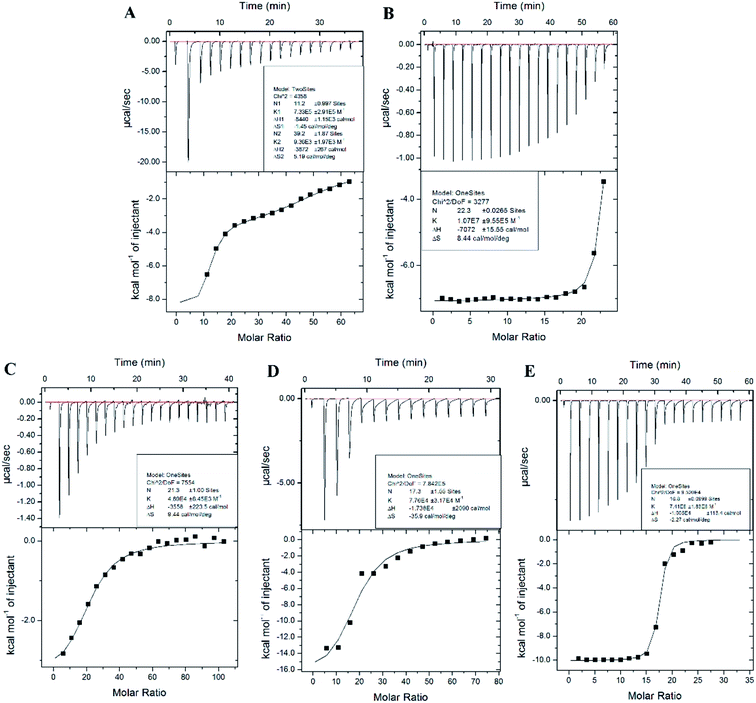
Isothermal titration calorimetry (ITC) has widely been used to measure interactions between metal ions and biomolecules, such as amino acids, peptides, and protein.41–43 ITC is the most quantitative technique used to determine the thermodynamic properties of any biochemical reaction. The raw data from Fig. 4 show an exothermic reaction of Cd2+, Pb2+, Hg2+, As3+, and Cr3+ binding to AjFER, as negative peaks correspond to an endothermic reaction. This finding was also reflected by the negative enthalpy value shown in Table 2. Gibbs free energy (ΔG) was calculated to represent the direction of the reaction. The value of ΔG in Table 1 was negative, indicating a spontaneous reaction favorable for the combination of protein with metal ions. Fig. 4A shows an apparent two-inflection point. Accordingly, a model of two sets of independent binding sites was used to fit a curve to the data for Cd-AjFER. For Cd-AjFER, both strong and weak binding classes were observed with AjFER (Fig. 4A, Table 2). The stoichiometry of the strong binding class was ∼11 molar equivalents per protein, and the weak binding class was ∼39 molar equivalents per protein. The other metal ions showed only one inflection point, and the data for the other metal ions were used to curve fit a model of one set of independent binding sites (Fig. 4B–E). The derived parameters for the binding sites of different metal ions are summarized in Table 2. The binding constant is used to characterize the affinity between metal ions and proteins. Table 2 shows obvious affinity differences of Mn+-FER, with the following order: Pb-Fer > Hg-Fer > Cd-Fer > As-Fer > Cr-Fer. These results suggest a strong affinity of AjFER to Pb2+ and a relatively weak affinity of AjFER to Cr3+. The binding constant of Cd-Fer was close to that of As-Fer. These results are consistent with the observed changes in secondary structure.| Protein | N | Ka (M−1) | ΔH0 (cal mol−1) | ΔS0a (cal mol−1 deg−1) | ΔG01b (kJ mol−1) |
|---|---|---|---|---|---|
a Calculated from ΔS = (ΔH − ΔG01)/T.b Calculated from ΔG01 = −RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K. K. |
|||||
| Cd-Fer | N1: 11.2 ± 0.997 | Ka1: 7.33 × 105 ± 2.91 × 105 | ΔH1:-8440 ± 1150 | ΔS1:-1.45 | −8007.68 ± 1150 |
| N2: 39.2 ± 1.87 | Ka2: 9.36 × 103 ± 1.97 × 103 | ΔH2:-3872 ± 267 | ΔS2:5.19 | −5419.4 ± 267 | |
| Pb-Fer | 22.3 ± 0.0265 | (1.07 ± 0.0955) × 107 | −7072 ± 15.55 | 8.44 | −9588.39 ± 15.55 |
| Cr-Fer | 21.3 ± 1.00 | (4.69 ± 0.845) × 105 | −3558 ± 223.5 | 9.44 | −6372.54 ± 223.5 |
| As-Fer | 17.3 ± 1.55 | (7.76 ± 3.17) × 104 | −1.738 × 104 ± 2090 | −35.9 | −6676.42 ± 2090 |
| Hg-Fer | 16.8 ± 0.0999 | (7.41 ± 1.83) × 106 | −1.005 × 104 ± 113 | −2.27 | −9373.2 ± 113 |
3.6 Mn+ binding to AjFER variants
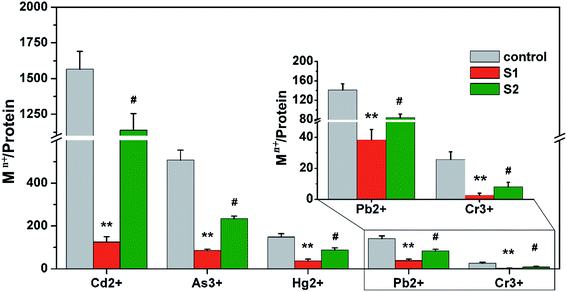
