DOI:
10.1039/C7RA06477B
(Paper)
RSC Adv., 2017,
7, 35290-35296
Inhibition of H1N1 influenza virus by selenium nanoparticles loaded with zanamivir through p38 and JNK signaling pathways
Received
9th June 2017
, Accepted 8th July 2017
First published on 13th July 2017
Abstract
Zanamivir is an effective drug for influenza virus infection, but strong molecular polarity and aqueous solubility limit its clinical application. In recent years, selenium nanoparticles (SeNPs) have attracted attention in the biological field. In this study, surface decoration of SeNPs using zanamivir (ZNV) with antiviral properties was demonstrated. SeNPs co-delivery of a zanamivir nanosystem was designed to reverse influenza virus infection. In breif, the MTT assay, cytopathic effect and nucleic acid level of the virus suggested that zanamivir modified SeNPs (Se@ZNV) resisted proliferation of H1N1 virus and MDCK cells achieved higher viability after treatment with this compound. Besides, both activation and expression of caspase-3 induced during H1N1 virus infection were depressed when treated with Se@ZNV. Furthermore, phosphorylation of p38 and JNK were down-regulated by Se@ZNV. Taken together, our study indicates that Se@ZNV is a novel promising pharmaceutical against H1N1 influenza virus infection.
1 Introduction
Influenza caused by influenza viruses poses a serious threat to society. It is associated with substantial morbidity among children every year.1,2 In 2009, the novel H1N1 influenza pandemic broke out in Mexico and America leading to more than 575![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 deaths worldwide.3,4 Zanamivir is a widely used, broad-spectrum and effective antiviral drug.5,6 Nevertheless, it has a strong molecular polarity, which leads to low absorbability by oral administration, making its administering inconvenient for children.7
400 deaths worldwide.3,4 Zanamivir is a widely used, broad-spectrum and effective antiviral drug.5,6 Nevertheless, it has a strong molecular polarity, which leads to low absorbability by oral administration, making its administering inconvenient for children.7
Selenium (Se) is an essential trace element in human body. Adequate Se helps to maintain activity of NK cells and promote proliferation of T cells, which are significant for antiviral immunity.8 Se possesses the ability to polarize the immune system toward a Th1 pattern and thereby increases the efficacy of vaccines against many viral pathogens.9 It is reported that Se supplementation has beneficial effect on immunity to influenza vaccine in older adults.8 A clinical trial demonstrated that Se supplementation significantly reduces the rate of CD4+ cells count decline among anti-retroviral therapy patients.9 Se yeast attenuates porcine circovirus type 2 (PCV2) infection through altering the systemic inflammation and maintaining the normal organ morphology.10 Diphenyl diselenide resists herpes simplex virus 2 infection in a mice model.11
Based on the distinctive physical and chemical property, nanomaterials have applied in medicine, agricultural technology and food industry.12–14 Among them, selenium nanoparticles (SeNPs) have attracted wide attention in biomedical field due to its weak toxicity compared to the metal nanoparticles, such as gold nanoparticles which may accumulate in the body and present potential toxicity.15–17 In our previous study, we used silver nanoparticles loading drugs to resist viral infection in vitro. But the compound had toxicity to the cells.18–20 Thus, SeNPs has an advantage for lower toxicity compared to those metal nanoparticles. Mahdavi M. reported that oral administration of synthetic selenium nanoparticles (SeNPs) induced robust Th1 cytokine pattern after hepatitis B virus vaccination in mouse model.21 Another paper stated that SeNPs showed anti-type-1 dengue virus activity by attenuating cytopathic effect.22 Due to their small diameter that making entrance to the cells easy, more and more nanophase materials are used as carriers of drugs or siRNA to construct novel nanostructured biomaterials in recent researches. For instance, SeNPs have been reported to promote apoptosis of various tumor cells, such as colorectal carcinoma, lymphoma, lung carcinoma and breast cancer, loading anti-tumor drugs or specific siRNAs.23–25 Infections of viruses lead to apoptosis of host cells. Therefore, in this study, we are aiming to explore whether zanamivir modified SeNPs antagonize MDCK cells apoptosis induced by H1N1 influenza virus.
