Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Profiling of BABA-induced differentially expressed genes of Zea mays using suppression subtractive hybridization†
Arun K. Shawa,
Pardeep K. Bhardwajb,
Supriya Ghoshc,
Ikbal Azaharc,
Sinchan Adhikaric,
Ayan Adhikaric,
Ang R. Sherpaa,
Samir K. Sahad and
Zahed Hossain *c
*c
aDepartment of Botany, West Bengal State University, Kolkata – 700126, West Bengal, India
bPlant Bioresources Division, Institute of Bioresources and Sustainable Development, Sikkim Centre, Gangtok – 737102, Sikkim, India
cDepartment of Botany, University of Kalyani, Nadia, Kalyani 741235, West Bengal, India. E-mail: zahed_kly@yahoo.com; Fax: +91-33-2582-8282; Tel: +91-33-2582-8750 ext. 217
dDepartment of Zoology, West Bengal State University, Kolkata – 700126, West Bengal, India
First published on 11th September 2017
Abstract
β-Aminobutyric acid (BABA), a potent chemical priming agent, induces plant resistance to a broad spectrum of biotic and abiotic stresses. Nevertheless, the molecular mechanisms underlying this phenomenon remain poorly understood. Therefore, this study aims to identify rare and differentially expressed transcripts in BABA-primed maize (Zea mays L.) using a suppression subtractive hybridization strategy. Forward subtraction was performed using the cDNA prepared from a BABA primed leaf as a tester and cDNA from the control leaf as the driver. For reverse subtraction, tester cDNA was synthesized from control leaf and driver cDNA from the BABA primed leaf. A total of 626 clones were picked randomly. In total 192 and 204 high quality expressed sequence tags (ESTs) were generated from forward (ZmSF) and reverse SSH library (ZmSR) respectively. Contigs analyses revealed 27 unigenes from the forward subtracted library and 4 unigenes from the reverse subtracted library. The ESTs encoding alpha amylase, inositol monophosphatase 3, tocopherol cyclase, thioredoxin H-type, aspartic proteinase, NADP-malic enzyme and PEP carboxykinase were expressed predominantly in BABA-primed leaf and found to be predominantly associated with major metabolic and biosynthesis events along with seed germination, developmental processes and adaptive responses to environmental stimuli such as drought, salinity, osmotic stress and pathogen attack. In this study, we present the first report of transcriptome analysis of maize leaf exposed to BABA. The expression profiles of candidate genes from subtracted libraries were checked using semi quantitative RT-PCR and further validated by qRT-PCR. Expression profiles largely corroborated the quality and functional diversity of both libraries. In ZmSF, 130 ESTs (67.70%) did not show any significant similarity to any protein in the NCBI database, indicating a significant fraction of novel and rare expressed transcripts that were rapidly and strongly induced in response to BABA. Further research needs to be done for these novel transcripts to elucidate their function in a BABA-potentiated stress adaptation mechanism. Overall, our study adds new insights into the BABA potentiated defense mechanisms in plants, and strengthens the available transcriptome database of maize.
Introduction
Plants are able to remember a previous exposure to stressful conditions, which helps them to overcome a subsequent harsh event, a phenomenon known as priming.1 The primed plants exhibit faster and stronger activation of defense responses upon renewed exposure to pathogen attack or in response to environmental stresses.2,3 Besides pathogens, various natural and synthetic compounds have emerged as potent inducers of priming, which is often associated with enhanced resistance to a broad range of environmental cues including abiotic and biotic factors.Among the various chemical inducers of plant resistance, β-aminobutyric acid (BABA), a non-protein amino acid has been known to induce plant defense for more than 50 years.4,5 A wealth of reports are available on the BABA-mediated enhanced defensive capacity of plants against pathogen infection,6–8 drought,1,9–12 salinity,12 heavy metal stress,13,14 and acid rain.15
Suppression subtractive hybridization (SSH) is one of the most powerful and effective method for generating subtracted cDNA libraries to compare two mRNA populations and obtain cDNAs representing genes that are either overexpressed or exclusively expressed in one population as compared to another.16 This PCR-based cDNA subtraction technique combines normalization and subtraction in a single procedure, which helps in identification of differentially expressed abundant genes and enrichment of rare transcripts to facilitate the identification of novel genes.17 In contrast to high throughput sequencing techniques, SSH is a more cost-effective approach for rapid discovery of novel genes as it facilitates enrichment of the rare sequences by more than 1000-fold in a single round of subtractive hybridization.18