Ferritin is composed of 24 identical subunits, forming a caged structure with an internal diameter of 8 nm, which can accommodate ∼3500 ions. The arrangement of subunits forms three small molecule channels, including a two-fold channel, a three-fold channel, and a four-fold channel, on the surface of the protein cage. In eukaryotes, the three-fold channel is the main protein cage for small molecules.44,45 To further verify the ion channel and location of heavy metal ion binding to protein, in the present study three-fold-symmetry channel variants (S1) and ferroxidase center variants (S2) were prepared. The overexpressed and purified variants and native AjFER (control group) were treated with five metal ions under the same conditions. As shown in Fig. 5, the metal ion enrichment of variant S1 was obviously decreased compared with the control group (P < 0.01), suggesting that three-fold channels are the main channels of heavy metal ion sequestration into the protein cage. The content of the five metal ions binding to variant S2 also decreased (P < 0.05), suggesting that the ferroxidase center can bind metal ions.4 Discussion
The binding activity of AjFER for different heavy metals was investigated using scanning electron microscopy (SEM) because of the sensitivity and selectivity of this technique.46–49 After purifying recombinant AjFER, the vast majority of subunits in AjFER were detected as free peptide chains. Inclusion bodies solubilized in the presence of urea were diluted with denaturation solution, and the bioactive recombinant AjFER was released. The morphology of recombinant AjFER and natural HSF was similar. The shapes of some ferritin aggregations were irregular, with a small amount of ferritin fragments. The addition of metal ions, as a result of combinations with metal ligands and interactions between amino acid side chains, such as hydrophobic bonding, hydrogen bonding, van der Waals force and electrostatic effects, significantly influenced ferritin folding.26,50,51 These factors gradually increased the number of subunits constituting ferritin, thereby increasing the thickness of the protein shell. Several space changes were observed in ferritin, with the surfaces forming several large gaps. However, metal ion hydrations may destroy the ferritin hydration layer, making ferritin intermediates easier to fold. The stronger the metal ion hydration, the greater the role in promoting the formation of ferritin metal aggregations. Eventually, the reaction systems formed ferritin-Cd, ferritin-As, ferritin-Hg, ferritin-Pb, and ferritin-Cr aggregations.The ICP-MS technique has widely been used for the determination of metal elements because of its high level of sensitivity and capability for multi-element analysis.52–54 ICP-MS has commonly been used to determine the geographic origin of medicinal herbs, minor and trace elements in aromatic spices, and agricultural crops.55–57 In this study, AjFER was expressed and purified based on a previous study, and then, the recombinant ferritin was characterized according to enrichment level after exposure to different heavy metals. The heavy metal elements enriched by AjFER were analyzed using ICP-MS to better understand the role of the AjFER in heavy metal detoxification.
