Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Endogenous omega-3 long-chain fatty acid biosynthesis from alpha-linolenic acid is affected by substrate levels, gene expression, and product inhibition†
Shun-he Wangab,
Yong Panab,
Jing Liab,
Hai-qin Chenab,
Hao Zhangab,
Wei Chen abc,
Zhen-nan Gu
abc,
Zhen-nan Gu *ab and
Yong Q. Chenabd
*ab and
Yong Q. Chenabd
aState Key Laboratory of Food Science and Technology, Jiangnan University, Wuxi, Jiangsu 214122, P. R. China. E-mail: zhennangu@jiangnan.edu.cn; Fax: +86 0510 8591 2155; Tel: +86 0510 8519 7096
bSchool of Food Science and Technology, Jiangnan University, Wuxi, Jiangsu 214122, P. R. China
cBeijing Innovation Centre of Food Nutrition and Human Health, Beijing Technology and Business University (BTBU), Beijing 100048, P. R. China
dDepartment of Cancer Biology, Wake Forest School of Medicine, Winston-Salem, NC 27157, USA
First published on 22nd August 2017
Abstract
Previous studies have suggested that dietary alpha-linolenic acid (ALA) increases the levels of omega-3 long-chain polyunsaturated fatty acids (ω-3 LC-PUFAs) in vivo, but the conversion procedure and the genes involved remain poorly understood. In the present work, we designed diets containing various concentrations of ALA and eicosapentaenoic acid (EPA) to feed to mice. Dietary ALA increased the ALA levels in the body in a linear manner and also increased the ω-3 LC-PUFA concentration, but higher ALA intake (above 5%) had no additional effect on ω-3 LC-PUFA levels in vivo. Dietary ALA at a moderate level increased the expression of genes such as Fads1, Fads2, and Elovl5, but higher levels of dietary ALA (above 5%) inhibited their expression in the liver. Further studies demonstrated that the converted EPA could also inhibit the expression of these genes in a concentration-dependent manner, which illustrated that Fads1, Fads2, and Elovl5 were the key genes involved in the conversion of ALA to ω-3 LC-PUFAs. Endogenous ω-3 LC-PUFA biosynthesis from ALA was affected by substrate level, gene expression, and product inhibition.
Introduction
As a major source of energy and structural components of membranes, fatty acids are essential for life. Omega-3 long-chain polyunsaturated fatty acids (ω-3 LC-PUFAs), such as alpha-linolenic acid (ALA, 18:3ω-3), eicosapentaenoic acid (EPA, 20:5ω-3), docosapentaenoic acid (DPA, 22:5ω-3), and docosahexaenoic acid (DHA, 22:6ω-3), are important for optimal health. These fatty acids have beneficial effects in reducing risk factors for obesity, diabetes, cardiovascular disease, and cancer.1,2 However, humans and mice are unable to synthesize ALA, the precursor of ω-3 LC-PUFAs, from palmitic acid because they are deficient in Δ12 and Δ15 desaturases.3 Therefore, ALA has to be obtained via dietary intake to then serve as a precursor for conversion to ω-3 LC-PUFAs.4,5Many factors affect ω-3 fatty acid biosynthesis. EPA, DPA, and DHA are synthesized by the sequential action of desaturase (Fads1 and Fads2) and elongase (Elovl1-7) enzymes,6,7 which essentially takes place in the liver.8,9 Meanwhile, the ω-3/6 fatty acids share the same biosynthetic pathways involving the same desaturases and elongases.10–12 Several studies have shown that a supply of ω-3 fatty acids in the diet decreased the mRNA abundance of Δ6 desaturase and Δ5 desaturase, while the expression of Elovl2 was increased in the mammalian liver.13–15 In contrast, fatty acid desaturase activities were enhanced in pigs subjected to a linseed diet, according to Smink.16 Moreover, the dietary supplementation of ALA in mice decreased the Δ6 desaturase expression in the liver of the offspring at weaning.17 Finally, the varieties of ω-3 LC-PUFAs ingested by animals can lead to different effects on gene expression and the activities of enzymes involved in lipid biosynthesis.