In plants, SSH has been widely used to untie the differentially expressed genes associated with organ-/tissue-specific biochemical pathways and various biotic and abiotic stress responses.19–23 Applications of SSH technique include identification of genes associated with salinity stress in tomato,24 mangrove Bruguiera cylindrica,25 and Suaeda maritima,26 cadmium stress in maize,27 calcium stress in amaranth leaves,28 drought in peanut root and Alternanthera philoxeroides,29,30 ozone stress in Pisum sativum,31 low temperature stress in maize,32 waterlogging stress in maize root,33 boron-deficiency in citrus rootstock root,34 and chilling stress in pepper plant (Capsicum annuum L.).35
The priming phenomenon has been known for decades. However, until recently the underlying molecular basis of chemical priming remains elusive due to insufficient knowledge of how this chemical is perceived in plants. However, a recent study revealed that perception of β-aminobutyric acid in plants is mediated by an aspartyl-tRNA synthetase.36 In Arabidopsis, BABA treatment has been reported to induce chromatin modifications associated with transcriptional activation of pattern-triggered immunity (PTI)-responsive genes during pathogen attack.37,38 A genome-wide microarray analysis of BABA-treated Arabidopsis at post-inoculation phase revealed higher expression of wide number of genes primarily responsive to salicylic acid and pathogen infection.39,40 Moreover, Van der Ent et al.41 reported enhanced expression of transcription factors during induced resistance by BABA against the bacterial pathogen. Recently, we reported that pretreatment of 7 day-old maize seedlings with 600 μM BABA for a consecutive period of 5 days significantly alleviates the drought stress effects.9 Our findings suggest that BABA induced drought tolerance in maize is primarily mediated through accelerated stomatal closure together with fine tuning of JA and ABA pathways and activation of antioxidant defense system. However, differential expression of BABA-responsive genes associated with plant stress tolerance has not been well explored in model crops including maize.
In the present experiment, an attempt was made to identify rare and differentially expressed transcripts in BABA-primed maize plants using suppression subtractive hybridization (SSH) strategy for better understanding the molecular basis of priming phenomenon. In addition, possible mechanisms underlying the BABA-potentiated stress tolerance in plants were discussed.
Materials and methods
Plant material and BABA treatment
Surface sterilized maize seeds (Zea mays L. inbred line HKI-161) were allowed to germinate in plastic cups containing steam-sterilized soil and farmyard manure (FYM) mixture (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). Seedlings were maintained at 25 °C in plant growth chamber (Panasonic, Japan, Model: MLR-352H-PE) illuminated with white fluorescent light (600 μmol m−2 s−1, 16 h light period per day) and 65% RH. Seven days old seedlings were randomly divided into two groups of 30 plants each. To induce chemical priming, one group of plants were treated with 600 μM β-aminobutyric acid (BABA, Sigma-Aldrich, USA) (20 mL per cup) for a consecutive period of 5 days and considered as BABA-primed. Soil drenching with BABA was performed as previously described.6,12 A parallel group of 7 day-old plants were maintained with equal amount of water (20 mL per cup) for next 5 days and considered as control. The leaves of both control and BABA-primed plants were randomly harvested for the preparation of suppression subtractive hybridization cDNA libraries (SSHL) and gene expression analysis. Soon after harvest, leaves were immediately freezed in liquid nitrogen, and stored at −80 °C till further use.
1). Seedlings were maintained at 25 °C in plant growth chamber (Panasonic, Japan, Model: MLR-352H-PE) illuminated with white fluorescent light (600 μmol m−2 s−1, 16 h light period per day) and 65% RH. Seven days old seedlings were randomly divided into two groups of 30 plants each. To induce chemical priming, one group of plants were treated with 600 μM β-aminobutyric acid (BABA, Sigma-Aldrich, USA) (20 mL per cup) for a consecutive period of 5 days and considered as BABA-primed. Soil drenching with BABA was performed as previously described.6,12 A parallel group of 7 day-old plants were maintained with equal amount of water (20 mL per cup) for next 5 days and considered as control. The leaves of both control and BABA-primed plants were randomly harvested for the preparation of suppression subtractive hybridization cDNA libraries (SSHL) and gene expression analysis. Soon after harvest, leaves were immediately freezed in liquid nitrogen, and stored at −80 °C till further use.