2 Experimental
2.1 Materials
The madin darby canine kidney cell line MDCK was gained from American Type Culture Collection (ATCC, CCL-34TM). Influenza A/Hubei/74/2009 (H1N1) was seperated and stored in Virus Laboratory, Guangzhou Women and Children's Medical Center. Fetal bovine serum (FBS) and dulbecco's modified eagle medium (DMEM) were purchased from Gibco. L-1-Tosylamido-2-phenylethyl chloromethyl ketone (TPCK), zanamivir, ascorbic acid (VC), Na2SeO3 and thiazolyl blue tetrazolium bromide (MTT) were acquired from Sigma. One step RT-PCR kit was supplied from Takara. Primers and probe were synthetized by Sangon Biotech, China. The BCA protein assay kit and caspase-3 activity assay kit were purchased from Beyotime, China. p-p38, p-JNK, PARP, caspase-3 and β-actin monoclone antibodies from Cell Signaling Technology were used for western blot.
2.2 Synthesis of SeNPs and Se@ZNV
SeNPs were prepared as previously described.26 Briefly, 2 ml of 0.5 mM freshly prepared VC solution and 0.25 ml fresh 0.1 M Na2SeO3 solution were added dropwise into 22.75 ml ultrapure water, followed with constant magnetic stirring for 30 min at room temperature. After that, zanamivir was added. The excess VC, Na2SeO3 and zanamivir were eliminated by dialysis for 24 h. Then concentration of Se@ZNV was detected by ICP-AES (inductively coupled plasma-atomic emission spectrometry). The Se@ZNV nanoparticles solution was ultrasonicated in a water bath before passed through with a 0.22 μm pore size filter. The SeNPS and Se@ZNV samples were stored at 4 °C.
2.3 Characterization of Se@ZNV
The morphology of Se@ZNV nanoparticles was characterized by a transmission electron microscopy (TEM). According to the protocol, samples were prepared by dispersing the powder particles onto a holey carbon film on copper grids before acquisition of micrographs.27 Elemental composition of Se@ZNV was detected by an energy dispersive X-ray spectroscopy (EDX).28 Size distribution and zeta potential of Se@ZNV were determined by a zetasizer nano ZS particle analyser.29 Fourier transform infrared spectroscopy (FTIR) was carried out using dried samples that crushed with KBr. X-ray photoelectron spectrom (XPS) analysis was performed using an Escalab 250 spectrometer.30–32
2.4 Cell culture and cell infection
MDCK cells were cultured in DMEM with 10% FBS, 100 U ml−1 penicillin and 50 U ml−1 streptomycin at 37 °C with 5% CO2.33 H1N1 virus infection was performed and 50% tissue culture infective dose (TCID50) was calculated as previously described. In brief, MDCK cells were seeded in culture dishes for 24 h. Then cells were washed twice with phosphate buffered saline (PBS) and adsorbed with H1N1 in DMEM without FBS for 2 h. The supernatant was removed, and cells were cultured with DMEM containing 2% FBS and 2 μg ml−1 TPCK-treated trypsin.34 The cytopathic effect (CPE) was observed and TCID50 was calculated using Reed-Muench method.35 H1N1 virus used in this study was at the titer of 100 TCID50/ml.
2.5 Cells viability with Se@ZNV
Cytotoxicity of Se@ZNV was detected by MTT assay. Briefly, MDCK cells seeded in a 96-well culture plate were infected with H1N1 virus for 24 h as previously mentioned. Then cells were respectively treated with Se@ZNV containing 15.6 μM SeNPs and 1 nM zanamivir, 15.6 μM SeNPs and 1 nM zanamivir. After 72 h, MTT solution was added at a final concentration of 0.5 mg ml−1 per well. 5 h later, solution in the wells was aspirated off and 150 μl DMSO was added per well. The absorbance values at 570 nm were recorded 20 min later so that cytotoxicity of Se@ZNV against MDCK cells was measured.36,37
2.6 Viral proliferation with Se@ZNV
Nucleic acid level of H1N1 virus was determined by RT-PCR. MDCK cells were infected by H1N1 virus and treated with zanamivir, SeNPs and Se@ZNV respectively as mentioned above. RNA from those MDCK cells was extracted with trizol and chloroform. Then amplification was performed using a one step RT-PCR kit on an Applied Biosystems (ABI) 7500 instrument at 50 °C for 15 min, 95 °C for 15 min, followed by 40 cycles at 94 °C for 15 s and 58 °C for 45 s, and a final extension step at 68 °C for 5 min.38,39 Primers and probe were designed based on the sequences from NCBI database. Primers for H1N1 virus were forward primer 5′-CTCAGCAAATCCTACATTA-3′ and reverse primer 5′-TAGTAGATGGATGGTGAAT-3′. Sequence of probe was CCATAGCACGAGGACTTCTT. Primers of GAPDH were forward primer 5′-CGCCAAGAAGGTGATCATTTC-3′ and reverse primer 5′-CAGGAGGCGTTCGAGATGAC-3′.38,40
2.7 TEM image of H1N1 treated with Se@ZNV
MDCK cells were infected by H1N1 influenza virus and treated with Se@ZNV at 37 °C for 24 h. Culture supernatant which contained the virus was gained and centrifugated to remove the sediment. Then the virus solution was dropped to copper grids as previously described. The grids were stained with 2% phosphotungstic acid and examined by TEM.27
2.8 Caspase-3 activity
Caspase-3 activity was mensurated using a caspase-3 activity detection kit.41 Firstly, MDCK cells were infected with H1N1 virus followed by treatments with zanamivir, SeNPs and Se@ZNV. Then proteins of the cells were extracted following concentration determination using a BCA protein assay kit. Equal quality of proteins mixed with specific caspase 3 substrate Ac-DEVD-pNA were added in a 96-well plate at 37 °C for 1 h. The absorbance value were recorded at 405 nm.