In this study, we focus on ALA as the precursor of ω-3 LC-PUFAs and systematically analyze the conversion progression and gene expression in mice fed diets containing different levels of ALA.
Materials and methods
1 Animals and diets
Six-week-old male C57BL/6J mice were obtained from the Shanghai Laboratory Animal Center (Shanghai, China). After one week of acclimatization with free access to standard mouse chow (commercial diet, 17.14% of energy from fat, 5.05 g/100 g) and water, the mice were randomly divided into nine groups each containing six mice and fed ALA series diets (1, 2.5, 5, or 7.5 wt%), 5% ALA and EPA series diets (0.25, 0.5, 1 wt%), EPA diet (2 wt%), or the control diet (Ctl diet: depleted in ω-3 PUFA) for seven weeks. The diet ingredients were shown in ESI Tables S1 and S2.† All animals were maintained in barrier cages and fed with the appropriate special diet restricted to 10 g per mouse per day. This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the P. R. China. The protocol was approved by the Institutional Animal Welfare and Ethics Committee of Jiangnan University (Wuxi, China).2 Sample collections
Blood samples were collected at 0, 1, 3, 5, and 7 weeks after starting the experimental dietary feeding and were snap frozen in liquid nitrogen and stored at −80 °C until analysis. To minimize any possible stress derived from blood sampling, approximately 50 μL of retro-orbital blood was collected each time according to techniques published in the literature.18 After seven weeks, the mice were sacrificed and liver tissue samples were collected.3 Lipid extraction and derivatization
Fatty acids from blood were methylated and analyzed as previously described.19 Briefly, blood lipids were extracted using the method of Bligh and Dyer,20 with heneicosanoic acid (Sigma-Aldrich) and pentadecanoic acid (Sigma-Aldrich) as internal standard. Lipid extract was dried under a steam of nitrogen at 30 °C, dissolved in 1 mL ethanol, and added with 0.1 mL of 50% aqueous KOH. Tubes were purged with nitrogen, and then heated to 75 °C for 1 h. After cooling, nonsaponifiable lipids were removed by extraction with 3 mL of hexane, and aqueous layer was acidified with HCl and extracted with 3 mL of hexane. The hexane layer was transferred to a new tube and dried in a stream of nitrogen. The residue was treated with 0.5 M methanolic NaOH at 100 °C for 5 min. After cooling, samples were treated with 1 mL of 14% BF3 in methanol (Sigma-Aldrich),21,22 flushed with nitrogen, and then heated to 100 °C for 5 min. After cooling, samples were treated with 4 mL of hexane and 4 mL of saturated, aqueous NaCl, and the hexane layer was transferred to a new tube, dried under a stream of nitrogen, dissolved with 200 μL hexane. Then 1 μL of hexane was analyzed on a GCMS-QP2010 Ultra (Shimadzu Co., Tokyo, Japan) with 30 m × 0.25 mm diameter Rtx-Wax column (Restek International, Bellefonte, PA, USA) with a 0.25 μm coating.4 RNA extraction and quantitative real-time polymerase chain reaction
Total RNA isolation, cDNA synthesis, and transcript levels were conducted as previously described.23 Specific primers for each gene were designed using Primer5 software (PREMIER Biosoft, Canada) and are listed in Table S5.†5 Statistical and multivariate data analysis
One-way analysis of variance (ANOVA; SPSS 20.0, Chicago, IL, USA) was carried out to determine any differences between groups. Those variables (FA) with a p value of less than 0.05 were considered significant.Result
1 Dietary ALA increased its levels in a linear way in vivo
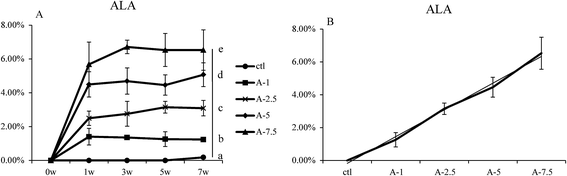
Several groups have reported that dietary ALA can lead to increased levels of ALA in the body in vivo;9,24 the aim of our study was to focus on the efficiency of dietary ALA intake and its connection to ALA levels in the blood. We therefore designed experimental diets containing different concentrations of ALA and the same level of linoleic acid (LA). There were no significant differences between the groups in terms of daily food intake and body weight (Fig. S3 and Table S6†). The lipid contents of these diets were verified by GC/MS analysis (Table S3†).The ALA levels in whole blood at different time points were analyzed by GC/MS after feeding the mice with the experimental diets. The relative amount of ALA content was analysed by the known amount of internal standard (heneicosanoic acid and pentadecanoic acid), and the percent composition of ALA was calculated. As seen in Fig. 1, the ALA content in whole blood increased and remained at a relatively stable level (Fig. 1B). The ALA levels were highest for the A-7.5 group, followed by the A-5, A-2.5, and A-1 groups (Fig. 1B), with significant differences (p < 0.05) between the different groups. Fig. 1B shows that there was a linear association between the dietary ALA content and the levels of ALA in whole blood.
