Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Genetically encoded conductive protein nanofibers secreted by engineered cells†
Ebuzer Kalyoncu,
Recep E. Ahan,
Tolga T. Olmez and
Urartu Ozgur Safak Seker *
*
UNAM – National Nanotechnology Research Center and Institute of Materials Science and Nanotechnology, Bilkent University, 06800 Bilkent, Ankara, Turkey. E-mail: urartu@bilkent.edu.tr
First published on 26th June 2017
Abstract
Bacterial biofilms are promising tools for functional applications as bionanomaterials. They are synthesized by well-defined machinery, readily form fiber networks covering large areas, and can be engineered for different functionalities. In this work, bacterial biofilms have been engineered for use as conductive biopolymers to interface with electrodes and connect bacterial populations to electronic gadgets. Bacterial biofilms are designed with different conductive peptide motifs, as the aromatic amino acid content of fused peptide motifs has been suggested to contribute to electronic conductivity by influencing monomer stacking behavior. To select the best candidates for constructing conductive peptide motifs, conductivity properties of aromatic amino acids are measured using two different fiber scaffolds, an amyloid-like fiber (ALF) forming peptide, and the amyloidogenic R5T peptide of CsgA protein. Three repeats of aromatic amino acids are added to fiber-forming peptide sequences to produce delocalized π clouds similar to those observed in conductive polymers. Based on the measurements, tyrosine and tryptophan residues provide the highest conductivity. Therefore, the non-conductive E. coli biofilm is switched into a conductive form by genetically inserted conductive peptide motifs containing different combinations of tyrosine and tryptophan. Finally, synthetic biofilm biogenesis is achieved with conductive peptide motifs using controlled biofilm production. Conductive biofilms on living cells are formed for bioelectronics and biosensing applications.
Introduction
Long range electron transfer has been a major topic of interest in peptide and protein research for decades.1–4 Investigations of electron transfer in biologically important molecules such as photolyases,5 cryptochoromes,6 phototropins7 and ribonucleotide reductase8,9 have demonstrated that aromatic amino acids such as tyrosine and tryptophan are vital for catalytic activity in many enzymes.10–13 Amino acid composition and structure of the peptides contribute to electronic conductivity,14 and many studies have focused on electron transfer between aromatic amino acids using peptide motifs. Conjugated molecules with low electron tunneling barriers typically exhibit superior conductivity, and peptide and protein based conductive materials are promising for several applications.15–18 In nature, electron transfer is necessary for the circulation of energy in the biosphere. Geobacter sulfurreducens and Shewanella oneidensis have conductive nanowire structures to transfer electrons to oxidized metals as final electron acceptors during anaerobic respiration.19,20 Conductivity in biology utilizes many of the design principles that are found in conductive polymers.21 For example, polyacetylene is a conjugated polymer with free mobile π electrons, the doping of which results in strong conductivity. Similarly, the C-terminal of the PilA protein takes part in the biofilm formation of G. sulfurreducens and has aromatic clusters playing important role in conductivity.22 It has been shown that, when aromatic amino acids are replaced by alanine (which lack functional side groups) in the PilA nanowire, G. sulfurreducens becomes unable to transfer electrons and to reduce oxidized metals;23 moreover, recent studies on genetic engineering of the PilA protein increased the conductivity of PilA nanofibers through the substitution of amino acids with tryptophan residues.24 This demonstrated the importance of the aromatic amino acids in conductivity and highlighted their potential to be incorporated in biofilm-forming protein structures to transfer electrons to electrodes.25–27 Due to the difficulties on the growth of G. sulfurreducens, which needs an electron acceptor under anaerobic conditions for growth28,29 and can only survive at oxygen concentrations below 10% in the environment,30 the development of novel interface systems utilizing well-documented model organisms would contribute greatly to the potential of bioconductive materials in device applications. In particular, interfacing cells with electrodes can provide many opportunities in microbial fuel cell applications, biosensors, electrogenetic applications, light-harvesting applications, and synthetic biology applications.31Escherichia coli is commonly used as the go-to organism in synthetic biology applications, as it expresses a natural fiber system (the curli group of proteins) that is capable of displaying specific protein sequences.32,33 In recent reports, E. coli was engineered to carry electrons from its electron pool to the cell membrane by the use of a pathway electron conduit borrowed from S. oneidensis.34,35 This non-native pathway was domesticated and coupled with a native E. coli cytochrome, NapC.34 Studies revealed the possibility of pumping out electrons from E. coli cytosol to the external environment.34 However, to enable such direct electron transfer with electrode surfaces, there is a need to interface cells with synthetic electrical functionalities with electrode surfaces. In addition to cell–cell communications in bacteria (i.e. quorum sensing),36 recent studies have shown that bacteria in biofilms can communicate using electrical signals. The presence of ion channel-mediated electrical communication was subsequently demonstrated in bacteria, and changes in bacterial membrane potential were found to alter their metabolism during these communication events.37 Given these recent developments, the design of a conductive interface would also be beneficial to control charge transfer into bacterial biofilm communities, potentially controlling the behavior of the entire bacterial population. In addition, the secretion of such an interfacing material from living cells will greatly simplify the design of biofilm-interfacing devices by eliminating additional surface modification steps that are necessary to assemble the bacteria on electrode surfaces.