The bioactive Apostichopus japonicus ferritin was obtained using a restructuring technique. AjFER absorbed several types of heavy metals, and the concentrations of different metals were all greater than those of horse spleen ferritin. The flexible adjustment of the ferritin protein shell and the combination of the close degree between protein subunits play important roles in the physiological functions of ferritin.58–60 Recombinant AjFER, without iron core framework support, may be more flexible and show increased plasticity, thus providing more enrichment of heavy metals. Different heavy metal ions have different atomic radius sizes, and the numbers of nuclei are also different. Moreover, when the metal ions pass through ferritin ion channels, the different valences and redox abilities may cause different changes in internal ferritin factors.33,50 These factors could lead to different metal ion concentration levels.
Iron absorption and ferritin release are controlled by the complex phosphorus iron structure on the surface of the iron core.61–63 The rate of iron release is associated with the reducing agent and formation of the iron core.61 Medium H+ and OH− not only participate in the composition of the iron core but also combine with amino acids on the inside and outside of the protein shell surface, thus affecting the conformation and the flexible adjustment ability of the protein shell.33 The phosphate groups distributed throughout the ferritin shell increase the electrostatic repulsion between protein molecules, enabling more favorable dispersion in solution.64 In addition, the metal phosphate seed is an auto-catalyst, and metal phosphate will be rapidly developed in the ferritin cage.23 Our results were consistent with the theory that recombinant AjFER, which is more flexible and shows more plasticity, is more attractive to heavy metal ions than HSF. Phosphate stimulation can increase the concentration of metal ions in AjFER compared with the ordinary dialysis control groups.
Circular dichroism is a spectroscopic technique that is broadly used for assessing macromolecular interactions and conformational changes in solution.65,66 In this study, we determined the secondary structure of ferritin after refolding, using standard HSF as a comparison sample. The results suggest that activated AjFER can be obtained using a stepwise decrease of urea. Because of its corrected secondary structure, Native-PAGE revealed that 24 subunits were successfully packed together through intermolecular forces. The metal-ferritin conformation was also examined using CD spectroscopy. The results showed that the α-helices in the metal ion-treated group decreased. It is likely that the metals bound to ferritin may shift the ferritin conformation toward disordered. Ferritin cages are formed by arrays of self-assembling four-helix bundles,67 and metal ion binding sites are primarily located on α-helices in ferritin, as revealed in the crystal structure.67–69 Heavy metal ion binding with amino acid residues on helices may be the main reason for the decrease in α-helices. For Pb-AjFER, the β-sheet showed an obvious increase. This inconsistency may be attributed to the properties and binding sites of Pb2+, as changes in the microenvironment near binding sites promote β-sheet formation. Harshith Subramanian et al.66 investigated the effect of Cu2+ and Mn2+ on horse spleen apoferritin, revealing that the number of β-sheets changed in response to treatments with different metal ion species and at different concentrations. We readily admit that only a secondary structure measurement, as found here, may not fully reflect the binding mechanism of metal ions and protein. Protein crystallization is needed to determine the binding sites in the protein cavity.
The stoichiometry of metal ion binding to AjFER determined using ITC is inconsistent with results from ICP-MS above. The stoichiometry of metal ions binding to AjFER determined using ITC is inconsistent with results from ICP-MS. In study of ITC, the stoichiometry of Cd, As, Hg, Pb and Cr binding to AjFER was 50.4, 17.3, 16.8, 22.3 and 21.3, respectively. While the results of ICP-MS indicated that the stoichiometry of Cd, As, Hg, Pb and Cr are 1139.5, 234.2, 87.8, 84 and 8.2, respectively. There are two possible reasons to account for this case. First, most metal ions entering the cavity are in a free state. These metal ions do not or only indirectly bind to amino acid residues, producing little heat change. Enthalpy changes from second- and third-class binding sites are too weak to be fitted to a curve. Thus, the stoichiometry shown in Table 2 may mainly represent first-class binding sites. Second, one binding site may capture more than one metal ion, such as the formation of a gold cluster.68,70 The stoichiometries of Pb2+ and Cr3+ are approximately 22 and 21, respectively. Thus, these binding sites may be located in 24 subunits, suggesting the existence of a binding site in each subunit. In addition, the ΔS from Pb-Fer and Cr-Fer showed positive values, suggesting that the change occurred due to a hydrophobic interaction. Thus, these interactions were most likely located in the 24 subunits of the ferroxidase center. The stoichiometry of Cd2+, As3+, and Hg2+ are 11, 17, and 17, respectively, implying metal ion binding to eight three-fold channels with relatively strong binding, and approximately two metal atoms in each three-fold channel. Bou-Abdallah et al.71 examined the affinity of ferritin to Zn(II) and Tb(III) using ITC, revealing that three-fold channels in ferritin strongly adsorb Zn(II) and Tb(III) and the ferroxidase center weakly adsorbs Zn(II) and Tb(III). This inconsistence may be caused by the different properties of different metal ions and different microenvironments of different proteins. Since ITC directly measures the total heat flow, all processes occurring upon the addition of the titrant will contribute to the measured enthalpy. Therefore, the details of heat contribution from specific amino acid residues should be described using protein crystallization technology and site-directed mutagenesis in future studies.