2 Effect of dietary ALA on the ω-3 LC-PUFA levels in vivo
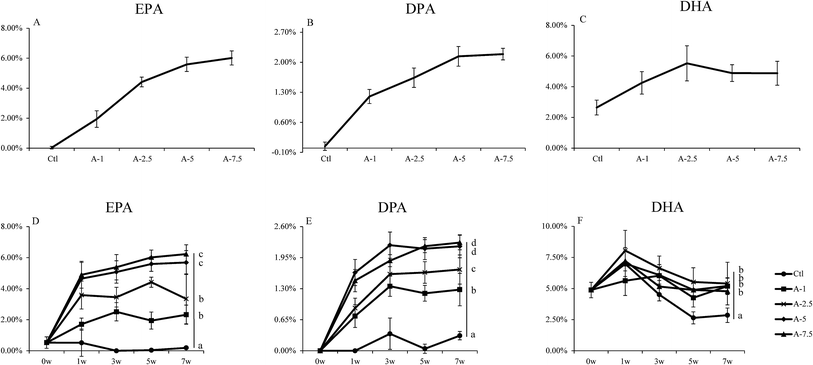
It has been reported that ALA can be converted to ω-3 LC-PUFA in vivo.9,16,24 We further analyzed the conversion efficiency by feeding mice with several diets that differed in ALA content (Table S3†).As seen in Fig. 2, the EPA and DPA levels in the A-7.5, A-5, A-2.5, and A-1 groups increased and the Student's t test showed that there were significant differences (p < 0.05) between the A-1, A-2.5, and A-5 groups; however, there was no statistical difference between the A-5 and A-7.5 groups. The DHA level in the blood increased slightly between the control diet group and the ALA-supplemented diet groups, although there were no significant changes between the ALA-supplemented diet groups. The EPA and DPA levels increased steadily with the increasing dietary ALA intake.
These results indicated that supplementation of certain amounts of dietary ALA could efficiently increase the levels of ω-3 LC-PUFAs (EPA and DPA), but further dietary intake of ALA above 5% had little additional effect on increasing the concentrations of ω-3 LC-PUFAs in vivo, which demonstrated the importance of substrate levels in the conversion.
3 Modulation of genes expression related to ALA/ω-3 LC-PUFA conversion by dietary ALA in liver
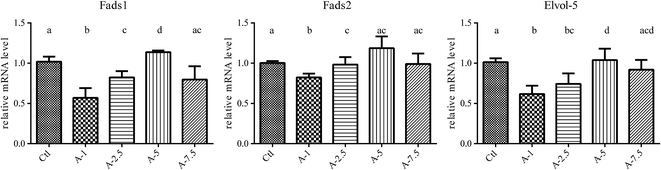
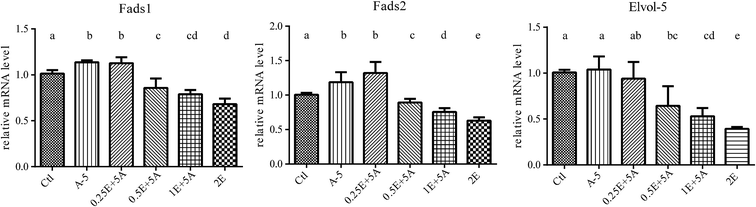
The liver is the main organ involved in the synthesis and metabolism of fatty acids.8,9 To further investigate the phenomenon that higher ALA intake (above 5%) had no additional effect on increasing the blood levels of EPA and DPA, we analyzed the expression of the key genes related to ALA/ω-3 LC-PUFA conversion in the liver, including Fads1, Fads2, Elovl5, Hacd3, Hacd4, Pecr, Tecr, and Hsd17b12 (the results are shown in the ESI†), using fluorescence quantitative PCR (qPCR).With the increase of dietary ALA, the gene expression of Fads1, Fads2, and Elovl5 first increased and then decreased, with 5% ALA in the diet representing the turning point (Fig. 3). These data agreed with our blood analysis results that showed that increasing dietary ALA intake above 5% had no additional effects on ALA to EPA conversion.
4 EPA inhibit the expression of genes related to ALA/ω-3 LC-PUFA conversion in vivo
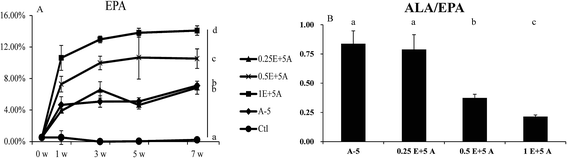
As dietary ALA increased the blood levels of both ALA and EPA, it was interesting to determine whether the EPA converted in vivo affected the expression of genes related to ALA/EPA conversion, including Fads1, Fads2, and Elovl5. Because 5% dietary ALA led to the highest ALA to EPA conversion efficiency, we designed experimental diets with different amounts of EPA plus 5% ALA (Table S4†). The supplementation of dietary EPA did increase the EPA levels, as shown in Fig. 4.As seen in Fig. 5, the gene expression levels of Fads1, Fads2, and Elovl5 in the liver were decreased when the EPA content in the blood was increased. As well as the decreased gene expression, the ratio of ALA/EPA also declined. These results indicated that while ALA activates the gene expression of Fads1, Fads2, and Elovl5 to drive ALA to EPA conversion, EPA downregulates the expression of these genes, and that the ALA/EPA ratio in vivo dictates the expression of genes related to ALA/EPA conversion.
Discussion
In this paper, we studied the conversion efficiency of ALA to ω-3 LC-PUFAs in vivo, and the main gene expression regulation involved in this conversion. We found that dietary ALA could increase ω-3 LC-PUFA levels in the blood and affect the expression of genes related to ALA to ω-3 LC-PUFA conversion in the liver.Several groups have reported that dietary ALA can increase the ALA and EPA levels in the body in humans, mice and rats.24–26 Our data also show that dietary ALA can increase ALA levels in vivo. In addition, our study indicated that the relationship between the dietary ALA content and the percent of ALA in whole blood was linearly dependent. This observation means that the in vivo absorption of dietary ALA from the gut to the blood was quick and efficient (Fig. 1).
Dietary ALA effectively increased the EPA and DPA levels but had a smaller effect on DHA levels in vivo, which has also been reported by other researchers.9,27 However, our research suggested that although dietary ALA did increase the EPA and DPA content within a certain range (from 1% to 5%), higher concentrations of ALA (above 5%) did not enhance the conversion of ALA to ω-3 LC-PUFAs. This illustrates the importance of substrate levels of ALA in the conversion. Compared to the control group that was fed a diet depleted in ω-3 PUFAs, the ALA supplementation significantly increased the DHA levels in vivo (Fig. 2). However, the transformation rate was relatively lower than other ω-3 LC-PUFAs, in agreement with the results of Zhong Fu et al.28 The phenomenon of lower DHA levels in vivo warrants further investigation.