Nanofibers exhibiting a broad range of biological functions have been synthesized using new nanoscale assembly methods.38 One particular technique entails the production of nanofibers through high speed gyration and the subsequent modification of the nanofiber surface by genetically functionalized fusion proteins, which allows the mass production of complex nanofiber structures. The secretion of nanofibers or their precursors directly from genetically engineered bacteria is another method for the mass production of functional fibers. Biofilm protein fibers are very promising interfacing materials between living cells and abiotic entities such as electrodes and surfaces.39 Many bacteria can form biofilms in response to different stimuli, including starvation, toxic substances, extracellular materials, social competition or other stress conditions.40–43 E. coli forms biofilms using a curli operon that is formed by csgBA and csgDEFG operons. csgBA codes for CsgA and CsgB proteins, which form the backbone of the biofilm structures, whereas csgDEFG codes the accessory proteins for biofilm production.44 E. coli has two structural curli proteins: the major subunit CsgA and minor subunit CsgB.45 Among these proteins, CsgB, serves as a nucleation site for CsgA protein to attach and form the nanofiber network.46,47 We aimed to engineer the CsgA protein for functional peptide fusion. Intermolecular interactions between curli subunits allow them to form large hierarchical networks, which makes them promising candidates as bionanomaterials.48 Repeating units of CsgA proteins form intermediate-length curli fibers49 and can be modified with various peptide tags for the surface expression of inorganic materials.50 Previously, CsgA was fused with various peptide motifs and the functionality of these peptides was demonstrated.51 Curli system also enables the cell populations to strongly adhere on surfaces, enabling a robust contact interface.49,52
Results and discussion
In this study, given the previous findings on the role of aromatic amino acids in conductivity,1,23,24,53–55 we first tested the electrical conductivity of four aromatic amino acids; tyrosine, typtophan, phenylalanine and histidine, on two different fiber-forming scaffold peptides; an amyloid-like fiber (ValGlyGlyLeuGly)56 peptide and the amyloidogenic R5T peptide (SerValAspValThrGlnValGlyPheGly) of CsgA protein,57 to form seven distinct, potentially conductive peptide sequences (Fig. 1 and Table 1). Synthetic peptide scaffolds helped us to screen the conductivity of aromatic amino-acids for best candidates to be used in protein engineering.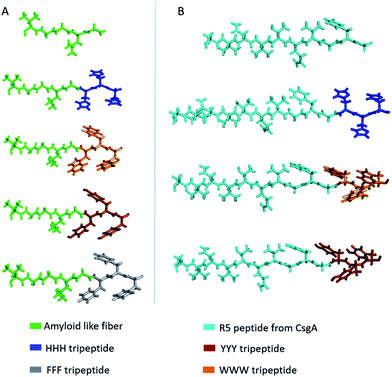 | ||
| Fig. 1 Molecular models of designed synthetic peptides. (A) Amyloid-like fiber templated peptides. (B) R5T peptide templated peptides. | ||
| Groups | Peptide | Sequence | Gravy indexc |
|---|---|---|---|
| a Amyloid like fiber described by del Mercato et al.56b R5 peptide from CsgA protein described by Lembre et al.57c GRAVY: grand average of hydropathicity. | |||
| 1 | ALFa | VGGLG | 1.360 |
| 1 | ALF3H | VGGLGHHH | −0.350 |
| 3 | ALF3W | VGGLGWWW | 0.512 |
| 2 | ALF3Y | VGGLGYYY | 0.362 |
| 2 | ALF3F | VGGLGFFF | 1.900 |
| 1 | R5Tb | SVNVTQVGFG | 0.610 |
| 1 | R5T3H | SVNVTQVGFGHHH | −0.262 |
| 3 | R5T3W | SVNVTQVGFGWWW | 0.262 |
| 2 | R5T3Y | SVNVTQVGFGYYY | 0.169 |
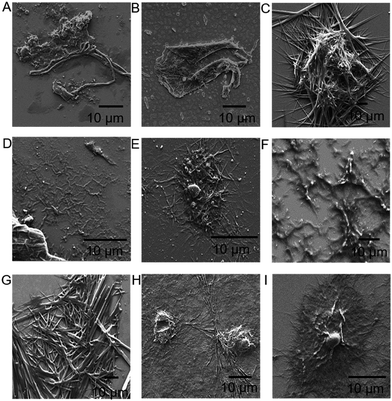
These aromatic amino acids had similar structures to conductive polymers; as such, all peptide constructs were expected to be conductive to some degree. To probe the effect of the addition of different aromatic amino acids on the assembly of the peptide fibers, the morphologies of the peptides were investigated with atomic force microscopy (AFM) (Fig. S1†) and scanning electron microscopy (SEM), as demonstrated in Fig. 2.
Samples for SEM imaging were prepared on silica wafers. The original scaffolds on Fig. 2A and F were observed to form fiber structures, as is expected from previous findings.56,57 The addition of aromatic residues resulted in different morphologies. We did not observe a general trend for morphology changes as a function of the type of the amino acids in different scaffold peptides. It is assumed that fiber-making scaffold peptide sequences, in conjugation with the added amino acids, are controlling the final aggregate/fiber morphology. In all of the cases, addition of the aromatic amino acids to the scaffolding peptides did not preventing the formation of large peptide fibers or aggregates.