Many mutagenesis studies in terms of ferritin have been performed.72–74 Wade et al. prepared glutamic acid residue mutagenesis in the shell of human ferritin and assessed their role in iron core development.73 In this study, the mutagenesis of key amino acid residues inside the ferritin cage was conducted, and their roles were explored. Here, we found that heavy metal ions penetrate into the ferritin shell via three-fold channels where amino acids H116, D129, and E132 are located. These amino acids residues play crucial roles in metal ion adsorption. The channel is composed of two three-fold highly conserved negative amino acid rings. The three Asp residues from three different subunits form the inner layer close to the shell interior, and three Glu residues form the middle layer in the ferritin shell before funnel opening. Calculations of the electrostatic surface potential indicated the existence of a negative electrostatic funnel that allows the attraction and insertion of many divalent ions, such as Fe, Ca, Zn, Mg, and Mn.20 Molecular diffusion research in human heavy chain ferritin and horse spleen ferritin verified this finding and elucidated the penetrability of the ferritin cavity to small molecules via the three-fold channels.44,45 A thermodynamic study in terms of three-fold channel mutants has verified an iron ion channel in the cavity, indicating that the binding ability for Fe2+ decreased.41 The ferroxidase center is composed of two iron sites, FeA, including Glu-27, Glu-65, and His-62, and FeB, including Glu-107 and Glu-62. In this study, theoretically, the amount of metal ions bound to the ferroxidase center variant should decrease by 48 metal atoms per mutant cage at most. Notably, the amount of metal ions was reduced by more than 48, with the exception of Cr3+. These results indicate that ferroxidase center sites may contribute to metal ion migration from the entrance of three-fold channels to the inner protein cavity.
5 Conclusions
We characterized recombinant AjFER from Apostichopus japonicus with heavy metal detoxification activity. This study indicated that AjFER showed an advantage in the enrichment capacity of Cd2+, Hg2+, Cr3+, Pb2+ and As3+ compared with standard HSF. In addition, the content of heavy metals showed a marked increase in phosphate-stimulated groups over the control group. The affinity of ferritin to different heavy metal ions was Pb-Fer > Hg-Fer > Cd-Fer > As-Fer > Cr-Fer. The heavy metal ions primarily entered the ferritin shell via three-fold channels composed of three key amino acid residues H116, D129, and E132. The ferroxidase center, including E25, Y32, E59, E60, H63, E105, and Q139, contribute to the enrichment of heavy metal ions. These studies provide a significant theoretical basis for the development of materials for heavy metal ion removal.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 41176123, No. 40776075 and No. 41676159) and the K. C. Wong Magna Fund of Ningbo University. Our ITC work was performed at the National Center for Protein Science Shanghai.References
- J. A. Cooke, S. M. Andrews and M. S. Johnson, Water, Air, Soil Pollut., 1990, 51, 43–54 CrossRef CAS.
- K. Pan and W. X. Wang, Sci. Total Environ., 2012, 421–422, 3–16 CrossRef CAS PubMed.
- G. Zhang, D. Liu, H. Wu, L. Chen and Q. Han, Ecotoxicology, 2012, 21, 1726–1733 CrossRef CAS PubMed.
- Y. Ku and I. L. Jung, Water Res., 2001, 35, 135–142 CrossRef CAS PubMed.