To further confirm the role of candidate genes in stress mitigation, an additional drought experiment was conducted. Seven days old maize seedlings were treated with BABA for 5 days and considered as BABA-primed. These BABA-primed plants were then exposed to drought stress by withholding watering for 9 days. A similar set of 7 days old seedlings were maintained with regular watering for 5 days followed by drought stress for 9 days. These plants without BABA treatment were considered as non-primed plants. Control plants were maintained under same growth condition with regular watering throughout the experimental period. On 9th day of stress treatment, leaves were harvested in liquid nitrogen and stored at −80 °C until further use.
Preparation of suppression subtractive hybridization cDNA libraries
Total RNA was isolated from the leaf tissues of control and BABA primed seedlings following RNA isolation method described by Ghawana et al.42 RNA quantity was analysed using NanoDrop™ 2000c spectrophotometer (Thermo Fisher Scientific Inc., USA) by recording absorbance at 260 and 280 nm. Also, the RNA was run on a denaturing agarose/formaldehyde gel and visualized using gel documentation system (Bio-Rad ChemiDoc™ XRS+ Imaging system, USA). Poly(A+) RNA (mRNA) was purified from total RNA of control and BABA primed leaves using Oligotex® Direct mRNA Mini Kit (Qiagen, Germany) as per the manufacturer's instructions. SSH was performed using PCR-select™ cDNA SSH kit (Clontech, USA) as described Bhardwaj et al.22 Forward subtraction was performed using the cDNA prepared from BABA primed leaf as tester and cDNA from control leaf as driver. For reverse subtraction, tester cDNA was synthesized from control leaf and driver cDNA from the BABA primed leaf. Two successive hybridizations were performed in order to equalize and enrich differentially expressed sequences. For first hybridization, excess driver cDNA was added to each of the tester samples, heat denatured and hybridized at 68 °C for 8 h. In the second hybridization, two tester samples from the first hybridization were mixed together and freshly denatured driver cDNA was added to the sample followed by hybridization at 68 °C for overnight. For exponential amplification of differentially expressed cDNA, two rounds of PCR amplification were performed with oligo nucleotides complementary to adapters 1 and 2R. The resulting PCR products enriched for differentially expressed cDNAs were separately ligated into TA cloning system (Invitrogen, USA) to make forward and reverse libraries. Then these ligated products were transformed into Escherichia coli strain DH5α-competent cells. The transformed recombinant white colonies were picked randomly from both (forward and reverse) libraries then cultured in Luria-Bertani broth supplemented with 100 μg mL−1 ampicillin, and plasmids were isolated using HiYield™ Plasmid Mini Kit (RBC, Taiwan).Sequencing, functional annotation and sequence assembly of ESTs
Single pass sequencing of plasmids was performed using the Big Dye terminator (version 3.1) cycle sequencing mix (Applied Biosystems, USA) on an automated DNA sequencer (3130xl, Genetic Analyzer-1203-019, Applied Biosystems, USA) with M13 forward primer (5′-GTAAAACGACGGCCAGTG-3′). For removal of regions of vector, adaptor and low complexity contamination, all the ESTs of both libraries were screened using National Center for Biotechnology Information (NCBI) VecScreen software. The resulting unique transcripts were functional annotated and analyzed using Blast2GO bioinformatics tool43 using default e-value 10−3 and highly similar segment pair wise (HSP) value of 15. Annotation and InterPro Scan (IPS) analysis were performed to detect the protein motifs and domains on the basis of quality hits. Also, ESTs were mapped for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation. On the basis of functional annotation, these ESTs were divided into proteins with known function and proteins with no match in the database. Based on redundancy, ESTs were clustered into singletons (i.e., unique clones) and contigs (i.e., 2 or more clones). Both of these together formed a nonredundant independent sequence termed as unigenes. In total 27 unigenes from forward SSH libraries were submitted to NCBI dbEST database with accession numbers JZ923580 to JZ923606.Semiquantitative RT-PCR analysis of the differentially expressed ESTs
To validate the quality of the control and BABA primed subtractive libraries, expression analysis of differentially expressed genes was performed by semi-quantitative RT-PCR. First, total RNA (2.0 μg) was treated with DNase I (Amplification grade, Invitrogen, USA) to remove contaminating genomic DNA and first-strand cDNA was synthesized using Superscript III (Invitrogen, USA) kit following manufacturer's protocol. GAPDH was used as an internal control. Primers for selected differentially expressed genes from both the libraries were designed using the primer3 (v.0.4.0) software (http://gmdd.shgmo.org/primer3). All the primers used in this study are listed in ESI Table 1.† The PCR reaction was performed on a programmable thermocycler (Applied Biosystems 2720 Thermal Cycler, USA) following this cycling conditions: initial denaturation at 94 °C for 5 min, followed by 30–35 cycles at 94 °C for 30 s, 60 °C for 40 s, 72 °C for 30 s, and then a final extension at 72 °C for 5 min. Each PCR reaction performed in triplicates. Final amplified PCR products were separated on 1.2% agarose gel stained with ethidium bromide and visualized using a gel documentation system (Bio-Rad ChemiDoc™ XRS+ Imaging system, USA).Gene expression analysis using qRT-PCR
Total RNA was extracted from frozen leaf tissue using TRIzol reagent (Thermo Scientific) following manufacturer's instructions. Total RNA (2 μg) was incubated for 5 min at 65 °C and then 20 μL of first strand cDNA was prepared following protocol of Verso cDNA synthesis kit (Thermo Scientific).All the maize gene sequences used for primer designing were downloaded from NCBI database (http://www.ncbi.nlm.nih.gov). The primers for quantitative real-time PCR (qRT-PCR) analysis were designed using the primer 3 v.0.4.0 software (http://gmdd.shgmo.org/primer3). Gene amplification and gel electrophoresis was performed to confirm that the primers amplified only a single product of expected size (data not shown). Each qRT-PCR reaction was performed with three biological replicates and two technical replicates. The reaction was performed in 10 μL reaction mixture containing 1 μL cDNA samples as template, 5 μL of 2× SYBR® Green Master Mix (Applied Biosystems, USA), 3 μL of DEPC-treated water and 1 μL of diluted forward and reverse primer mix (10 pmol). The reactions were performed in StepOne™ Real-Time PCR System (Applied Biosystems, USA) using the following programme: initial denaturation at 95 °C for 10 min, followed by 40 cycles of amplification (95 °C for 15 s, 57 °C for 30 s and 72 °C for 15 s) and final melt curve analysis was performed to verify specificity of each amplicons. Transcript level of all the genes was normalized with an internal reference, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene from maize. The relative expression ratio of each gene was calculated using comparative Ct value method as described previously by Livak and Schmittgen.44 Data represented here are mean values of relative fold change. Expression is shown after normalization to GAPDH. Values were calculated using ΔΔCT method, and the error bar showing standard deviation. All the primers used in this study are listed in ESI Table 2.†
Results
Cloning, sequencing and clustering of ESTs by constructing suppression subtractive hybridization cDNA library
Suppression subtractive cDNA library (SSHL) was prepared using the RNA isolated from control (CO) and BABA-primed (BP) maize leaves. A total of 626 expressed sequence tags (ESTs) were generated from forward (ZmSF, 286 ESTs) and reverse (ZmSR, 340 ESTs) SSHL (Table 1). Depending upon the quality of sequences in ZmSF, 192 ESTs were analyzed for annotation and contigs assembly and clustered into 10 singletons and 17 contigs, constituting a total of 27 unigenes. Similarly, gene function analysis grouped 204 high quality ESTs from reverse library (ZmSR) into 0 singletons and 4 contigs, constituting a total of 4 unigenes (Table 1). ESTs in ZmSF represent the genes up-regulated in response to BABA-priming (BP) whereas reverse library (ZmSR) represents the genes which are down-regulated under BABA treatment.| Parameter | Forward library | Reverse library |
|---|---|---|
| Total ESTs sequenced | 286 | 340 |