The ω-3 LC-PUFA compositional studies indicated that dietary ALA contributes to ω-3 LC-PUFAs in vivo. However, the data also revealed that the conversion became saturated at a dietary ALA content of 5%, which was the same turning point observed in the gene expression analysis. The trend was that with the increase of dietary ALA, the expression levels of Fads1, Fads2, and Elovl5 first increased and then decreased. However, for other genes, including Hacd3, Hacd4, Pecr, Tecr, and Hsd17b12, it was difficult to reach an exact conclusion for the ALA/ω-3 LC-PUFA conversion (Fig. S1 and S2†). Many factors may influence the expression of genes involved in lipid biosynthesis.14,29 For instance, there are four reactions in the long-chain fatty acid elongation cycle, in which every step may involve many isozymes.14,30 Moreover, many desaturase genes participate in the synthesis of ω-3 LC-PUFAs from the precursor essential PUFAs,31 such as LA and ALA, which share the same enzyme system.16,32
Our data demonstrated that the expression of lipogenic enzymes could be affected by different dietary levels of ALA (Fig. 3). The study of Tonnac et al.13 also showed that the Fads2 gene was downregulated with a ω-3 LC-PUFA diet. In addition, our study further investigated the effects of diets supplemented with the same content of ALA and different levels of EPA. The results indicated that dietary EPA inhibits the expression of Fads1, Fads2, and Elovl5 in the liver and the inhibition of the gene expression was dependent on the ALA/EPA ratio. The converted EPA from dietary ALA and the ALA/EPA ratio determined the conversion of ALA/EPA in the liver and proved that the aforementioned genes play a decisive role in the conversion.
In summary, we analyzed the conversion of ALA to ω-3 LC-PUFAs and the expression of genes related to this conversion. We found that different levels of ALA in the diet could increase the EPA and DPA levels but had a smaller effect on DHA levels in vivo, and proved that Fads1, Fads2, and Elovl5 were the key genes related to the conversion of ALA to ω-3 LC-PUFAs.
Conclusions
In the present work, we have studied the conversion efficiency of ALA to ω-3 LC-PUFAs in vivo, as well as the expression of genes involved in this conversion. We found that dietary ALA increased ALA levels in vivo in a linear manner and increased the levels of EPA and DPA in vivo within a certain range (1–5%), although higher levels of dietary ALA did not lead to a significant increase in EPA and DPA levels. Moreover, our data indicated that the genes Fads1, Fads2, and Elovl5 play a key role in the conversion of ALA to ω-3 LC-PUFAs. This study illustrates that the endogenous biosynthesis of ω-3 LC-PUFAs from ALA is affected by substrate level, gene expression, and product inhibition.Conflict of interest
The authors declare no competing financial interests.Acknowledgements
This work was supported by the State key research and development plan (No. 2017YFD0400200), the National Natural Science Foundation of China (No. 31471128, No. 31571827), the Fundamental Research Funds for the Central Universities (JUSRP51702A) and the program of “Collaborative innovation center of food safety and quality control in Jiangsu Province”.References
- U. Schwab, L. Lauritzen, T. Tholstrup, T. Haldorssoni, U. Riserus, M. Uusitupa and W. Becker, Food Nutr. Res., 2014, 58, 25145 CrossRef PubMed
.
- Z. Gu, J. Suburu, H. Chen and Y. Q. Chen, BioMed Res. Int., 2013, 2013, 824563 Search PubMed
.
- J. G. Wallis, J. L. Watts and J. Browse, Trends Biochem. Sci., 2002, 27, 467–473 CrossRef CAS PubMed
.
- A. A. Spector, Lipids, 1999, 34(Suppl), S1–S3 CrossRef CAS PubMed
.
- D. B. Jump, J. Biol. Chem., 2002, 277, 8755–8758 CrossRef CAS PubMed
.
- A. Voss, M. Reinhart, S. Sankarappa and H. Sprecher, J. Biol. Chem., 1991, 266, 19995–20000 CAS
.
- H. Sprecher, Biochim. Biophys. Acta, 2000, 1486, 219–231 CrossRef CAS
.
- L. A. Afman and M. Muller, Prog. Lipid Res., 2012, 51, 63–70 CrossRef CAS PubMed
.
- J. Suburu, Z. Gu, H. Chen, W. Chen, H. Zhang and Y. Q. Chen, Food Biosci., 2013, 4, 1–12 CrossRef CAS PubMed
.
- R. T. Holman, J. Nutr., 1988, 118, 535–540 CAS
.