- S. Y. Kang, J. U. Lee, S. H. Moon and K. W. Kim, Chemosphere, 2004, 56, 141–147 CrossRef CAS PubMed.
- A. Jusoh, L. S. Shiung, N. A. Ali and M. J. M. M. Noor, Desalination, 2007, 206, 9–16 CrossRef CAS.
- J. Landaburu-Aguirre, V. García, E. Pongrácz and R. L. Keiski, Desalination, 2009, 240, 262–269 CrossRef CAS.
- A. E. Samrani, B. S. Lartiges and F. Villiéras, Water Res., 2008, 42, 951–960 CrossRef PubMed.
- F. Tessele, M. Misra and J. Rubio, Miner. Eng., 1998, 11, 535–543 CrossRef CAS.
- I. Heidmann and W. Calmano, J. Hazard. Mater., 2008, 152, 934–941 CrossRef CAS PubMed.
- F. Kaczala, M. Marques and W. Hogland, Bioresour. Technol., 2009, 100, 235–243 CrossRef CAS PubMed.
- R. Apiratikul and P. Pavasant, Bioresour. Technol., 2008, 99, 2766–2777 CrossRef CAS PubMed.
- M. Tuzen, K. O. Saygi, C. Usta and M. Soylak, Bioresour. Technol., 2008, 99, 1563–1570 CrossRef CAS PubMed.
- J. Zhang, T. Gui, J. Wang and J. Xiang, Int. J. Biol. Macromol., 2015, 72, 320–325 CrossRef CAS PubMed.
- Q. N. Liu, Z. Z. Xin, Y. Liu, Z. F. Wang, Y. J. Chen, D. Z. Zhang, S. H. Jiang, X. Y. Chai, C. L. Zhou and B. P. Tang, Fish Shellfish Immunol., 2017, 63, 297–303 CrossRef CAS PubMed.
- G. C. Ford, P. M. Harrison, D. W. Rice, J. M. A. Smith, A. Treffry, J. L. White and J. Yariv, Philos. Trans. R. Soc., B, 1984, 304, 551–565 CrossRef CAS.
- S. C. Andrews, P. M. Harrison, S. J. Yewdall, P. Arosio, S. Levi, W. Bottke, M. V. Darl, J. F. Briat, J. P. Laulhère and S. Lobreaux, J. Inorg. Biochem., 1992, 47, 161–174 CrossRef CAS PubMed.
- P. D. Hempstead, S. J. Yewdall, A. R. Fernie, D. M. Lawson, P. J. Artymiuk, D. W. Rice, G. C. Ford and P. M. Harrison, J. Mol. Biol., 1997, 268, 424–448 CrossRef CAS PubMed.
- R. R. Crichton and J. P. Declercq, Biochim. Biophys. Acta, 2010, 1800, 706–718 CrossRef CAS PubMed.
- F. Bouabdallah, G. Zhao, G. Biasiotto, M. Poli, P. Arosio and N. D. Chasteen, J. Am. Chem. Soc., 2012, 130, 17801–17811 CrossRef PubMed.
- F. C. Meldrum, B. R. Heywood and S. Mann, Science, 1992, 257, 522–523 CAS.
- T. Douglas, D. P. Dickson, S. Betteridge, J. Charnock, C. D. Garner and S. Mann, Science, 1995, 269, 54–57 CrossRef CAS PubMed.
- G. Liu, H. Wu, A. Dohnalkova and Y. Lin, Anal. Chem., 2007, 79, 5614–5619 CrossRef CAS PubMed.
- F. C. Meldrum, V. J. Wade, D. L. Nimmo, B. R. Heywood and S. Mann, Nature, 1991, 349, 684–687 CrossRef CAS.
- S. Pead, E. Durrant, B. Webb, C. Larsen, D. Heaton, J. Johnson and G. D. Watt, J. Inorg. Biochem., 1995, 59, 15–27 CrossRef CAS PubMed.
- K. Y. Kim, Y. L. Sang, Y. S. Cho, I. C. Bang, K. H. Kim, S. K. Dong and Y. K. Nam, Fish Shellfish Immunol., 2007, 23, 1043–1059 CrossRef CAS PubMed.
- T. Rakshit and R. Mukhopadhyay, J. Colloid Interface Sci., 2012, 388, 282–292 CrossRef CAS PubMed.