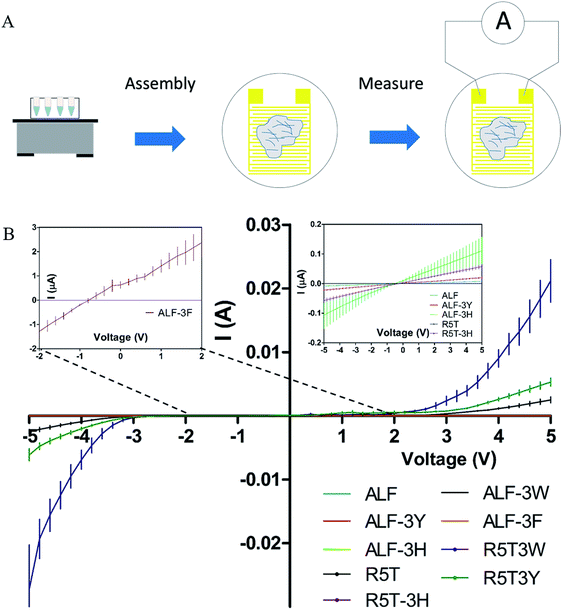
Conductivity of peptide fibrils were investigated using probe station measurements under the configuration shown in Fig. 3A. In order to explore the conductive properties of peptide fibrils, their solutions were deposited on gold electrodes. Electrical conductivities of the peptide samples are provided in Fig. 3A. High-current voltages were obtained for R5T3W, R5T3Y, and ALF3W samples, which also exhibited non-linear I–V characteristics. The rest of the samples showed a linear ohmic behavior in the range of −5 V to +5 V, suggesting that their Schottky barrier I/V may be higher or that they are more insulating in this region (Fig. 3A, insets). It is clear that tryptophan and tyrosine insertions have superior conductivities in comparison to other insertions (Fig. 3B).11,53,58
According to these findings, tyrosine and tryptophan were selected for the functionalization of the curli nanofibers to convert them into conductive fibers. The C-terminal of the PilA protein (Fig. 4A) features a conductive motif composed of 5 aromatic amino acids and 6 charged amino acids, both of which are crucial for fiber conductivity.24,59 In a recent study, the insertion of tryptophan residues in the C-terminal of G. sulfurreducens PilA protein resulted in increased nanofiber conductivity.24 One of the critical points in the engineering of conductive protein fibers is to increase conductivity through the insertion of additional amino acid residues. Consequently, we mimicked the conductive peptide motif of PilA as presented on Fig. 4A, and modified this initial sequence with four different peptide designs shown in Fig. 4B. These peptides were genetically fused with the 3′ end of the csgA gene. Charged amino acids were kept at the same positions in the modified CsgA design for conductive nanofiber formation, while the aromatic amino acids were replaced by tyrosine or tryptophan (Fig. 4B).
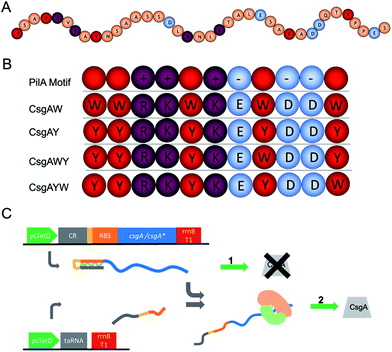 | ||
| Fig. 4 Genetic engineering of curli subunits used to form conductive nanofibers. (A) C-terminal amino acid sequence of G. sulfurreducens PilA protein. Orange balls indicate non-charged, non-aromatic amino acids. (B) Addition of conductivity-enhancing and aromatic amino acids to the E. coli CsgA protein sequence, mimicking the motifs found in G. sulfurreducens PilA protein. The conductive motif is composed of 5 aromatic amino acids (red), 3 positively charged amino acids (purple) and 3 negatively charged amino acids (blue). CsgAW, CsgAY, CsgAWY, and CsgAYW versions of conductive motifs are designed and fused to the C-terminal of csgA. The rationale behind this design is that aromatic amino acids are well-recognized to be important in electron transfer, while recent studies have also demonstrated the importance of charged amino acids for electron transfer in PilA conductivity.2,59 Consequently, we decided to exclude linker amino acids and used a combination of aromatic and charged aminoacids in adjacent position. (C) Genetic elements of the translational riboregulator system used for the strict control of the CsgA and conductive motifs fused to the CsgA monomer. Translational riboregulator (cis-repressor (CR)) is a ribonucleic acid (RNA) that binds to the ribosomal binding site and blocks translation. taRNA is a trans-activator RNA which binds to CR and opens the ribosome binding site to allow the translation of the fusion protein. The system is controlled at the transcription level. | ||
Gene fragments used in the biosynthesis of conductive nanofibers were amplified from the E. coli genome. DNA sequences of the designed conductive motifs were added to the 3′ end of gene fragments by PCR in conjunction with overlap extension. Recombinant genes were cloned into pZa-tetO-CmR using the cut ligate method, vector maps can be seen in the ESI document.† The inducible expression of recombinant proteins was therefore achieved through the pZa-tetO-CmR vector. Expressions of CsgAW, CsgAY, CsgAWY and CsgAYW proteins are under tight regulation by an anhydrotetracycline (aTc) inducer-responsive riboregulator, the working principle of the riboregulator is given in Fig. 4C. The riboregulators were used to prevent any leakage from the uninduced cells, preventing their contribution to the curli network of the biofilm. The riboregulator enables the control of the expression of the target gene at the transcript level.60
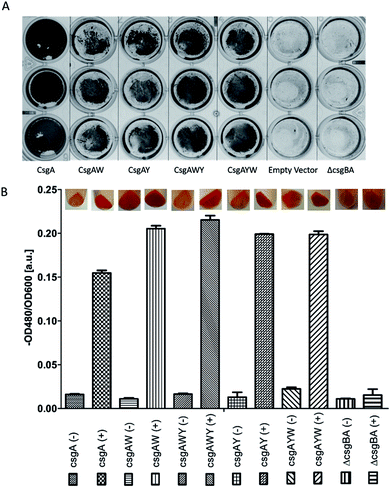
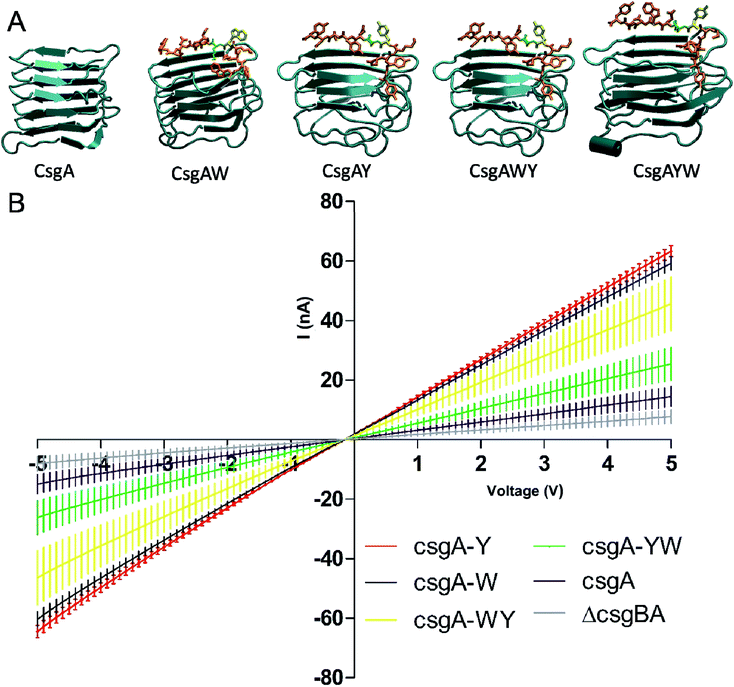
In order to verify that all conductive curli subunits formed thick, adherent biofilms in the presence of aTc, crystal violet biofilm assay was performed in triplicate. Biofilm formation of all the constructs was confirmed by crystal violet staining (Fig. 5A). Designed conductive curli subunits enable bacteria to attach to surfaces and can readily be observed in 24-well plates. In contrast, negative controls (E. coli MG1655 PRO ompR234 ΔcsgBA and empty pZA vector-transformed ΔcsgBA cells) show little attachment on the surface of 24-well plates. The formation of curli fibrils produced by recombinant bacteria were also characterized by Congo red (CR) assay. The absorbance of 25 μg ml−1 CR was subtracted from the absorbance of supernatant at 480 nm and normalized against OD600. The amount of CR-bound cells and curli fibers were calculated as −OD480/OD600 and represented in Fig. 5B. Congo red assay results demonstrated that C-terminal modified CsgA variants retain their ability to form amyloid fibrils and bind ∼1.33-fold more Congo red than unmodified CsgA protein (Fig. 5B). CR staining results also suggest that there might be a slight change in the final protein folding of CsgA, which may have an effect on the structure of the final fiber assemblies.
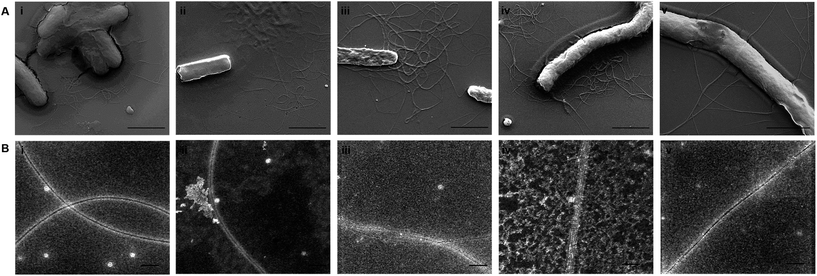
To acquire an idea about the morphology of curli fibers produced by recombinant cells, biofilms were analyzed with scanning electron microscopy (SEM). In Fig. 6A, SEM images reflect the assembly and morphology of conductive fibers around the engineered bacteria on silica surfaces. SEM data showed that conductive fibers were 2–2.5 μm in length and 25–35 nm in thickness. This length is consistent with 5 to 7 individual curli fibers coming together to form fiber bundles through hydrophobic interactions between aromatic amino acids.61
Synthetic conductive curli biofilms were further investigated by probe station analysis. Fig. 7B shows the I/V curve of the engineered biofilms. Conductivity of CsgAW, CsgAY, CsgAWY, CsgAYW and CsgA protein expressing cells, as well as csgBA KO cells were compared for the determination of optimally conducting fiber motifs. Conductivity curves revealed that biofilms exhibit a linear ohmic behavior (Fig. 7B). CsgAW, CsgAY, CsgAWY and CsgAYW motifs exhibited conductivity values about 7.61, 8.13, 5.86, and 3.27 fold higher than E. coli ΔcsgBA cells and 4.08, 4.37, 3.14, and 1.76 fold higher than unmodified CsgA biofilms, respectively (Fig. 7B). The increase in the conductivity of synthetic curli fibers compared to native curli fibers is attributable to the conductive motifs that are fused to CsgA monomers, as the expression systems of native CsgA and synthetic CsgAs are the same. However, differences were observed between the abilities of aromatic amino acids to influence conductive properties in synthetic peptide networks and curli fibers, due to differences in expression systems and self-assembly behavior. Nevertheless, it should be noted that the optimization of aromatic amino acid sequences through peptide systems was the rationale behind the use of tryptophan and tyrosine residues in the initial design of conductive curli fibers. According to a recent study, aromatic amino acid substitution into hepta-alanine peptides increases the conductance of the peptide chain,4 which is also observed in our engineered CsgA proteins. Substitution of tryrosine with tryptophan in CsgWY and CsgAYW slightly decreases the conductivity of the proteins. Conductivity is observed as CsgWY > CsgAYW (Fig. 7B) and their tryptophan positions can be seen in Fig. 4B.
To gain insights into the conductivity of engineered CsgA proteins containing conductive peptide motifs, template-based tertiary structure modelling was performed using RaptorX, a web-based server, to obtain their 3D structure, because homology modelling is not available to model CsgA protein (Fig. 7A).61,62 3D structure of conductive CsgA obtained from RaptorX server is consistent with previous models of CsgA.
According to the predicted structures, a coil motif has been observed between 133th–135th residues, whereas the residues between 136th–141th amino acids were predicted to fold as a beta-sheet structure on top of the last beta-sheet turn of CsgA protein in all engineered CsgA models where glycine residue after the periplasmic targeting sequence of CsgA is taken as first residue. The 136th and 137th residues are assigned as the most disordered structures in the model. Interestingly, the solvent accessibility of lysines on 137th shows similar trend with conductance of engineered CsgA proteins. Due to non-aromatic nature of lysine amino acid side chains, it is not likely that it participates to the electron movement through bundle structure, yet it might strengthen the lateral structure of CsgA fibers.
The conductance of proteinaceous nanowires, such as the pili of G. sulfurreducens, is explained by two different hypotheses, π–π stacking of aromatic amino acids and electron hopping.63,64 π–π stacking requires the tight packing of aromatic amino acids within 3.2 Å distances to establish a metal-like conductivity profile similar to that seen in conductive polymers with delocalized π electrons, such as polyacetlyene.63 While CsgA fibrils were previously proposed to grow in perpendicularly oriented β-strands,61 our TEM results (Fig. 6B) have revealed fiber bundles that are much thicker than single CsgA monomers (∼3–4 nm for single CsgA fibers65 against ∼15–20 nm in TEM images). Consequently, CsgA fibrils appear to interact with each other laterally to form thicker bundles, and even under optimal packing, recombinant CsgA proteins are not expected to establish long-range π–π stacking interactions. In contrast, electron hopping is a more likely scenario due to the presence of tyrosine amino acids, which have oxidizable phenol side chains.66 We therefore suggest that both short-range π–π interactions and electron hopping between stacks may be responsible for electron transfer.
Materials and methods
Peptide assembly into fibers
Peptide monomers of self-assembled fiber networks were synthesized using Fmoc and Boc chemistry. The designed peptides were synthesized (LifeTein, USA) with the addition of homotripeptide containing aromatic amino acids to the C-terminal end of the scaffold peptides in Table 1. Peptide solutions in group 1 were prepared by either dissolving the pellet directly in ddH2O, in group 2 dissolving the peptide pellet in DMSO and diluting the peptide solution in ddH2O at a concentration of 4.0 mg ml−1 (for highly hydrophobic peptides), or in group 3 dissolving the pellet in DMF and diluting in ddH2O (for tryptophan-containing peptides). The self-assembly of the peptides was performed by placing the peptide solution in a vial tube for three weeks at 27 °C for the maturation of amyloid-like fibrils as suggested previous work.56AFM characterization of peptides
20 μl of the peptide solutions were dropped on mica surfaces and left for overnight drying. Samples were characterized in tapping mode using Si probes with a nominal spring constant of 40 N m−1 and a resonance frequency of 300 kHz (TED PELLA INC, USA).SEM characterization of peptides
10 μl of the peptide solutions were deposited on silica surfaces. After evaporation, samples were sputter-coated with 5 nm of Au/Pd alloy using a precision coating system (Gatan Inc, USA).Preparation of gold electrodes
Interdigitated gold electrodes were prepared for the measurement of conductivity. In order to create the electrodes represented in Fig. 3A, we used a custom shadowing mask with 175 μm holes. All preparation was performed in a clean room environment. Thermonax coverslips were placed on glass slides and shadowing masks were placed on top of the coverslips. Shadowing masks were adhered to coverslip surfaces with small thermal sticky tapes. 8 gold electrodes can be prepared per run of thermal evaporator. 0.5 g of gold was used per run and gold electrodes were obtained with a thickness of ∼50 nm.Conductivity measurement of peptides
For the conductivity measurement, 20 μl of mature fibrils were drop-casted on interdigitated electrodes and left for drying in a home-made high-humidity environment. Measurements were obtained from three electrodes for each peptide assembly. Current–voltage (I–V) curves were obtained from −5 to +5 volts under a two-electrode configuration.Clonning of the CsgA conductive peptides
csgAW, csgAY, csgAWY and csgAYW gene fragments listed in ESI Table 1† were amplified from pZa-tetO-csgA-CmR (primers were listed in ESI Table 2†). Recombinant genes were cloned into pZa-tetO-CmR using the cut ligate method using kpnI/mluI restriction sites and the plasmid constructs are represented in Fig. S2, S4, S6, and S8† and sequence alignments of the constructs are shown in Fig. S3, S5, S7, and S9.†Strain and expression of curli proteins
Our inducible synthetic circuits were transformed into an E. coli MG1655 PRO ΔcsgBA ompR234 host strain. This strain has higher expression of curli genes from native curli operon at 30 °C in growth medium.67,68 In order to produce curli proteins, starting cultures were inoculated from a single colony and grown in LB medium at 37 °C for 16 h. For the experimental culture, 20 μl cell volume from overnight cultures were transferred to 2 ml M63 minimal medium supplemented with 1 mM MgSO4 and 0.2% glucose and grown at 30 °C in 24 well plates without shaking for 3 days. For the induction of biofilm formation, aTc was used in a concentration of 200 ng μl−1.Crystal violet assay
E. coli MG1655 PRO ompR234 ΔcsgBA and empty pZA vector transformed ΔcsgBA cells were used as negative control strains. For crystal violet staining, 24 well plates were stained with 400 μl of 0.1% crystal violet (Sigma) and incubated for 10 minutes at room temperature. After staining, wells were washed three times with 1 ml ddH2O. Images were taken using a ChemiDoc MP imaging system (BioRad).Congo red assay
A single colony was inoculated into 10 ml LB including relevant antibiotic and grown at 37 °C for 16 h. Then, 0.5 ml overnight culture were inoculated into 15 ml LB. When the cultures reach OD600 between 0.3 and 0.5, aTc (final concentration of 200 ng μl−1) were added to induce fibril formation for 18 hours. 1 ml of cell culture were used for OD600 cell normalization and 800 μl of cell culture were mixed with 5 × Congo red solution for a final concentration of 25 μg ml−1 and incubated 30 min at room temperature. The cells and curli were centrifuged down at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min and supernatants were taken for quantification of CR. The absorbance of CR in supernatant were measured at 480 nm.
000 rpm for 5 min and supernatants were taken for quantification of CR. The absorbance of CR in supernatant were measured at 480 nm.
SEM characterization of biofilms
For sample preparation, biofilm formation was induced by aTc under static culture conditions. Samples were washed with ddH2O and fixed with 2.5% glutaraldehyde overnight at +4 °C, following by dehydration steps with increasing ethanol concentrations, followed by sputtering 5 nm Au/Pd alloy.TEM characterization of biofilms
Production of the curli biofilms were performed in 24 well plate using minimal medium supplemented with aTc (200 ng μl−1), 1 mM MgSO4 and 0.2% glucose without shaking for 3 days 10 ul of sample were placed on a TEM grid (Ni grid with formvar/C support, 300 mesh) and incubated for 30 seconds. Then the fluid were removed from the sample and three rounds of wash with distilled water were performed on the grid for 30 seconds. Samples were incubated with negative stain dye (3% uranyl acetate) for 30 s and excess fluid was removed from the grid. Samples were analyzed by FEI Tecnai TEM at 200 keV.Conductivity measurement of biofilms
Biofilms were formed in M63 minimal medium without shaking for 3 days following induction with aTc. Biofilms were then washed twice with ddH2O to remove medium salts and drop-casted on interdigitated gold electrodes to measure the conductivity of biofilm.Conclusion
Conductive self-assembled peptide systems are of considerable interest because of the well-recognized ability of the self-assembly process to enhance the conductivity of polymers,69,70 and the promising applications of conductive self-assembled peptides for a range of disciplines.56,57,71 In this work, we report the first demonstration of a curli-based self-assembling system that is secreted directly by E. coli cells and exhibits higher conductivity compared to native biofilms. Sequence optimization of the PilA-inspired peptide molecule revealed that aromatic amino acids were responsible for conductivity, possibly due to π–π stacks and electron hopping. Polytyrosine and polytryptophan motifs were strongly capable of promoting biofilm conductivity; however, mixed tyrosine–tryptophan sequences were less effective for this purpose. This phenomenon may derive from the differential pH responses associated with these amino acids, which may prevent their combination from conducting effectively in the biofilm environment. Nevertheless, monotypic repeats of aromatic amino acids are shown to impart a strong conductivity when incorporated into a CsgA-derived, self-assembling peptide sequence, and may be further optimized for various applications in energy and materials design.Author contributions
All authors have given approval to the final version of the manuscript.Acknowledgements
We are thankful to TUBITAK for financial support (Grant number 114M163). UOSS thanks to TUBA-GEBIP Distinguished Young Scientist Award program. EK and TTO are supported by TUBITAK-BIDEB scholarship program. We thank Professor Hilmi Volkan Demir, Berkay Bozok and Shahab Akhavan for probe station measurements.References
- H. B. Gray and J. R. Winkler, Chem. Phys. Lett., 2009, 483, 1–9 CrossRef CAS PubMed.
- A. Shah, B. Adhikari, S. Martic, A. Munir, S. Shahzad, K. Ahmad and H. B. Kraatz, Chem. Soc. Rev., 2015, 44, 1015–1027 RSC.
- S. Saen-Oon, M. F. Lucas and V. Guallar, Phys. Chem. Chem. Phys., 2013, 15, 15271–15285 RSC.
- C. L. Guo, X. Yu, S. Refaely-Abramson, L. Sepunaru, T. Bendikov, I. Pecht, L. Kronik, A. Vilan, M. Sheves and D. Cahen, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 10785–10790 CrossRef CAS PubMed.
- T. Biskup, B. Paulus, A. Okafuji, K. Hitomi, E. Getzoff, S. Weber and E. Schleicher, J. Biol. Chem., 2013, 288, 9249–9260 CrossRef CAS PubMed.
- D. Nohr, S. Franz, R. Rodriguez, B. Paulus, L. O. Essen, S. Weber and E. Schleicher, Biophys. J., 2016, 111, 301–311 CrossRef CAS PubMed.
- U. V. Pedmale, R. B. Celaya and E. Liscum, Arabidopsis Book, 2010, vol. 8, p. e0125 Search PubMed.
- W. Oehlmann and G. Auling, Microbiology, 1999, 145, 1595–1604 CrossRef CAS PubMed.
- G. Bleifuss, M. Kolberg, S. Potsch, W. Hofbauer, R. Bittl, W. Lubitz, A. Graslund, G. Lassmann and F. Lendzian, Biochemistry, 2001, 40, 15362–15368 CrossRef CAS PubMed.
- M. Defelippis, M. Faraggi and M. Klapper, J. Am. Chem. Soc., 1990, 112, 5640–5642 CrossRef CAS.
- O. Morozova and A. Yurkovskaya, J. Phys. Chem. B, 2015, 119, 140–149 CrossRef CAS PubMed.
- C. Hoganson and C. Tommos, Biochim. Biophys. Acta, Bioenerg., 2004, 1655, 116–122 CrossRef CAS PubMed.
- V. Davidson, G. Huether, W. Kochen, T. Simat and H. Steinhart, Tryptophan, Serotonin and Melatonin, 1999, vol. 467, pp. 587–595 Search PubMed.
- L. Sepunaru, S. Refaely-Abramson, R. Lovrinčić, Y. Gavrilov, P. Agrawal, Y. Levy, L. Kronik, I. Pecht, M. Sheves and D. Cahen, J. Am. Chem. Soc., 2015, 137, 9617–9626 CrossRef CAS PubMed.
- D. Lovley, S. Gottesman, C. Harwood and O. Schneewind, Annu. Rev. Microbiol., 2012, 66, 391–409 CrossRef CAS PubMed.
- N. Malvankar and D. Lovley, Chemsuschem, 2012, 5, 1039–1046 CrossRef CAS PubMed.
- R. Croce and H. van Amerongen, Nat. Chem. Biol., 2014, 10, 492–501 CrossRef CAS PubMed.
- Y. Hayamizu, C. R. So, S. Dag, T. S. Page, D. Starkebaum and M. Sarikaya, Sci. Rep., 2016, 6, 33778 CrossRef CAS PubMed.
- G. Reguera, K. D. McCarthy, T. Mehta, J. S. Nicoll, M. T. Tuominen and D. R. Lovley, Nature, 2005, 435, 1098–1101 CrossRef CAS PubMed.
- Y. Gorby, S. Yanina, J. McLean, K. Rosso, D. Moyles, A. Dohnalkova, T. Beveridge, I. Chang, B. Kim, K. Kim, D. Culley, S. Reed, M. Romine, D. Saffarini, E. Hill, L. Shi, D. Elias, D. Kennedy, G. Pinchuk, K. Watanabe, S. Ishii, B. Logan, K. Nealson and J. Fredrickson, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 11358–11363 CrossRef CAS PubMed.
- R. Balint, N. Cassidy and S. Cartmell, Acta Biomater., 2014, 10, 2341–2353 CrossRef CAS PubMed.
- P. Reardon and K. Mueller, J. Biol. Chem., 2013, 288, 29260–29266 CrossRef CAS PubMed.
- M. Vargas, N. S. Malvankar, P. L. Tremblay, C. Leang, J. A. Smith, P. Patel, O. Snoeyenbos-West, O. Synoeyenbos-West, K. P. Nevin and D. R. Lovley, mBio, 2013, 4, e00105–e00113 CrossRef PubMed.
- Y. Tan, R. Y. Adhikari, N. S. Malvankar, S. Pi, J. E. Ward, T. L. Woodard, K. P. Nevin, Q. Xia, M. T. Tuominen and D. R. Lovley, Small, 2016, 12, 4481–4485 CrossRef CAS PubMed.
- H. Kim, H. Park, M. Hyun, I. Chang, M. Kim and B. Kim, Enzyme Microb. Technol., 2002, 30, 145–152 CrossRef CAS.
- D. Bond and D. Lovley, Appl. Environ. Microbiol., 2003, 69, 1548–1555 CrossRef CAS PubMed.
- K. Rabaey, N. Boon, S. Siciliano, M. Verhaege and W. Verstraete, Appl. Environ. Microbiol., 2004, 70, 5373–5382 CrossRef CAS PubMed.
- A. Esteve-Nunez, C. Nunez and D. Lovley, J. Bacteriol., 2004, 186, 2897–2899 CrossRef CAS PubMed.
- A. Esteve-Núñez, M. Rothermich, M. Sharma and D. Lovley, Environ. Microbiol., 2005, 7, 641–648 CrossRef PubMed.
- W. C. Lin, M. V. Coppi and D. R. Lovley, Appl. Environ. Microbiol., 2004, 70, 2525–2528 CrossRef CAS PubMed.
- N. S. Malvankar and D. R. Lovley, Curr. Opin. Biotechnol., 2014, 27, 88–95 CrossRef CAS PubMed.
- V. Sivanathan and A. Hochschild, Genes Dev., 2012, 26, 2659–2667 CrossRef CAS PubMed.
- N. Van Gerven, P. Goyal, G. Vandenbussche, M. De Kerpel, W. Jonckheere, H. De Greve and H. Remaut, Mol. Microbiol., 2014, 91, 1022–1035 CrossRef CAS PubMed.
- H. Jensen, A. Albers, K. Malley, Y. Londer, B. Cohen, B. Helms, P. Weigele, J. Groves and C. Ajo-Franklin, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 19213–19218 CrossRef CAS PubMed.
- C. Goldbeck, H. Jensen, M. TerAvest, N. Beedle, Y. Appling, M. Hepler, G. Cambray, V. Mutalik, L. Angenent and C. Ajo-Franklin, ACS Synth. Biol., 2013, 2, 150–159 CrossRef CAS PubMed.
- C. M. Waters and B. L. Bassler, Annu. Rev. Cell Dev. Biol., 2005, 21, 319–346 CrossRef CAS PubMed.
- A. Prindle, J. T. Liu, M. Asally, S. Ly, J. Garcia-Ojalvo and G. M. Suel, Nature, 2015, 527, 59–63 CrossRef CAS PubMed.
- S. Zhang, B. Karaca, S. VanOosten, E. Yuca, S. Mahalingam, M. Edirisinghe and C. Tamerler, Macromol. Rapid Commun., 2015, 36, 1322–1328 CrossRef CAS PubMed.
- C. Beloin, A. Roux and J. Ghigo, Bacterial Biofilms, 2008, vol. 322, pp. 249–289 Search PubMed.
- M. Weiss-Muszkat, D. Shakh, Y. Zhou, R. Pinto, E. Belausov, M. Chapman and S. Sela, Appl. Environ. Microbiol., 2010, 76, 1545–1554 CrossRef CAS PubMed.
- M. Winfield and E. Groisman, Appl. Environ. Microbiol., 2003, 69, 3687–3694 CrossRef CAS PubMed.
- H. C. Flemming, J. Wingender, U. Szewzyk, P. Steinberg, S. A. Rice and S. Kjelleberg, Nat. Rev. Microbiol., 2016, 14, 563–575 CrossRef CAS PubMed.
- M. Davey and G. O'toole, Microbiol. Mol. Biol. Rev., 2000, 64, 847–867 CrossRef CAS PubMed.
- M. Hammar, A. Arnqvist, Z. Bian, A. Olsen and S. Normark, Mol. Microbiol., 1995, 18, 661–670 CAS.