2.9 Western blot analysis
Proteins related to apoptosis were examined by western blot as previously described.42,43 MDCK cells treated with zanamivir, SeNPs and Se@ZNV for 24 h after H1N1 infection were lysed with RIPA buffer. The protein concentrations were quantified using a BCA protein assay kit. Then equivalent amount of proteins were separated on sodium dodecyl sulfate (SDS)–polyacrylamide gel followed by transfer onto PVDF membranes. The membranes were incubated with specific primary antibodies, such as p-p38, p-JNK, c-PARP, caspase-3 and β-actin at 4 °C overnight before combining with horseradish peroxidase (HRP)-linked secondary antibodies. Images were gained using a Tannon gel imaging system after reaction with ECL solution.
2.10 Statistical analysis
GraphPad Prism 5.0 software was used for data analysis.44 Data were analyzed using two-tailed Student's t-test to evaluate differences between two groups or one-way analysis of variance (ANOVA) for multiple group comparisons.45 Differences were considered statistically significant with P < 0.05 (*) or P < 0.01 (**).
3 Results and discussions
3.1 Preparation and characterization of Se@ZNV
The morphology of SeNPs and Se@ZNV was firstly characterized by TEM. As was shown in Fig. 1A, SeNPs and Se@ZNV presented monodisperse and spherical structure. Size distribution in Fig. 1E further verified that the average sizes of Se@ZNV and SeNPs were 82 nm and 142 nm respectively. Moreover, Fig. 1C suggested that the size of Se@ZNV nanoparticles slightly increased, but kept 80 nm to 90 nm in 30 days. The small size of Se@ZNV contributed to the highly stable nanostructures and made it easy to cross cell membrane. In Fig. 1B, surface elemental composition analysis of Se@ZNV by EDX showed a strong signal of Se atoms (31.46%), with 12.49% C atom and 0.99% O atom. 55.06% Cu atom might come from the copper screen used in the experiment. In Fig. 1D, the zeta potential of SeNPs alone was −24.5 mV and increased to −34.8 mV after loading zanamivir, which explained the higher stability of Se@ZNV.
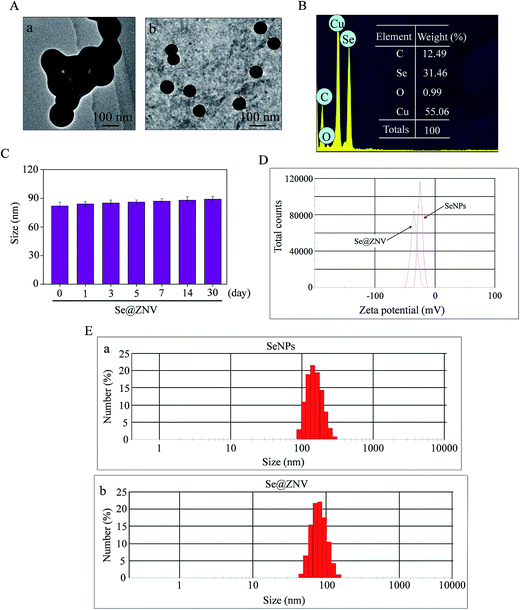 |
| | Fig. 1 Characterization of SeNPs and Se@ZNV. (A) TEM images of SeNPs (a) and Se@ZNV (b). (B) Elemental composition analysis of Se@ZNV by EDX. (C) Size distribution of Se@ZNV in aqueous solutions in 30 days. (D) Zeta potentials of SeNPs and Se@ZNV. (E) Size distributions of SeNPs (a) and Se@ZNV (b). | |
3.2 FTIR spectrum and XPS detection
In Fig. 2A, the FTIR spectrum of Se@ZNV testified that zanamivir ligand formed part of the nano-composite. Zanamivir displayed IR absorbance peaks at 1750 cm−1 and 755 cm−1, corresponding to the conjugated C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C
O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N covalent bonds, which might due to the formation of Se@ZNV. In Fig. 2B, the XPS spectra were also recorded to examine the interaction between ZNV and Se@ZNV. The O 1s peak in the spectrum of Se@ZNV further confirmed that ZNV had been successfully conjugated to the SeNPs.