- C. Li, Z. Li, Y. Li, J. Zhou, C. Zhang, X. Su and T. Li, PLoS One, 2012, 7, e51428 CAS.
- Y. Lu, A. Zhang, C. Li, P. Zhang, X. Su, Y. Li, C. Mu and T. Li, Fish Shellfish Immunol., 2013, 35, 271–277 CrossRef CAS PubMed.
- A. Treffry, P. M. Harrison, A. Luzzago and G. Cesareni, FEBS Lett., 1989, 247, 268–272 CrossRef CAS PubMed.
- D. M. Lawson and J. M. Smith, Nature, 1991, 349, 541 CrossRef CAS PubMed.
- P. Arosio, R. Ingrassia and P. Cavadini, Biochim. Biophys. Acta, 2009, 1790, 589–599 CrossRef CAS PubMed.
- R. J. Hilton, B. Zhang, L. N. Martineau, G. D. Watt and R. K. Watt, J. Inorg. Biochem., 2012, 108, 8–14 CrossRef CAS PubMed.
- H. Q. Huang, Q. M. Lin and Z. B. Lou, Protein J., 2000, 19, 441–447 CrossRef CAS.
- S. Lobreaux, S. J. Yewdall, J. F. Briat and P. M. Harrison, Biochem. J., 1992, 288, 931–939 CrossRef CAS PubMed.
- H. Q. Huang, Z. Q. Xiao, X. Chen, Q. M. Lin, Z. W. Cai and P. Chen, Biophys. Chem., 2004, 111, 213–222 CrossRef CAS PubMed.
- S. Yun, S. Yang, L. Huang, X. Qi, P. Mu and G. Zhao, Food Res. Int., 2012, 48, 271–276 CrossRef CAS.
- L. I. Cheng-Hua, J. Cui, L. I. Ye, J. Zhou, S. U. Xiu-Rong and L. I. Tai-Wu, Oceanol. Limnol. Sin., 2011, 42, 567–572 Search PubMed.
- L. Chen, J. Zhou, Y. Zhang, S. Chu, W. He, L. Ye and X. Su, PLoS One, 2015, 10, e0119427 Search PubMed.
- T. Wiseman, S. Williston, J. F. Brandts and L. N. Lin, Anal. Biochem., 1989, 179, 131–137 CrossRef CAS PubMed.
- F. Bouabdallah, P. Arosio, P. Santambrogio, X. Yang, C. Januschandler and N. D. Chasteen, Biochemistry, 2002, 41, 11184–11191 CrossRef CAS.
- Y. Zhang, A. Shreeram Akilesh and D. E. Wilcox, Inorg. Chem., 2000, 39, 3057–3064 CrossRef CAS PubMed.
- Y. Zhang and D. E. Wilcox, JBIC, J. Biol. Inorg. Chem., 2002, 7, 327–337 CrossRef CAS PubMed.
- X. Yang, P. Arosio and N. D. Chasteen, Biophys. J., 2000, 78, 2049–2059 CrossRef CAS PubMed.
- X. Yang and N. D. Chasteen, Biophys. J., 1996, 71, 1587–1595 CrossRef CAS PubMed.
- S. Z. Abdullah, P. R. Bérubé and D. J. Horne, J. Membr. Sci., 2014, 463, 113–125 CrossRef CAS.
- M. Ananyev, A. Gavrilyuk, S. Mitri, A. Demin, V. Malkov and P. Tsiakaras, Electrochim. Acta, 2014, 125, 371–379 CrossRef CAS.
- U. Trdan and J. Grum, Corros. Sci., 2014, 82, 328–338 CrossRef CAS.
- L. Zhang, J. Liu, H. Xiao, D. Liu, Y. Qin, H. Wu, H. Li, D. Na and W. Hou, Chem. Eng. J., 2014, 250, 1–5 CrossRef CAS.
- H. Q. Huang, F. Z. Zhang, L. S. Xu, Q. M. Lin, J. W. Huang and D. Zeng, Bioelectrochem. Bioenerg., 1998, 44, 301–307 CrossRef CAS.
- J. Y. Zhu, H. Q. Huang, X. D. Bao, Q. M. Lin and Z. Cai, Aquat. Toxicol., 2006, 78, 127–135 CrossRef CAS PubMed.