- M. M. Barnhart and M. R. Chapman, Annu. Rev. Microbiol., 2006, 60, 131–147 CrossRef CAS PubMed.
- N. Hammer, J. Schmidt and M. Chapman, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 12494–12499 CrossRef CAS PubMed.
- N. Hammer, B. McGuffie, Y. Zhou, M. Badtke, A. Reinke, K. Brannstrom, J. Gestwicki, A. Olofsson, F. Almqvist and M. Chapman, J. Mol. Biol., 2012, 422, 376–389 CrossRef CAS PubMed.
- T. Knowles and M. Buehler, Nat. Nanotechnol., 2011, 6, 469–479 CrossRef CAS PubMed.
- M. Dueholm, S. Nielsen, K. Hein, P. Nissen, M. Chapman, G. Christiansen, P. Nielsen and D. Otzen, Biochemistry, 2011, 50, 8281–8290 CrossRef CAS PubMed.
- A. Y. Chen, Z. Deng, A. N. Billings, U. O. Seker, M. Y. Lu, R. J. Citorik, B. Zakeri and T. K. Lu, Nat. Mater., 2014, 13, 515–523 CrossRef CAS PubMed.
- P. Nguyen, Z. Botyanszki, P. Tay and N. Joshi, Nat. Commun., 2014, 5, 4945 CrossRef CAS PubMed.
- C. Beloin, A. Houry, M. Froment, J. M. Ghigo and N. Henry, PLoS Biol., 2008, 6, e167 Search PubMed.
- N. Amdursky, Phys. Chem. Chem. Phys., 2013, 15, 13479–13482 RSC.
- C. Shih, A. K. Museth, M. Abrahamsson, A. M. Blanco-Rodriguez, A. J. Di Bilio, J. Sudhamsu, B. R. Crane, K. L. Ronayne, M. Towrie, A. Vlcek, J. H. Richards, J. R. Winkler and H. B. Gray, Science, 2008, 320, 1760–1762 CrossRef CAS PubMed.
- K. Takematsu, H. Williamson, A. M. Blanco-Rodríguez, L. Sokolová, P. Nikolovski, J. T. Kaiser, M. Towrie, I. P. Clark, A. Vlček, J. R. Winkler and H. B. Gray, J. Am. Chem. Soc., 2013, 135, 15515–15525 CrossRef CAS PubMed.
- L. L. del Mercato, P. P. Pompa, G. Maruccio, A. Della Torre, S. Sabella, A. M. Tamburro, R. Cingolani and R. Rinaldi, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 18019–18024 CrossRef CAS PubMed.
- P. Lembre, C. Vendrely and P. Di Martino, Protein Pept. Lett., 2013, 20, 942–946 CrossRef CAS PubMed.
- O. Morozova, R. Kaptein and A. Yurkovskaya, J. Phys. Chem. B, 2012, 116, 12221–12226 CrossRef CAS PubMed.
- R. Y. Adhikari, N. S. Malvankar, M. T. Tuominen and D. R. Lovley, RSC Adv., 2016, 6, 8363–8366 Search PubMed.
- F. J. Isaacs, D. J. Dwyer, C. Ding, D. D. Pervouchine, C. R. Cantor and J. J. Collins, Nat. Biotechnol., 2004, 22, 841–847 CrossRef CAS PubMed.
- P. Tian, W. Boomsma, Y. Wang, D. Otzen, M. Jensen and K. Lindorff-Larsen, J. Am. Chem. Soc., 2015, 137, 22–25 CrossRef CAS PubMed.
- M. Kallberg, H. P. Wang, S. Wang, J. Peng, Z. Y. Wang, H. Lu and J. B. Xu, Nat. Protoc., 2012, 7, 1511–1522 CrossRef CAS PubMed.
- N. S. Malvankar, M. Vargas, K. Nevin, P. L. Tremblay, K. Evans-Lutterodt, D. Nykypanchuk, E. Martz, M. T. Tuominen and D. R. Lovley, mBio, 2015, 6, e00084-15 CrossRef PubMed.
- G. T. Feliciano, R. J. Steidl and G. Reguera, Phys. Chem. Chem. Phys., 2015, 17, 22217–22226 RSC.
- F. Shewmaker, R. P. McGlinchey, K. R. Thurber, P. McPhie, F. Dyda, R. Tycko and R. B. Wickner, J. Biol. Chem., 2009, 284, 25065–25076 CrossRef CAS PubMed.
- B. Giese, M. Graber and M. Cordes, Curr. Opin. Chem. Biol., 2008, 12, 755–759 CrossRef CAS PubMed.
- C. Prigent-Combaret, G. Prensier, T. Le Thi, O. Vidal, P. Lejeune and C. Dorel, Environ. Microbiol., 2000, 2, 450–464 CrossRef CAS PubMed.
- O. Vidal, R. Longin, C. Prigent-Combaret, C. Dorel, M. Hooreman and P. Lejeune, J. Bacteriol., 1998, 180, 2442–2449 CAS.
- N. Li and M. Guiver, Macromolecules, 2014, 47, 2175–2198 CrossRef CAS.
- L. Wu, Z. Zhang, J. Ran, D. Zhou, C. Li and T. Xu, Phys. Chem. Chem. Phys., 2013, 15, 4870–4887 RSC.
- J. Lerner Yardeni, M. Amit, G. Ashkenasy and N. Ashkenasy, Nanoscale, 2016, 8, 2358–2366 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: AFM images for designed peptide polymers, cloning of the gene fragments, list of primers used in this study, plasmid maps of cloned genes, SEM images for a biofilm on the surface of electrodes and empty interdigitated gold electrode, and current voltage measurement of the empty electrode. See DOI: 10.1039/c7ra06289c |
| This journal is © The Royal Society of Chemistry 2017 |