N covalent bonds, which might due to the formation of Se@ZNV. In Fig. 2B, the XPS spectra were also recorded to examine the interaction between ZNV and Se@ZNV. The O 1s peak in the spectrum of Se@ZNV further confirmed that ZNV had been successfully conjugated to the SeNPs.
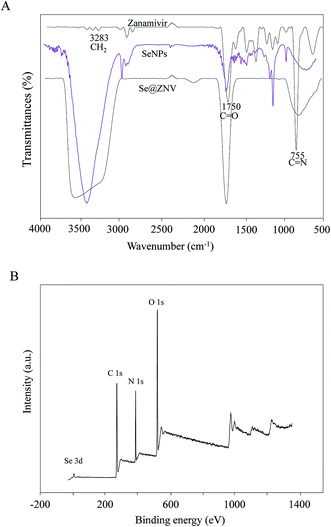 |
| | Fig. 2 FTIR spectrum and XPS detection of SeNPs and Se@ZNV. (A) FTIR spectrums. (B) XPS detection. | |
3.3 Cells viability and viral proliferation with Se@ZNV
MDCK cells were treated with zanamivir, SeNPs and Se@ZNV respectively. The cytotoxic effect of MDCK cells infected by H1N1 virus was observed and antiviral activity of Se@ZNV was investigated by MTT assay. As was seen in Fig. 3A, viability of MDCK cells without any treatment post infection was 45.7%, while cells treated with zanamivir and SeNPs reached to 62.2% and 41.4% respectively. Moreover, cells treated with Se@ZNV reached to 72.7% significantly. It suggested that antiviral activity of SeNPs effectively enhanced with zanamivir on the surface. Total RNA of cells in different groups were extracted and nucleic acid level of H1N1 was detected. As Fig. 3B presented, cells treated with Se@ZNV got a lower level of H1N1 virus than zanamivir alone, indicating that zanamivir was more effective after being coated by SeNPs. In Fig. 3C, cells infected by H1N1 without treatments showed enlarged intercellular space, cell swelling and lysis. These H1N1-induced cytopathic effect were attenuate when treated with Se@ZNV after infection. These assays suggested that Se@ZNV effectively suppressed the H1N1 virus proliferation.
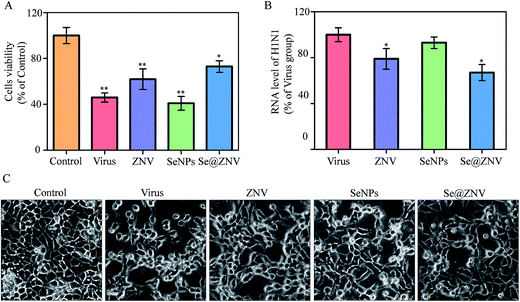 |
| | Fig. 3 Cells viability and viral proliferation with Se@ZNV. (A) Cells viability by MTT assay. (B) RNA level of H1N1. (C) Cytotoxic effect of MDCK cells. | |
3.4 Morphology change of H1N1 virus by Se@ZNV
As was shown in Fig. 4, the untreated H1N1 virus particles presented a spherical shape. After interaction with Se@ZNV for 24 h, the virus particles turned misty and the edge was unclear. This result demonstrated that Se@ZNV could directly destroy the H1N1 virus. It was similar to the previous report that silver nanoparticles destroyed H3N2 influenza virus structure and led to disruption of viral function.46 The Se@ZNV compound may directly interact with H1N1 viral capsid proteins and thereby inhibit the viral binding and endocytosis to the host cells.
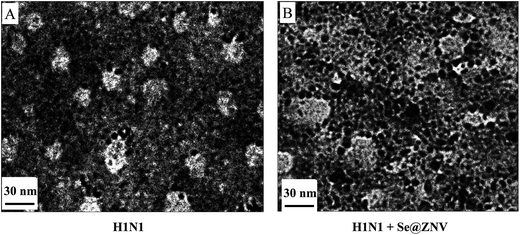 |
| | Fig. 4 TEM images of H1N1 virus. (A) H1N1 virus without any treatment. (B) H1N1 treated with Se@ZNV. | |
3.5 Inhibition of caspase-3 by Se@ZNV
It has been reported that H1N1 influenza virus induces apoptosis of cells and caspase-3 participates in the process.47,48 Caspase-3 plays as a pivotal role in apoptosis, which can be activated during virus infection. It is responsible for the proteolytic cleavage of many critical proteins, such as the poly-ADP-ribose polymerase (PARP). Cleavage of PARP (c-PARP) facilitates cellular disassembly and serves as a marker of apoptosis.49 In this study, MDCK cells were treated with zanamivir, SeNPs or Se@ZNV after H1N1 virus infection. Then activity of caspase-3 was determined using a detection kit and expression of caspase-3 or c-PARP were detected by western blot. As was indicated in Fig. 5A, caspase-3 activity of cells infected by H1N1 virus without treatment was 273% of the control group that uninfected while the Se@ZNV treated group substantially dropped to 161%. In Fig. 5B, protein expressions of both caspase-3 and c-PARP in Se@ZNV group were decreased obviously compared with the untreated group which increased after infection. That is to say, caspase-3 and PARP protein were regulated when cells were treated with Se@ZNV. These results suggested that Se@ZNV inhibited apoptosis of MDCK cells during H1N1 infection by regulating caspase-3 protein.
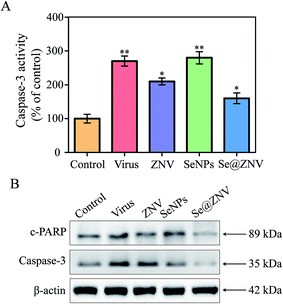 |
| | Fig. 5 Activation and expression of caspase-3. (A) Activation of caspase-3. (B) Expression of caspase-3 protein. | |
3.6 Signaling pathways
Jun-amino-terminal kinase (JNK) and p38 mitogen-activated protein kinases (MAPK) signalings are potently and preferentially activated by a variety of environmental stresses including virus infection. Phosphorylation of JNK and p38 are signs of apoptosis.50 These two signaling ways were investigated by western blot in our study. As was displayed in Fig. 6B, expressions of p-JNK and p-p38 raised in untreated H1N1 infection group, consistent with the previous report.51 But both the expression evidently decreased after treated with Se@ZNV. The result reflected that p38 MAPK and JNK signaling pathways participated in the inhibition of cells apoptosis with Se@ZNV. Se@ZNV might inhibit apoptosis of MDCK cells induced by H1N1 influenza infection through regulating the expressions of p38 and JNK.
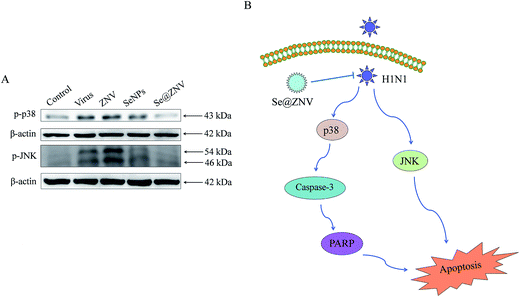 |
| | Fig. 6 Signaling pathways participated in the inhibition of H1N1 by Se@ZNV. (A) Regulation of p38 and JNK by Se@ZNV. (B) Sketch of p38 and JNK signaling pathways. | |
4 Conclusions
In conclusion, our study synthetized a new nano-compound comprised by selenium nanoparticles loading the antiviral drug zanamivir (Se@ZNV). The Se@ZNV revealed good biological activity to restrain proliferation of H1N1 virus in MDCK cells. Cytopathic effect induced by H1N1 virus was markedly alleviated, which led to higher cells viability. Activation of caspase-3 and cleavage of PARP during H1N1 virus infection were depressed by Se@ZNV. Besides, p38 and JNK signaling pathways were referred to the inhibition of H1N1 virus infection using Se@ZNV. Altogether, this study illustrates that Se@ZNV can effectively prevent MDCK cells from H1N1 influenza virus infection.
Conflict of interest
All the authors report no conflicts of interest in this work.
Authors' contributions
BZ designed the study and supervised the work. ZL, YL, MG, MX, CW, MZ, TX and YX carried out the experiments. ZL and YL analyzed the data and drafted the manuscript. ZL and BZ refined the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by the China Postdoctoral Science Foundation (No. 2015M582366), the Technology Planning Project of Guangdong Province (No. 2014A020212697), the Technology Planning Project of Guangzhou (No. 201607010120).
References
- L. Boes, B. Boedeker, P. Schmich, M. Wetzstein, O. Wichmann and C. Remschmidt, Vaccine, 2017, 35, 3789–3796 CrossRef PubMed.
- Z. F. Lin, M. Q. Zhao, M. Guo, L. Kuang, C. B. Wang, G. W. Lian and B. Zhu, Clin. Lab., 2015, 61, 917–924 CAS.
- F. S. Dawood, A. D. Iuliano, C. Reed, M. I. Meltzer, D. K. Shay, P. Y. Cheng, D. Bandaranayake, R. F. Breiman, W. A. Brooks, P. Buchy, D. R. Feikin, K. B. Fowler, A. Gordon, N. T. Hien, P. Horby, Q. S. Huang, M. A. Katz, A. Krishnan, R. Lal, J. M. Montgomery, K. Molbak, R. Pebody, A. M. Presanis, H. Razuri, A. Steens, Y. O. Tinoco, J. Wallinga, H. Yu, S. Vong, J. Bresee and M. A. Widdowson, Lancet Infect. Dis., 2012, 12, 687–695 CrossRef PubMed.
- M. Rahman, S. A. Hoque, M. A. Islam and S. R. Rahman, Virus Genes, 2017, 53, 377–385 CrossRef CAS PubMed.
- F. M. Marty, J. Vidal-Puigserver, C. Clark, S. K. Gupta, E. Merino, D. Garot, M. J. Chapman, F. Jacobs, E. Rodriguez-Noriega, P. Husa, D. Shortino, H. A. Watson, P. J. Yates and A. F. Peppercorn, Lancet Respir. Med., 2017, 5, 135–146 CrossRef CAS PubMed.
- D. Y. Oh, J. Panozzo, S. Vitesnik, R. Farrukee, D. Piedrafita, J. Mosse and A. C. Hurt, Antiviral Ther., 2017 DOI:10.3851/IMP3135..
- D. Dholakia, S. Goyal, S. Jamal, A. Singh, A. Das and A. Grover, BMC Bioinf., 2016, 17, 239–249 CrossRef PubMed.
- K. Ivory, E. Prieto, C. Spinks, C. N. Armah, A. J. Goldson, J. R. Dainty and C. Nicoletti, Clin. Nutr., 2017, 36, 407–415 CrossRef CAS PubMed.
- H. Steinbrenner, S. Al-Quraishy, M. A. Dkhil, F. Wunderlich and H. Sies, Adv. Nutr., 2015, 6, 73–82 CrossRef CAS PubMed.
- G. Liu, G. Yang, G. Guan, Y. Zhang, W. Ren, J. Yin, Y. M. Aguilar, W. Luo, J. Fang, X. Yu, T. Li and Y. Yin, PLoS One, 2015, 10, e0115833 Search PubMed.
- G. Sartori, N. S. Jardim, M. H. Marcondes Sari, F. Dobrachinski, A. P. Pesarico, L. C. Rodrigues Jr, J. Cargnelutti, E. F. Flores, M. Prigol and C. W. Nogueira, J. Cell. Biochem., 2016, 117, 1638–1648 CrossRef CAS PubMed.
- S. A. C. Carabineiro, Molecules, 2017, 22, 1–16, DOI:10.3390/molecules22050857.
- N. A. Anjum, V. Adam, R. Kizek, A. C. Duarte, E. Pereira, M. Iqbal, A. S. Lukatkin and I. Ahmad, Environ. Res., 2015, 138, 306–325 CrossRef CAS PubMed.
- D. Li, D. Y. Lv, Q. X. Zhu, H. Li, H. Chen, M. M. Wu, Y. F. Chai and F. Lu, Food Chem., 2017, 224, 382–389 CrossRef CAS PubMed.
- X. D. Zhang, J. Chen, Z. Luo, D. Wu, X. Shen, S. S. Song, Y. M. Sun, P. X. Liu, J. Zhao, S. Huo, S. Fan, F. Fan, X. J. Liang and J. Xie, Adv. Healthcare Mater., 2014, 3, 133–141 CrossRef CAS PubMed.
- X. D. Zhang, Z. Luo, J. Chen, X. Shen, S. Song, Y. Sun, S. Fan, F. Fan, D. T. Leong and J. Xie, Adv. Mater., 2014, 26, 4565–4568 CrossRef CAS PubMed.
- X. D. Zhang, Z. Luo, J. Chen, H. Wang, S. S. Song, X. Shen, W. Long, Y. M. Sun, S. Fan, K. Zheng, D. T. Leong and J. Xie, Small, 2015, 11, 1683–1690 CrossRef CAS PubMed.
- Z. F. Lin, Y. H. Li, M. Guo, T. T. Xu, C. B. Wang, M. Q. Zhao, H. Z. Wang, T. F. Chen and B. Zhu, RSC Adv., 2017, 7, 742–750 RSC.
- Y. H. Li, Z. F. Lin, M. Q. Zhao, M. Guo, T. T. Xu, C. B. Wang, H. M. Xia and B. Zhu, RSC Adv., 2016, 6, 89679–89686 RSC.
- Y. H. Li, Z. F. Lin, T. T. Xu, C. B. Wang, M. Q. Zhao, M. S. Xiao, H. Z. Wang, N. Deng and B. Zhu, RSC Adv., 2017, 7, 1453–1463 RSC.
- M. Mahdavi, F. Mavandadnejad, M. H. Yazdi, E. Faghfuri, H. Hashemi, S. Homayouni-Oreh, R. Farhoudi and A. R. Shahverdi, Journal of Infection and Public Health, 2017, 10, 102–109 CrossRef PubMed.
- S. Ramya, T. Shanmugasundaram and R. Balagurunathan, J. Trace Elem. Med. Biol., 2015, 32, 30–39 CAS.
- M. Kumari, L. Ray, M. P. Purohit, S. Patnaik, A. B. Pant, Y. Shukla, P. Kumar and K. C. Gupta, Eur. J. Pharm. Biopharm., 2017 DOI:10.1016/j.ejpb.2017.05.003.
- S. Kumar, M. S. Tomar and A. Acharya, Colloids Surf., B, 2015, 126, 546–552 CrossRef CAS PubMed.
- L. Kamrani Moghaddam, S. Ramezani Paschepari, M. A. Zaimy, A. Abdalaian and A. Jebali, Cancer Gene Ther., 2016, 23, 321–325 CrossRef CAS PubMed.
- Y. Li, Z. Lin, M. Zhao, T. Xu, C. Wang, H. Xia, H. Wang and B. Zhu, Int. J. Nanomed., 2016, 11, 3065–3076 CrossRef CAS PubMed.
- Y. Li, Z. Lin, M. Zhao, T. Xu, C. Wang, L. Hua, H. Wang, H. Xia and B. Zhu, ACS Appl. Mater. Interfaces, 2016, 8, 24385–24393 CAS.
- Y. Xia, P. You, F. Xu, J. Liu and F. Xing, Nanoscale Res. Lett., 2015, 10, 349–362 CrossRef PubMed.
- Y. Li, M. Guo, Z. Lin, M. Zhao, M. Xiao, C. Wang, T. Xu, T. Chen and B. Zhu, Int. J. Nanomed., 2016, 11, 6693–6702 CrossRef PubMed.
- S. Yu, W. Zhang, W. Liu, W. Zhu, R. Guo, Y. Wang, D. Zhang and J. Wang, Nanotechnology, 2015, 26(145703), 1–15 CAS.
- G. Gonzalez-Gil, P. N. Lens and P. E. Saikaly, Front. Microbiol., 2016, 7, 571–584 Search PubMed.
- S. N. Nadig, S. K. Dixit, N. Levey, S. Esckilsen, K. Miller, W. Dennis, C. Atkinson and A. M. Broome, RSC Adv., 2015, 5, 43552–43562 RSC.
- B. Gilbertson, W. C. Ng, S. Crawford, J. L. McKimm-Breschkin and L. E. Brown, J. Virol., 2017, 91, 1–33 CrossRef PubMed.
- S. Shahsavandi, M. M. Ebrahimi, S. Masoudi and H. Izadi, Adv. Virol., 2015, 2015(675921), 1–6 Search PubMed.
- K. Ming, Y. Chen, F. Yao, J. Shi, J. Yang, H. Du, X. Wang, Y. Wang and J. Liu, Int. J. Biol. Macromol., 2017, 94, 28–35 CrossRef CAS PubMed.
- S. Besbes, S. Shah, I. Al-Dybiat, S. Mirshahi, H. Helfer, H. Najah, C. Fourgeaud, M. Pocard, I. Ghedira, J. Soria and M. Mirshahi, Int. J. Cell Biol., 2017, 2017, 1873834 Search PubMed.
- M. H. El-Dakdouki, K. El-Boubbou, D. C. Zhu and X. Huang, RSC Adv., 2011, 1, 1449–1452 RSC.
- Z. Yang, G. Mao, Y. Liu, Y. C. Chen, C. Liu, J. Luo, X. Li, K. Zen, Y. Pang, J. Wu and F. Liu, Virol. Sin., 2013, 28, 24–35 CrossRef CAS PubMed.
- J. R. Yang, C. Y. Kuo, H. Y. Huang, F. T. Wu, Y. L. Huang, C. Y. Cheng, Y. T. Su, F. Y. Chang, H. S. Wu and M. T. Liu, J. Clin. Microbiol., 2014, 52, 76–82 CrossRef CAS PubMed.
- G. A. Maroniche, M. Sagadin, V. C. Mongelli, G. A. Truol and M. del Vas, Virol. J., 2011, 8, 308–315 CrossRef CAS PubMed.
- F. Bai, B. Ni, M. Liu, Z. Feng, Q. Xiong and G. Shao, Vet. Microbiol., 2015, 175, 58–67 CrossRef CAS PubMed.
- B. Zhu, T. Xu, Z. Lin, C. Wang, Y. Li, M. Zhao, L. Hua, M. Xiao and N. Deng, Arch. Virol., 2017, 162, 1649–1660 CrossRef CAS PubMed.
- B. Zhu, Y. Li, Z. Lin, M. Zhao, T. Xu, C. Wang and N. Deng, Nanoscale Res. Lett., 2016, 11, 1–8 CrossRef CAS PubMed.
- B. G. Kipre, N. K. Guessennd, M. W. Kone, V. Gbonon, J. K. Coulibaly and M. Dosso, BMC Complementary Altern. Med., 2017, 17, 170–174 CrossRef CAS PubMed.
- T. Lei, R. Manchanda, A. Fernandez-Fernandez, Y. C. Huang, D. Wright and A. J. McGoron, RSC Adv., 2014, 4, 17959–17968 RSC.
- D. Xiang, Y. Zheng, W. Duan, X. Li, J. Yin, S. Shigdar, M. L. O'Connor, M. Marappan, X. Zhao, Y. Miao, B. Xiang and C. Zheng, Int. J. Nanomed., 2013, 8, 4103–4113 CrossRef PubMed.
- S. Kluge, Y. Genzel, K. Laus, A. Serve, A. Pflugmacher, B. Peschel, E. Rapp and U. Reichl, Biotechnol. J., 2016, 11, 1332–1342 CrossRef CAS PubMed.
- X. Wang, J. Tan, S. Biswas, J. Zhao, K. Devadas, Z. Ye and I. Hewlett, Viruses, 2016, 8, 33–43 CrossRef PubMed.
- O. Julien and J. A. Wells, Cell Death Differ., 2017 DOI:10.1038/cdd.2017.44.
- X. Sui, N. Kong, L. Ye, W. Han, J. Zhou, Q. Zhang, C. He and H. Pan, Cancer Lett., 2014, 344, 174–179 CrossRef CAS PubMed.
- F. Zang, Y. Chen, Z. Lin, Z. Cai, L. Yu, F. Xu, J. Wang, W. Zhu and H. Lu, Immunology, 2016, 148, 56–69 CrossRef CAS PubMed.
Footnote |
| † Authors contributed equally to the work. |
|
| This journal is © The Royal Society of Chemistry 2017 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article ,
Min Guo,
Misi Xiao,
Changbing Wang,
Mingqi Zhao,
Tiantian Xu,
Yu Xia and
Bing Zhu
,
Min Guo,
Misi Xiao,
Changbing Wang,
Mingqi Zhao,
Tiantian Xu,
Yu Xia and
Bing Zhu *
*
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 deaths worldwide.3,4 Zanamivir is a widely used, broad-spectrum and effective antiviral drug.5,6 Nevertheless, it has a strong molecular polarity, which leads to low absorbability by oral administration, making its administering inconvenient for children.7
400 deaths worldwide.3,4 Zanamivir is a widely used, broad-spectrum and effective antiviral drug.5,6 Nevertheless, it has a strong molecular polarity, which leads to low absorbability by oral administration, making its administering inconvenient for children.7
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C
O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N covalent bonds, which might due to the formation of Se@ZNV. In Fig. 2B, the XPS spectra were also recorded to examine the interaction between ZNV and Se@ZNV. The O 1s peak in the spectrum of Se@ZNV further confirmed that ZNV had been successfully conjugated to the SeNPs.
N covalent bonds, which might due to the formation of Se@ZNV. In Fig. 2B, the XPS spectra were also recorded to examine the interaction between ZNV and Se@ZNV. The O 1s peak in the spectrum of Se@ZNV further confirmed that ZNV had been successfully conjugated to the SeNPs.