- H. P. Cho, M. T. Nakamura and S. D. Clarke, J. Biol. Chem., 1999, 274, 471–477 CrossRef CAS PubMed
.
- H. P. Cho, M. Nakamura and S. D. Clarke, J. Biol. Chem., 1999, 274, 37335–37339 CrossRef CAS PubMed
.
- A. De Tonnac, E. Labussiere, A. Vincent and J. Mourot, Br. J. Nutr., 2016, 116, 7–18 CrossRef CAS PubMed
.
- G. Manohar, S. Elena, T. Alan and C. M. Thomas, Biochem. J., 1988, 249, 351–356 CrossRef
.
- A. Lamaziere, C. Wolf, U. Barbe, P. Bausero and F. Visioli, Prostaglandins, Leukotrienes Essent. Fatty Acids, 2013, 88, 149–154 CrossRef CAS PubMed
.
- W. Smink, W. J. Gerrits, M. Gloaguen, A. Ruiter and J. van Baal, Animal, 2012, 6, 262–270 CrossRef CAS PubMed
.
- L. Shomonov-Wagner, A. Raz and A. Leikin-Frenkel, Lipids Health Dis., 2015, 14, 14 CrossRef PubMed
.
- K. Yurko-Mauro, J. Kralovec, E. Bailey-Hall, V. Smeberg, J. G. Stark and N. Salem Jr, Lipids Health Dis., 2015, 14, 99 CrossRef PubMed
.
- Q. Yang, S. Wang, Y. Ji, H. Chen, H. Zhang, W. Chen, Z. Gu and Y. Q. Chen, Lipids Health Dis., 2017, 16, 10 CrossRef PubMed
.
- W. J. Dyer and E. G. Bligh, Can. J. Biochem. Physiol., 1959, 37, 911–917 CrossRef PubMed
.
- L. R. Hartman, Lab. Pract., 1973, 22, 475–476 CAS
.
- D. C. Schwenke, J. Lipid Res., 2002, 43, 1927–1938 CrossRef CAS
.
- I. J. Edwards, H. Sun, Y. Hu, I. M. Berquin, J. T. O'Flaherty, J. M. Cline, L. L. Rudel and Y. Q. Chen, J. Biol. Chem., 2008, 283, 18441–18449 CrossRef CAS PubMed
.
- D. Sugasini and B. R. Lokesh, Lipids, 2012, 47, 1155–1167 CrossRef CAS PubMed
.
- K. B. Kim, Y. A. Nam, H. S. Kim, A. W. Hayes and B. M. Lee, Food Chem. Toxicol., 2014, 70, 163–178 CrossRef CAS PubMed
.
- J. T. Brenna, N. Salem Jr, A. J. Sinclair and S. C. Cunnane, A. International Society for the Study of Fatty and I. Lipids, Prostaglandins, Leukotrienes Essent. Fatty Acids, 2009, 80, 85–91 CrossRef CAS PubMed
.
- J. R. Gingrich, R. J. Barrios, R. A. Morton, B. F. Boyce, F. J. DeMayo, M. J. Finegold, R. Angelopoulou, J. M. Rosen and N. M. Greenberg, Cancer Res., 1996, 56, 4096–4102 CAS
.
- Z. Fu and A. J. Sinclair, Lipids, 2000, 35, 395–400 CrossRef CAS PubMed
.
- A. Kihara, J. Biochem., 2012, 152, 387–395 CrossRef CAS PubMed
.
- A. Jakobsson, R. Westerberg and A. Jacobsson, Prog. Lipid Res., 2006, 45, 237–249 CrossRef CAS PubMed
.
- M. T. Nakamura and T. Y. Nara, Annu. Rev. Nutr., 2004, 24, 345–376 CrossRef CAS PubMed
.
- E. Zamberletti, F. Piscitelli, V. De Castro, E. Murru, M. Gabaglio, P. Colucci, C. Fanali, P. Prini, T. Bisogno, M. Maccarrone, P. Campolongo, S. Banni, T. Rubino and D. Parolaro, J. Lipid Res., 2017, 58, 301–316 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra06728c |
| This journal is © The Royal Society of Chemistry 2017 |