- A. Gonzalvez, S. Armenta and M. D. L. Guardia, TrAC, Trends Anal. Chem., 2009, 28, 1295–1311 CrossRef CAS.
- V. S. Selih, M. Sala and V. Drgan, Food Chem., 2014, 153, 414–423 CrossRef CAS PubMed.
- Y. H. Wei, J. Y. Zhang, D. W. Zhang, L. G. Luo and T. H. Tu, Food Chem., 2014, 159, 507 CrossRef CAS PubMed.
- A. Bibak, S. Stürup, V. Haahr, A. Pernille Gundersen and V. Gundersen, J. Agric. Food Chem., 1999, 47, 2678–2684 CrossRef CAS PubMed.
- N. Khan, Y. C. Ji, E. Y. Nho, N. Jamila, G. Habte, J. H. Hong, I. M. Hwang and K. S. Kim, Food Chem., 2014, 158, 200–206 CrossRef CAS PubMed.
- Y. K. Kwon, Y. S. Bong, K. S. Lee and G. S. Hwang, Food Chem., 2014, 161, 168–175 CrossRef CAS PubMed.
- L. Huang, Z. C. Lin, Q. Lin, L. Z. Luo and H. Q. Huang, Chem. Res. Chin. Univ., 2008, 24, 550–556 CrossRef CAS.
- H. Q. Huang, H. U. Xiao-Hui, X. P. Fang, T. M. Cao and B. Kong, Chin. J. Anal. Chem., 2009, 37, 631–636 CAS.
- L. Z. Luo, H. W. Jin, Z. W. Cai and H. Q. Huang, Chin. J. Anal. Chem., 2011, 39, 155–162 CAS.
- H. Huang, R. K. Watt, R. B. Frankel and G. D. Watt, Biochemistry, 1993, 32, 1681–1687 CrossRef CAS.
- H. Q. Huang, Q. M. Lin, B. Kong, R. Y. Zeng, Y. H. Qiao, C. H. Chen, F. Z. Zhang and L. S. Xu, Protein J., 1999, 18, 497–504 CrossRef CAS.
- B. Kong, H. Q. Huang, Q. M. Lin, W. S. Kim, Z. Cai, T. M. Cao, H. Miao and D. M. Luo, Protein J., 2003, 22, 61–70 CrossRef CAS.
- G. Matheis, Food Chem., 1991, 39, 13–26 CrossRef CAS.
- R. Xing, X. Wang, C. Zhang, Y. Zhang, Q. Wang, Z. Yang and Z. Guo, J. Inorg. Biochem., 2009, 103, 1039–1044 CrossRef CAS PubMed.
- H. Subramanian, M. Jaganathan and A. Dhathathreyan, Nano Hybrids, 2016, 12, 33–41 CrossRef.
- X. Liu and E. C. Theil, Acc. Chem. Res., 2005, 38, 167–175 CrossRef CAS PubMed.
- B. Maity, S. Abe and T. Ueno, Nat. Commun., 2017, 8, 14820 CrossRef CAS PubMed.
- C. Pozzi, S. Ciambellotti, C. Bernacchioni, F. D. Pisa, S. Mangani and P. Turano, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 2580–2585 CrossRef CAS PubMed.
- C. Sun, Y. Yuan, Z. Xu, T. Ji, Y. Tian, S. Wu, J. Lei, J. Li, N. Gao and G. Nie, Bioconjugate Chem., 2015, 26, 193–198 CrossRef CAS PubMed.
- F. Bou-Abdallah, P. Arosio, S. Levi, C. Janus-Chandler and N. D. Chasteen, JBIC, J. Biol. Inorg. Chem., 2003, 8, 489–497 CAS.
- P. Santambrogio, P. Pinto, S. Levi, A. Cozzi, E. Rovida, A. Albertini, P. Artymiuk, P. M. Harrison and P. Arosio, Biochem. J., 1997, 322, 461–468 CrossRef CAS PubMed.
- V. J. Wade, S. Levi, P. Arosio, A. Treffry, P. M. Harrison and S. Mann, J. Mol. Biol., 1991, 221, 1443–1452 CrossRef CAS PubMed.
- M. A. Kilic, S. Spiro and G. R. Moore, Protein Sci., 2003, 12, 1663–1675 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |