Open Access Article
Open Access ArticleColorimetric detection of streptomycin in milk based on peroxidase-mimicking catalytic activity of gold nanoparticles†
Jing Zhao,
Yuangen Wu *,
Han Tao,
Huayun Chen,
Wenping Yang and
Shuyi Qiu
*,
Han Tao,
Huayun Chen,
Wenping Yang and
Shuyi Qiu
Guizhou Province Key Laboratory of Fermentation Engineering and Biopharmacy, School of Liquor and Food Engineering, Guizhou University, Huaxi District, Guiyang 550025, Guizhou Province, P. R. China. E-mail: wuyg1357@163.com
First published on 7th August 2017
Abstract
A novel colorimetric aptamer sensor (aptasensor) for the detection of streptomycin (STR) is developed based on the peroxidase-like activity of gold nanoparticles (AuNPs) and their interaction with STR-aptamer complex. It is known that AuNPs can easily oxidize substrates in the presence of hydrogen peroxide in a manner identical to peroxidase and 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) is a commonly used peroxidase substrate with green oxidation product and a characteristic absorption peak at 733 nm. In the absence of STR, STR1 aptamer is adsorbed on the surface of AuNPs, and restrains its catalytic activity due to shield effect of ssDNA sequence against substrates. STR first binds with STR1 aptamer to form an STR-aptamer complex; thus, aptamer sequences do not get absorbed on the surface of the nanoparticles and their peroxidase activity is further greatly enhanced by the STR-aptamer complex, which enables AuNPs to catalyse the oxidation of the substrates. To achieve a higher level of sensitivity, several key parameters of the proposed aptasensor such as pH, concentrations of STR1 aptamer and AuNPs, and incubation and reaction temperature have been investigated. Under optimal conditions, the proposed aptasensor can detect STR in a linear range from 0.1 μM to 0.5 μM with a limit of detection (LOD) as low as 86 nM and exhibits good selectivity. Moreover, further studies also validate the applicability of the proposed aptasensor in milk samples, revealing that it may have enormous potential utility for practical STR detection in food products in the future.
Introduction
Streptomycin (STR) is a broad-spectrum aminoglycoside anti-microbial agent obtained from Streptomyces griseus for Gram-negative bacterial infection treatment and is used not only in human health care, but also in agriculture, veterinary medicine and other fields.1 Excess of STR could result in presence of STR residues in animal derived food products, causing serious side effects to human health such as allergic reactions, loss of hearing and toxicity to the kidneys.2 Considering the hazard of STR excess, most countries have set a standard for STR residues in animal products. According to the European Commission, the maximum residue limit of STR in milk and meat is 200 and 500 μg kg−1, respectively, and a similar standard is established by the Ministry of Agriculture of China as well. To ensure food safety and human health, the development of sensitive and feasible ways for STR detection is in significant demand.Recently, a series of analytical approaches have been reported for quantitative determination of STR residue in food products. High-performance liquid chromatography (HPLC) can detect STR with high sensitivity but it requires post-column derivatization owing to the lack of a chromophore group in STR.3 Excessive cost and tedious pre-treatment restrict the applications of liquid chromatography-mass spectrometry (LC-MS), although it can detect STR accurately and sensitively.4,5 Immunochemical assays like radioimmunoassay, enzyme-linked immunosorbent assay (ELISA) and fluorescence immunoassay have been employed to detect STR residues, but in real samples, cross-reactions with other substances can be a barrier for efficient determination of the target.6–8 Therefore, it is favourable to develop a more economical and facile method for STR detection.
Aptamers are short single-stranded DNA (ssDNA) or RNA molecules selected from random-sequence nucleic acid libraries by SELEX (systematic evolution of ligands by exponential enrichment) and have attracted considerable attention in the field of analytical chemistry.9–11 They not only possess great binding ability with high specificity, sensitivity and remarkable affinity, but also offer an edge over traditional antibodies, including cost-effectiveness, simplicity of synthesis and modification, low toxicity and immunogenicity.12–14 Considering the above advantages, aptamers have potential application as recognition elements in biosensors for antibiotics. Zhou and his team have screened an ssDNA aptamer (named STR1) against STR with a dissociation constant (Kd) of 199.1 nM, and they further employed this aptamer and quantum dot tags to detect STR with LOD of 10 nM.15,16 Some electrochemical aptasensors are constructed for STR detection using STR1 aptamer as the recognition element. Taghdisi and co-workers have developed an electrochemical aptasensor with excellent LOD, but the modification of the gold electrode is rather tedious.17 The electrochemical aptasensor constructed by Guo and his team can detect STR sensitively, but the method requires complicated composite material preparation and complex construction.18 The mentioned methods for STR detection depend on electrochemical apparatus, which cannot meet the demand of rapid and on-site STR detection in actual samples. It is therefore necessary to develop a more convenient way to determine STR.
Colorimetric assay is a common technique for analytical applications since recognition events can be easily monitored by the naked eye.19 AuNPs is a widely used nanomaterial in the construction of colorimetric aptasensors due to their simplicity, ease of synthesis and extremely high absorption coefficient as well as strong distance-dependent optical properties.20,21 The aggregation of AuNPs has been widely applied for colorimetric detection; however, the presence of a non-specific target results in the lack of accuracy in colorimetric detection. Catalytic activity of AuNPs mimics peroxidase activity in that AuNPs can oxidize substrates to form coloured products, which can offer a substitutable signal output for aptamer-based colorimetric sensors. But owing to lack of a molecular recognition element, biosensors utilizing the enzyme-mimicking activity of AuNPs are limited to those used for non-specific detection of glucose and hydrogen peroxide, and AuNPs' intrinsic peroxidase-like activity for antibiotic detection has been rarely reported.22,23 Recently, Sharma and co-workers built a novel aptasensor for kanamycin detection using the catalytic activity of AuNPs as signal output.24 Unfortunately, peroxidase-mimicking catalytic activity of AuNPs has not been applied for STR detection to date.
Herein, we explore a colorimetric aptasensor for STR utilizing intrinsic peroxidase-like activity of AuNPs and their interaction with an STR-aptamer complex. The blocking of the AuNPs surface through conjugation with STR1 aptamer leads to the inhibition of peroxidase activity, resulting in no colour change of the sensing solution. In the presence of STR, STR1 aptamer first binds with STR molecules to form an STR-aptamer complex; thus, aptamer sequences do not get absorbed on the surface of the nanoparticles and nanoparticle peroxidase activity is further enhanced greatly by the STR-aptamer complex. As a result, the substrate of ABTS is catalysed to produce a green product that exhibits an obvious absorption peak at 733 nm. The sensing strategy of the proposed aptasensor utilizing catalytic ability of AuNPs is first employed to detect STR. In addition, the aptasensor is cost-effective, easy to fabricate, and may be extended to detect biological and environmental samples by replacing corresponding aptamers.
Experimental
Reagents and chemicals
STR-binding single-stranded DNA (STR1 aptamer sequence: 5′-TAGGGAATTCGTCGACGGATCCGGGGTCTGGTGTTCTGCTTTGTTCTGTCGGGTCGTCTGCAGGTCGACGCATGCGCCG-3′, 79-mer) was synthesized and HPLC-purified by Sangon Biotechnology Co., Ltd. (Shanghai, China) and dissolved in Tris–HCl buffer solution (20 mM Tris–HCl, 50 mM NaCl, pH 7.2) before use. Materials for AuNP preparation, HAuCl4, sodium borohydride, cysteamine, and antibiotics like tetracycline, streptomycin sulfate, amoxicillin sodium, diclofenac sodium, oxytetracycline dihydrate, carbamazepine, and penicillin G potassium salt were obtained from Aladdin Incorporated (Shanghai, China). ABTS was procured from Sigma-Aldrich Co. LLC. (Milwaukee, WI, USA). 96-well microplates (Costar) were purchased from Corning Incorporated (NY, USA). All reagents were of analytical grade, and bi-distilled water was used throughout the experiments.Apparatus
A microplate spectrophotometer Multiskan GO (Thermo Scientific, USA) was used to record UV/vis absorption spectra and absorbance values of each sample. The size and morphology of AuNPs were determined by a transmission electron microscope (TEM) (Hitachi, LTD. Japan). Nanoparticle size and zeta potential analyzer (Beckman Coulter, USA) was used to measure the zeta potential of nanoparticles.Preparation of AuNPs
All glassware used was thoroughly cleaned with freshly prepared aqua regia solution (HCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HNO3, 3
HNO3, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), and rinsed with bi-distilled water and then dried in an oven. Citrate-coated AuNPs (CC-AuNPs) were obtained by classical citrate reduction of HAuCl4 according to previously published protocol.25 Briefly, 10 mL of 38.8 mM sodium citrate solution was rapidly injected to a boiling 100 mL of 1.0 mM HAuCI4 solution under vigorous stirring. The mixed solution was boiled for 15 min and further stirred for another 15 min then cooled to room temperature and filtered using an ultrafiltration membrane (250 nm aperture). In the case of cysteamine-stabilized AuNPs (CS-AuNPs), 1.2 mL of 213 mM cysteamine and 1.42 mM HAuCI4 were mixed, and then the mixture was blended under ambient temperature for 20 min.26 Subsequently, 30 μL of 10 mM NaBH4 was added to the above solution, and the mixture was stirred for another 25 min at room temperature in the dark. As for unmodified AuNPs (umAuNPs), according to the borohydride reduction method, 1.5 mL of 29.43 mM HAuCI4 solution was diluted with 118.5 mL bi-distilled water.27 Afterward, 3 mL of 264.34 mM NaBH4 solution were added dropwise under vigorous stirring. The resulting wine-red colloidal solution was further stirred for 30 min and then left undisturbed overnight. All the AuNP solutions were stored in a refrigerator at 4 °C, and their amounts were determined according to Bouguer–Lambert–Beer law.
1), and rinsed with bi-distilled water and then dried in an oven. Citrate-coated AuNPs (CC-AuNPs) were obtained by classical citrate reduction of HAuCl4 according to previously published protocol.25 Briefly, 10 mL of 38.8 mM sodium citrate solution was rapidly injected to a boiling 100 mL of 1.0 mM HAuCI4 solution under vigorous stirring. The mixed solution was boiled for 15 min and further stirred for another 15 min then cooled to room temperature and filtered using an ultrafiltration membrane (250 nm aperture). In the case of cysteamine-stabilized AuNPs (CS-AuNPs), 1.2 mL of 213 mM cysteamine and 1.42 mM HAuCI4 were mixed, and then the mixture was blended under ambient temperature for 20 min.26 Subsequently, 30 μL of 10 mM NaBH4 was added to the above solution, and the mixture was stirred for another 25 min at room temperature in the dark. As for unmodified AuNPs (umAuNPs), according to the borohydride reduction method, 1.5 mL of 29.43 mM HAuCI4 solution was diluted with 118.5 mL bi-distilled water.27 Afterward, 3 mL of 264.34 mM NaBH4 solution were added dropwise under vigorous stirring. The resulting wine-red colloidal solution was further stirred for 30 min and then left undisturbed overnight. All the AuNP solutions were stored in a refrigerator at 4 °C, and their amounts were determined according to Bouguer–Lambert–Beer law.
Procedure of STR determination
In a typical experiment, 5 μL of 100 μM STR stock solutions was added into a 1.5 mL centrifuge tube and mixed with 5 μL of 5 μM STR1 aptamer solutions and 10 μL of binding buffer (10 mM Tris–HCl) thoroughly, and then the mixture was incubated at 30 °C for 30 min. Next, 200 μL of AuNPs was added to the centrifuge tube, and the mixture was incubated at 30 °C for another 30 min. Finally, 260 μL of 5 mM NaAc solution and 20 μL substrate solution (10 μL of 4 M H2O2 and 10 μL of 25 mM ABTS blended right before adding) was added into the mixture. The total volume of the sensing solution was 500 μL; 200 μL of it was taken into a 96-well microplate for UV/vis assay at 30 °C. Colour changes were observed by the naked eye, the visual readout was quantitatively monitored by measuring spectral absorbance, and spectral changes were observed by the UV/vis spectrophotometer. The absorbance values at 733 nm (ABTS+) of sample (A) and the blank sample (A0) were recorded to demonstrate the situation of AuNPs catalysing the oxidation of ABTS. The value of absorbance variations (ΔA = A0 − A) was calculated to evaluate the ability of the proposed aptasensor in colorimetric detection of STR.Sensitivity and selectivity of the colorimetric aptasensor
Under optimal sensing conditions, a varying amount of STR with final concentrations ranging from 0.1 μM to 1.2 μM were added into the proposed aptasensor to determine LOD following STR determination. To investigate selectivity of the colorimetric aptasensor, some antibacterial agents widely used in animal husbandry including amoxicillin, tetracycline, oxytetracycline, carbamazepine, diclofenac and penicillin were added to the sensing solution and the absorbance values at 733 nm were recorded.Analysis of STR in spiked actual sample
Considering the actual application of antibiotics, milk was chosen as the real sample for analysis. Milk was bought from local supermarkets and stored at 4 °C before use. STR-spiked real sample solutions were prepared by adding STR to the five times diluted milk samples, and the final concentrations of STR were 0.4, 0.6, 0.7, and 1 μM.21,28,29 To remove interferences, EDTA and trifluoracetic acid were added and the samples were defatted and deproteinized by centrifugation at 6000 rpm for 30 min at 4 °C and the supernatant was collected for detection. The detection of STR in milk samples was validated by the designed aptasensor as described above.Results and discussion
Sensing mechanism of the colorimetric aptasensor
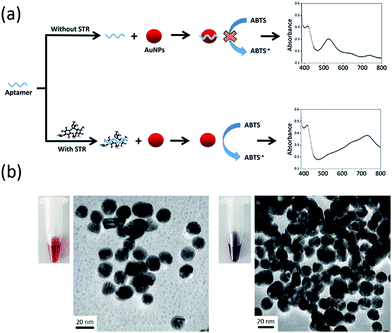
The presented colorimetric aptasensor is based on intrinsic peroxidase-like activity of AuNPs and their interaction with the STR-aptamer complex. AuNPs can catalyse oxidation of peroxidase substrate (ABTS) by hydrogen peroxide, thus producing the ABTS radical cation (ABTS+) with green colour. ABTS radical cation (ABTS+) displays a characteristic absorption peak at 733 nm. Hence, the detection results can also be demonstrated by measuring UV/vis absorption spectrum.Fig. 1a displays the concept of the catalysis-based colorimetric aptasensor. In the absence of STR, STR1 aptamer absorbed on the surface of AuNPs and increased their negative charge density, which prevented the nanoparticles from interacting with negatively charged ABTS and resulted in a reduction of catalytic abilities of AuNPs. Thus, oxidation of ABTS–H2O2 could not be catalysed and there was no colour change in the sensing solution. In the presence of STR molecules, STR1 aptamers first specifically bound with STR molecules to form the STR-aptamer complex; thus, the aptamer sequences no longer absorbed on the surface of AuNPs and their catalytic activity was further enhanced by the STR-aptamer complex. Consequently, AuNPs could oxidize substrates to generate green products (ABTS radical cations), and then the evident absorption peak at 733 nm could be observed. To reveal the catalytic property of nanoparticles in STR detection, TEM was used to characterize their morphology (Fig. 1b). In the absence of STR, the aptamer sequences absorbed on the nanoparticles and AuNPs dispersed evenly, exhibiting a red colour. In the presence of STR, the target molecules first bound with STR1 aptamers to form the STR-aptamer complex and further aggregated AuNPs to some extent; thus, the whole solution displayed a violet-blue colour.
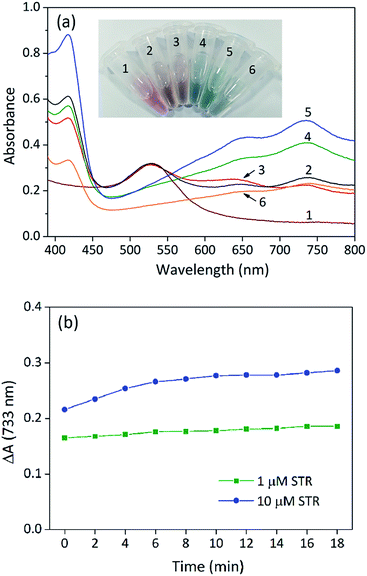
To confirm the feasibility of our sensing strategy, the spectral responses of the proposed colorimetric aptasensor were observed under different conditions (Fig. 2a). AuNPs alone had an evident absorption peak at 530 nm and displayed a red colour (sample 1), suggesting that the nanoparticles were in a well dispersed state. After the addition of substrates (ABTS and H2O2), the absorption value at 530 nm remained almost unchanged, while an absorption peak appeared at 733 nm (sample 2), which made the solution seem orange. These results demonstrated that gold nanoparticles were well dispersed and exhibited peroxidase-mimicking catalytic activity. When STR1 aptamer sequences absorbed on the surface of AuNPs (sample 3), the absorption peak at 530 nm was almost coincident with that of sample 1 and sample 2, indicating that most nanoparticles were dispersed. However, the absorbance value at 733 nm was slightly lower than that of sample 2, which testified that the catalytic ability of AuNPs was indeed inhibited by STR1 aptamer as mentioned above. When AuNPs were added into the incubation solution of STR molecules and STR1 aptamers, the absorption peaks at 530 nm disappeared, while the absorbance values at 650 nm and 733 nm increased remarkably (sample 4–5), and the absorbance variations were proportional to STR amounts, which makes it possible to detect STR by our colorimetric aptasensor. Under such conditions, the colour of sensing solutions changed from initial red to greyish green (sample 4) or green (sample 5). The probable reason for this colour change is that STR molecules could aggregate AuNPs (right photos in Fig. 1b) and AuNPs catalytic activity was enhanced greatly in the presence of STR molecules and STR1 aptamers. To find the actual reason for enhanced catalytic activity of AuNPs, STR molecules were added into AuNPs solution as the peroxidase to catalytically oxidize substrates and the absorption spectrum was recorded (sample 6). STR molecules could aggregate AuNPs, so the absorption peaks at 530 nm disappeared and the whole solution displayed a colour of light greyish green. It is noteworthy that the absorbance value at 733 nm was lower than that of sample 2, which indicated that the aggregation indeed reduced peroxidase catalytic activity of AuNPs. In the case of sample 4–5, the enhanced catalytic activity of AuNPs was probably ascribed to the STR-aptamer complex. We speculated the possible reason for catalytic enhancement is that the STR-aptamer complex changed the surface properties of the nanoparticles and could enhance the affinity of AuNPs toward substrates. Neither STR1 aptamer sequences nor STR molecules could oxidize substrates to produce a green product (Fig. S1, ESI†). We further measured the sensing signals in response to 1 μM and 10 μM of STR under different detection times. As shown in Fig. 2b, the absorbance variations (ΔA) of colorimetric aptasensor tended to be stable after 10 min. Therefore, the absorbance value at 10 min was assumed for subsequent experiments.
Optimization of sensing conditions
To achieve better performance of the proposed colorimetric aptasensor, some experimental parameters including concentration of aptamer and AuNPs, amount of hydrogen peroxide and ABTS, temperature for incubation and reaction, and buffer conditions were investigated.Effect of buffer
Like HRP and other peroxidase mimetics, the catalytic activity of AuNPs is strongly dependent on pH.30 The pH of the reaction medium is a critical factor in ABTS oxidation and a weak acidic condition is more favoured.31,32 Consequently, the effect of solution pH was optimized by using acetate buffer with a pH ranging from 3.0 to 5.0 and a fixed STR concentration at 1 μM. It is revealed in Fig. S2a (ESI†) that AuNPs exhibited higher catalytic activity under slightly acidic conditions, which was also observed for V2O5 nanowires and HRP with ABTS as the substrate.33 The highest colorimetric signal was obtained at pH 4.5, so it was chosen for further study.To test the effect of ionic strength on the sensing system, varying amounts of NaAc were employed to construct the colorimetric aptasensor. Fig. S2b (ESI†) describes the trend of absorbance variation (ΔA) with increasing NaAc concentrations. The sensing signal (ΔA) of the aptasensor increased with increasing concentration of salt until 5 mM, after which it decreased sharply. The likely reason is that a higher amount of salt would aggregate AuNPs due to electrostatic screening effect, so the catalytic activity of gold nanoparticles was reduced and the signal of our aptasensor became weak. Therefore, the optimal concentration of NaAc of 5 mM was for the aptasensor.
Effect of substrate concentration
Hydrogen peroxide (H2O2) plays a crucial role in both catalysis and colour change of the aptasensor. Therefore, the effect of hydrogen peroxide concentration on STR detection was studied. At lower concentrations of hydrogen peroxide, ΔA increased sharply (Fig. S3a, ESI†). With further increase of hydrogen peroxide concentration, the growth of absorbance variations (ΔA) slowed down and reached a maximum at 80 mM of H2O2. Hence, the concentration of H2O2 was selected to be 80 mM for further research. The overall trend is similar to that of Fe3O4 peroxidase mimetics.34ABTS is a chromogenic agent of the reaction and its concentration strongly influences colour development. The current sensing signal depends on the absorption value of ABTS radical cation at 733 nm, so the concentration of ABTS may somehow affect the performances of the aptasensor. Herein, the effect of ABTS concentration on the STR determination was investigated. Fig. S3b (ESI†) reflects a common tendency that absorbance variations (ΔA) enhanced gradually with increasing ABTS concentration (from 20 μM to 500 μM) at lower levels until ΔA reached a maximum. With further increasing of ABTS concentration from 500 μM to 1000 μM, the absorbance variations (ΔA) declined rapidly, suggesting that one-electron oxidation of ABTS occurs when the amount of ABTS is lower than a certain value.35 When the concentration of ABTS was greater than 1000 μM, the absorbance variations (ΔA) did not change, indicating that ABTS was saturated for AuNPs catalysis. Thus, 500 μM of ABTS was employed to ensure high sensitivity for STR detection.
Effect of AuNPs
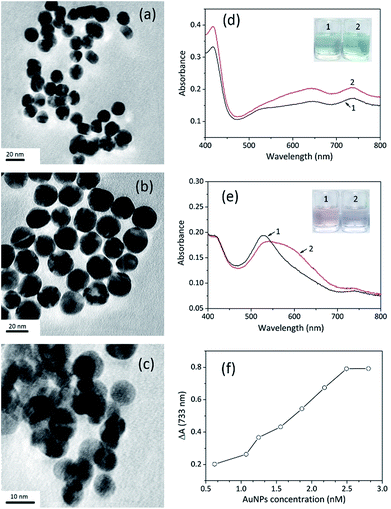
The catalytic activity of AuNPs is strongly related to the size, morphology and surface properties of the nanoparticles, and spherical AuNPs exhibit higher peroxidase activity. Herein, some spherical AuNPs with varied sizes and surface properties were prepared to investigate their influence on detection (Fig. 3a–c). They were compared as catalysts using ABTS oxidation as the reference. The comparative tests were conducted following the procedure of STR determination using the same volume of AuNPs and other sensing conditions were controlled to be consistent. The zeta potential of unmodified AuNPs (umAuNPs), citrate-coated AuNPs (CC-AuNPs) and cysteamine-stabilized AuNPs (CS-AuNPs) was measured to be −10.38 mV, −3.78 mV and 11.20 mV, respectively.When STR was added, the absorbance value of the CC-AuNP aptasensor increased significantly at 733 nm, which demonstrated that the catalytic activity of the AuNPs was greatly enhanced. These results are consistent with above sensing strategy, suggesting that CC-AuNPs are an appropriate catalyst to fabricate the proposed colorimetric aptasensor. According to the absorption spectra displayed in Fig. 3d, the positively charged CS-AuNPs aptasensor showed grey-green colour and had weak absorption peaks at 650 nm and 733 nm in the presence of STR1 aptamer, which may be ascribed to some aggregation of nanoparticles caused by negatively charged ssDNAs and the oxidation of ABTS. After adding 1 μM of STR into the CS-AuNP aptasensor, the absorbance values at 650 nm and 733 nm had increased to some extent, while the colour of sensing solutions had changed to light green. However, considering that the increase in absorbance intensity at 733 nm was negligible and the colour difference was indistinct, CS-AuNPs are not suitable for the construction of the colorimetric aptasensor. As shown in Fig. 3e, the aptamer absorbed on the umAuNPs, exhibiting a pink colour with a clear absorption peak at 530 nm, suggesting the AuNPs were well dispersed. An extremely weak absorption peak at 733 nm was also observed, but the absorbance value was much lower than that of CC-AuNPs. This phenomenon indicates that umAuNPs exhibited weaker catalytic activity compared to CC-AuNPs. The probable reason may be that the electron density of umAuNPs was higher (−10.38 mV), which was unfavourable for oxidation of negatively charged ABTS. The absorbance value at 530 nm decreased to a certain extent after the addition of 1 μM STR, and the sensing solution became colourless owing to some aggregation of nanoparticles. Meanwhile, the absorbance value at 733 nm increased slightly, which might be ascribed to the slight enhancement of umAuNPs' catalytic ability. Therefore, umAuNPs are not an ideal material for the fabrication of the proposed colorimetric aptasensor.
CC-AuNPs provided the highest sensing signal, so they were selected to construct the aptasensor in the subsequent experiments. The amount of CC-AuNPs had a major influence on STR detection and the concentration of the nanoparticles from 0.623 nM to 2.804 nM was investigated. The absorbance variations gradually increased with increasing concentration of CC-AuNPs. When the concentration was greater than 2.804 nM, the absorbance variations (ΔA) stopped increasing and maintained a relatively stable level. Low concentration of gold nanoparticles was insufficient to catalyse ABTS oxidation. When the concentration of nanoparticles reached a certain level, ABTS would be thoroughly oxidized. Therefore, the concentration of CC-AuNPs was chosen as 2.804 nM in subsequent experiments.
Effect of aptamer concentration
STR1 aptamer sequences could occupy the surface of the AuNPs and inhibit their catalytic ability, so their concentration was crucial for the detection of STR. Fig. S4 (ESI†) shows the effects of aptamer concentration on sensing signal. The lower concentration of STR1 aptamer was inadequate to inhibit the catalytic ability of AuNPs, resulting in higher background signals. Once aptamer concentration was over 50 nM, almost all nanoparticles were covered with aptamers. Therefore, the target of STR was insufficient to remove excess aptamer sequences, leading to the slow decrease in sensing signals. Thus, the amount of STR1 aptamer used was 50 nM in the subsequent experiments to achieve higher sensitivity.Effect of temperature
The effect of incubation temperature and catalytic reaction temperature was studied. It can be seen from Fig. S5a (ESI†) that the sensing signal first increased and then reduced with an increase in incubation temperature from 20 to 50 °C. The maximum absorbance value was obtained at 30 °C, so it was chosen as the incubation temperature for this work. In contrast to incubation temperature, the catalytic reaction temperature had a minor influence on sensing signal. As shown in Fig. S5b (ESI†), the absorbance variations increased slowly as the catalytic reaction temperature increased, which was in accordance with the trend observed for H2O2-mediated ABTS oxidation catalysed by Ag+ ion, Prussian blue nanoparticles and Fe3O4 nanoparticles.36,37 Considering the practical application of the aptasensor, 30 °C was chosen to be the catalytic reaction temperature.Sensitivity of the aptasensor
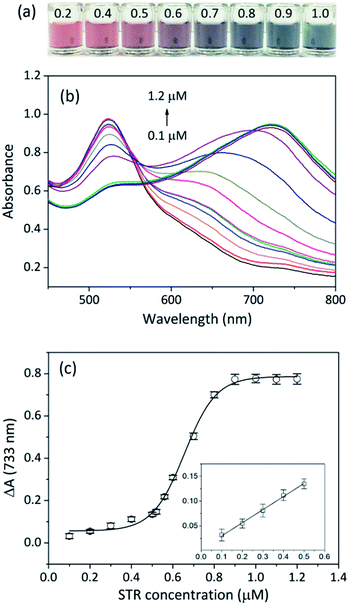
To evaluate the sensitivity of the aptasensor, a range of STR concentrations from 0.1 μM to 1.2 μM were added to the sensing solution, and absorbance variations (ΔA) at 733 nm were recorded. With increasing STR amounts, the colour of the sensing solution changed from red to grey then to deep green, suggestive of STR concentration-dependent catalysis (Fig. 4a). Interestingly, we could identify the solution colour change by the naked eye when the concentration of STR was higher than 0.6 μM. The absorption spectra of the sensing solutions (Fig. 4b) showed that the absorbance value at 733 nm increased with increasing concentration of STR, indicating gradually increasing catalysis. To quantify STR, the absorbance variation (ΔA) of the sensing solution was plotted; the calibration curve appeared like a sigmoid (Fig. 4c), which was fitted to the Hill plot with a correlation coefficient of 0.9942. Notably, the absorbance variation at 733 nm barely ever changed when STR concentration exceeded 0.9 μM, which might be because the binding interaction between the STR-aptamer complex and CC-AuNPs reached a saturation point. The absorbance variation at low STR concentration was fitted to linear equation (inset of Fig. 4c), y = 0.0036 + 0.2640x. Based on our previous reports, 3σ/slope was employed to determine the limit of detection (LOD) of this work to be as low as 86 nM, which is much lower than the mandatory standard set by European Commission and China Ministry of Agriculture (200 μg kg−1 in milk).38,39Traditional methods can detect STR sensitively, but most of them require sophisticated instruments and professional maintenance. An aptasensor is portable and inexpensive and can be easily applied for on-site and real-time detection. Up to now, aptasensors for STR detection can be divided into three major categories according to the signal output mode, and their principles and performances are summarized in Table S1 (ESI†). Although electrochemical aptasensors exhibit excellent sensitivity, they usually require tedious electrode modification and are susceptible to surrounding conditions.16–18,40,41 Moreover, some sophisticated instruments are mandatory for the measurement of electrochemical signals. Fluorescent aptasensors often need modification of aptamer sequences with organic dyes or heavy metals as fluorescence sources, which may be harmful to humans and the environment.42,43 Colorimetry can provide visual results and usually does not require expensive instruments, which makes it an ideal signal output mode for the construction of aptasensors. So far, the aggregation of AuNPs relied on the binding of targets and aptamers have been widely used in the fabrication of colorimetric aptasensors.15,44 Our colorimetric aptasensor is based on the enhancement of peroxidase catalytic activity of AuNPs, so it can avoid unexpected AuNP aggregation caused by non-specific targets. Besides, the current aptasensor only contains AuNPs and aptamers, and such materials do not require any modification.
Selectivity of the aptasensor
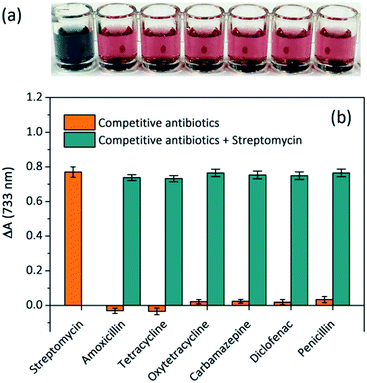
To investigate the specificity of the proposed colorimetric aptasensor, potentially interfering competitive antibiotics such as tetracycline, oxytetracycline, carbamazepine, penicillin, amoxicillin and diclofenac were individually added to the sensing solutions. The corresponding visual images of each sample are shown in Fig. 5a, and the solution containing STR showed a greyish green colour, while other solutions appeared red. As shown in Fig. 5b, the absorbance variations of sensing solutions with 1 μM of other competitive antibiotics were far lower than that with an equal amount of STR, and their values increased sharply when appending STR at the same time. These results clearly indicate that other competitive antibiotics display negligible interferences in the proposed aptasensor. Owing to the strong affinity between STR molecules and STR1 aptamers, the proposed aptasensor exhibits excellent selectivity.Detection of STR in spiked real samples
Considering the widespread use of antibiotics in animal husbandry, milk was selected as the real sample for evaluation. The applications of the present method were evaluated for STR determination in several spiked real samples, and the results are summarized in Table 1. It could be found that the spiked recoveries for STR by the proposed aptasensor were in the range of 90.8 to 98.9%, and the relative standard deviation (RSD) was calculated to be 1.4 to 5.6%. These results indicate the precision and reproducibility of the aptasensor, demonstrating that it has promising feasibility for rapid and sensitive STR detection in real samples.Conclusions
In conclusion, a simple and rapid colorimetric aptasensor has been successfully developed for the sensitive and selective detection of STR using enhanced peroxidase-mimicking catalytic activity of AuNPs. Upon addition of STR, the absorption value of the sensing solution at 733 nm increases, and its variation is directly associated with STR concentration. This assay not only gives a rapid visual readout with high selectivity against other competitive antibiotics, but also allows highly sensitive quantitative detection of STR with LOD of 86 nM. With recoveries ranging from 90.8 to 98.9%, the proposed aptasensor can be well applied in real sample detections. In general, the proposed colorimetric aptasensor exhibits two major advantages. First, the current work is simple and rapid. It is easy to fabricate and can favour large-scale production and application. Second, the accuracy of the aptasensor is guaranteed to some extent and the results can even be distinguished by the naked eye, which can be potentially beneficial to the detection of antibiotic residues in food products. This advantage comes from that the current aptasensor is based on the enhanced peroxidase-mimicking catalytic activity of AuNPs and can avoid inaccurate results caused by non-specific aggregation. Furthermore, the proposed sensing strategy may offer a general approach to detect a range of small molecules by employing their specific aptamers.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (21565009 and 21205020), the Natural Science Foundation of Guizhou Province ([2016]1403), the Cooperation Project of Guizhou Province Science and Technology Department (LH [2015]7682), the Natural Science Project of Guizhou Provincial Education Department (KY [2014]261), the Talent Research Project of Guizhou University ([2014]09), and the Innovation Fund of Guizhou University for Postgraduate (2017024).Notes and references
- M. Lan, W. Liu, J. Ge, J. Wu, J. Sun, W. Zhang and P. Wang, Spectrochim. Acta, Part A, 2015, 136, 871–874 CrossRef CAS PubMed.
- R. C. de Oliveira, J. A. Paschoal, M. Sismotto, F. P. Airoldi and F. G. Reyes, J. Chromatogr. Sci., 2009, 47, 756–761 Search PubMed.
- P. Vinas, N. Balsalobre and M. Hernández-Córdoba, Talanta, 2007, 72, 808–812 CrossRef CAS PubMed.
- R. H. Granja, A. M. M. Niño, R. A. Zucchetti, R. E. M. Niño, R. Patel and A. G. Salerno, Anal. Chim. Acta, 2009, 637, 64–67 CrossRef CAS PubMed.
- M. van Bruijnsvoort, S. J. Ottink, K. M. Jonker and E. de Boer, J. Chromatogr. A, 2004, 1058, 137–142 CrossRef CAS PubMed.
- Y. Qin, L. Bao and L. Zhu, J. South China Agric. Univ., 2002, 24, 88–91 Search PubMed.
- B. G. Knecht, A. Strasser, R. Dietrich, E. Märtlbauer, R. Niessner and M. G. Weller, Anal. Chem., 2004, 76, 646–654 CrossRef CAS PubMed.
- N. Pastor-Navarro, Á. Maquieira and R. Puchades, Anal. Bioanal. Chem., 2009, 395, 907–920 CrossRef CAS PubMed.
- R. Stoltenburg, C. Reinemann and B. Strehlitz, Biomol. Eng., 2007, 24, 381–403 CrossRef CAS PubMed.
- K.-M. Song, M. Cho, H. Jo, K. Min, S. H. Jeon, T. Kim, M. S. Han, J. K. Ku and C. Ban, Anal. Biochem., 2011, 415, 175–181 CrossRef CAS PubMed.
- Z. Chen, Y. Tan, C. Zhang, L. Yin, H. Ma, N. Ye, H. Qiang and Y. Lin, Biosens. Bioelectron., 2014, 56, 46–50 CrossRef CAS PubMed.
- C. Tuerk and L. Gold, Science, 1990, 249, 505–510 CAS.
- K. Zhang, K. Sefah, L. Tang, Z. Zhao, G. Zhu, M. Ye, W. Sun, S. Goodison and W. Tan, ChemMedChem, 2012, 7, 79–84 CrossRef CAS PubMed.
- P. Luo, Y. Liu, Y. Xia, H. Xu and G. Xie, Biosens. Bioelectron., 2014, 54, 217–221 CrossRef CAS PubMed.
- N. Zhou, J. Wang, J. Zhang, C. Li, Y. Tian and J. Wang, Talanta, 2013, 108, 109–116 CrossRef CAS PubMed.
- J. Xue, J. Liu, C. Wang, Y. Tian and N. Zhou, Anal. Methods, 2016, 8, 1981–1988 RSC.
- N. Mohammad Danesh, M. Ramezani, A. Sarreshtehdar Emrani, K. Abnous and S. M. Taghdisi, Biosens. Bioelectron., 2016, 75, 123–128 CrossRef CAS PubMed.
- J. Yin, W. Guo, X. Qin, J. Zhao, M. Pei and F. Ding, Sens. Actuators, B, 2017, 241, 151–159 CrossRef CAS.
- Y. S. Kwon, N. H. A. Raston and M. B. Gu, Chem. Commun., 2014, 50, 40–42 RSC.
- L. Li, B. Li, Y. Qi and Y. Jin, Anal. Bioanal. Chem., 2009, 393, 2051–2057 CrossRef CAS PubMed.
- Y. Luo, J. Xu, Y. Li, H. Gao, J. Guo, F. Shen and C. Sun, Food Control, 2015, 54, 7–15 CrossRef CAS.
- Y. Jv, B. Li and R. Cao, Chem. Commun., 2010, 46, 8017–8019 RSC.
- H. Wei and E. Wang, Chem. Soc. Rev., 2013, 42, 6060–6093 RSC.
- T. K. Sharma, R. Ramanathan, P. Weerathunge, M. Mohammadtaheri, H. K. Daima, R. Shukla and V. Bansal, Chem. Commun., 2014, 50, 15856–15859 RSC.
- G. Mayer, Nucleic acid and peptide aptamers: methods and protocols, Humana Press, New York, 2009 Search PubMed.
- J. Zheng, H. Zhang, J. Qu, Q. Zhu and X. Chen, Anal. Methods, 2013, 5, 917–924 RSC.
- H.-H. Deng, G.-W. Li, L. Hong, A.-L. Liu, W. Chen, X.-H. Lin and X.-H. Xia, Food Chem., 2014, 147, 257–261 CrossRef CAS PubMed.
- M. Ramezani, D. N. Mohammad, P. Lavaee, K. Abnous and T. S. Mohammad, Biosens. Bioelectron., 2015, 70, 181 CrossRef CAS PubMed.
- L. He, Y. Luo, W. Zhi and P. Zhou, Food Anal. Methods, 2013, 6, 1704–1711 CrossRef.
- W. Zhang, D. Ma and J. Du, Talanta, 2014, 120, 362–367 CrossRef CAS PubMed.
- X.-H. Zhou, D.-M. Kong and H.-X. Shen, Anal. Chem., 2009, 82, 789–793 CrossRef PubMed.
- Y. Li, X. Liu and R. Zhang, Spectrochim. Acta, Part A, 2017, 173, 880–885 CrossRef CAS PubMed.
- R. André, F. Natálio, M. Humanes, J. Leppin, K. Heinze, R. Wever, H. C. Schröder, W. E. Müller and W. Tremel, Adv. Funct. Mater., 2011, 21, 501–509 CrossRef.
- L. Gao, J. Zhuang, L. Nie, J. Zhang, Y. Zhang, N. Gu, T. Wang, J. Feng, D. Yang and S. Perrett, Nat. Nanotechnol., 2007, 2, 577–583 CrossRef CAS PubMed.
- M. R. Clausen, L. H. Skibsted and J. Stagsted, J. Agric. Food Chem., 2008, 56, 8692–8698 CrossRef CAS PubMed.
- X. H. Zhou, D. M. Kong and H. X. Shen, Anal. Chem., 2010, 82, 789–793 CrossRef CAS PubMed.
- Y. Ma, Z. Zhang, C. Ren, G. Liu and X. Chen, Analyst, 2012, 137, 485–489 RSC.
- Y. Wu, S. Zhan, L. Xu, W. Shi, T. Xi, X. Zhan and P. Zhou, Chem. Commun., 2011, 47, 6027–6029 RSC.
- Y. Wu, S. Zhan, F. Wang, L. He, W. Zhi and P. Zhou, Chem. Commun., 2012, 48, 4459–4461 RSC.
- Y. Yin, X. Qin, Q. Wang and Y. Yin, RSC Adv., 2016, 6, 39401–39408 RSC.
- J. Yin, W. Guo, X. Qin, M. Pei, L. Wang and F. Ding, New J. Chem., 2016, 40, 9711–9718 RSC.
- A. S. Emrani, N. M. Danesh, P. Lavaee, M. Ramezani, K. Abnous and S. M. Taghdisi, Food Chem., 2016, 190, 115–121 CrossRef CAS PubMed.
- S. M. Taghdisi, N. M. Danesh, M. A. Nameghi, M. Ramezani and K. Abnous, Food Chem., 2016, 203, 145–149 CrossRef CAS PubMed.
- V. Soheili, S. M. Taghdisi, M. H. Khayyat, B. S. F. Bazzaz, M. Ramezani and K. Abnous, Microchim. Acta, 2016, 183, 1687–1697 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Supplementary Table S1 and Fig. S1–S5. See DOI: 10.1039/c7ra06434a |
| This journal is © The Royal Society of Chemistry 2017 |