| High quality ESTs | 192 | 204 |
| Average read length (bp) | 205 | 371 |
| Highest sequence length (bp) | 764 | 669 |
| Unknown ESTs | 110 | 88 |
| No. of contigs | 17 | 4 |
| No. of singleton | 10 | 0 |
| No. of unigene | 27 | 4 |
Sequence annotation and functional categorization
Blast2GO gene annotation tool43 was used to further analyze the high quality ESTs. In ZmSF, 62 ESTs (32.29%) out of 192 ESTs showed significant homology to proteins whereas rest of the 130 ESTs (67.70%) did not show any significant homology to any protein which might suggest their novel functions. In ZmSR, 130 ESTs (63.72%) showed significant homology to proteins and rest of the 74 ESTs (36.27%) did not show any homology to any protein. In ZmSF, ESTs were classified into different categories viz. DNA repair, single organism signaling, L-ascorbic acid biosynthetic process, inositol biosynthetic process, signal transduction, salt and oxidative stress, response to cold and defense, xanthophyll, chlorophyll and fatty acid metabolic processes etc. (ESI Table 3†) whereas ESTs from ZmSR were classified into transport (ESI Table 4†). All the ESTs from ZmSF were found to be present in different cellular components like nucleus, extracellular region, mitochondria, cytosol, organelle envelope, plasma membrane (ESI Table 5†) and ESTs from ZmSR were predominantly found in mitochondria and integral component of membrane (ESI Table 6†). ESTs for alpha amylase (30) were found to be most abundant in ZmSF. This was followed by ESTs encoding inositol monophosphatase 3 (12), tocopherol cyclase (11), hypothetical protein (11), thioredoxin H-type (3), and aspartic proteinase (2) etc. respectively (Table 2). In ZmSR, metal transporter Nramp5 was found to be most abundant followed by unknown and hypothetical protein (Table 3). In ZmSF, most of the ESTs were homologous to genes from different species, with highest homology from Zea mays (92) followed by Lotus japonicas (16), Oryza sativa (2), Genlisea aurea (2), Sorghum bicolor (2), Heuchera sanguine (1), and Setaria italica (1) (Fig. 1A, ESI Table 7†). Most of the ESTs from ZmSR were homologous to genes from several species with maximum homology of genes from Glycine max (121), Lotus japonicus (36), Vigna angularis var. angularis (34), Morus notabilis (5), Medicago truncatula (3), Cajanus cajan (3), and Jatropha curcas (1) (Fig. 1B; ESI Table 8†).| Sr. no. | Sequence name | Homology | Redundancy | Seq. length | Min. E-value | Mean similarity | Accession no. |
|---|---|---|---|---|---|---|---|
| 1 | Zm_FL171 | Alpha-amylase | 30 | 239 | 3.61 × 10−17 | 93.05% | JZ923597 |
| 2 | Zm_FL147 | NA | 16 | 181 | — | — | JZ923595 |
| 3 | Zm_FL4 | NA | 16 | 163 | — | — | JZ923582 |
| 4 | Zm_FL16 | NA | 16 | 120 | — | — | JZ923584 |
| 5 | Zm_FL18 | NA | 15 | 187 | — | — | JZ923585 |
| 6 | Zm_FL3 | Inositol monophosphatase 3 | 12 | 222 | 1.44 × 10−17 | 89.65% | JZ923581 |
| 7 | Zm_FL531 | Tocopherol cyclase | 11 | 365 | 2.84 × 10−42 | 90.40% | JZ923605 |
| 8 | Zm_FL83 | Hypothetical protein | 11 | 198 | 3.54 × 10−14 | 96.00% | JZ923592 |
| 9 | Zm_FL142 | NA | 11 | 194 | — | — | JZ923594 |
| 10 | Zm_FL2 | NA | 09 | 216 | — | — | JZ923580 |
| 11 | Zm_FL78 | NA | 08 | 181 | — | — | JZ923591 |
| 12 | Zm_FL45 | NA | 08 | 175 | — | — | JZ923586 |
| 13 | Zm_FL61 | NA | 08 | 178 | — | — | JZ923590 |
| 14 | Zm_FL6 | NA | 04 | 216 | — | — | JZ923583 |
| 15 | Zm_FL166 | Thioredoxin H-type | 03 | 245 | 4.73 × 10−17 | 92.00% | JZ923596 |
| 16 | Zm_FL221 | Aspartic proteinase | 02 | 237 | 6.17 × 10−34 | 96.55% | JZ923601 |
| 17 | Zm_FL58 | NA | 02 | 142 | — | — | JZ923589 |
| 18 | Zm_FL48 | H aca ribonucleoprotein complex subunit 3-like protein | 01 | 108 | 3.04 × 10−17 | 98.05% | JZ923587 |
| 19 | Zm_FL554 | NADP-dependent malic enzyme | 01 | 764 | 3.08 × 10−149 | 91.35% | JZ923606 |
| 20 | Zm_FL174 | Phosphoenolpyruvate carboxykinase [ATP] | 01 | 192 | 1.34 × 10−38 | 98.00% | JZ923598 |
| 21 | Zm_FL216 | NA | 01 | 216 | — | — | JZ923600 |
| 22 | Zm_FL517 | Unknown | 01 | 171 | 4.08 × 10−32 | 94.85% | JZ923604 |
| 23 | Zm_FL99 | NA | 01 | 187 | — | — | JZ923593 |
| 24 | Zm_FL300 | NA | 01 | 180 | — | — | JZ923602 |
| 25 | Zm_FL209 | NA | 01 | 199 | — | — | JZ923599 |
| 26 | Zm_FL51 | NA | 01 | 172 | — | — | JZ923588 |
| 27 | Zm_FL481 | NA | 01 | 72 | — | — | JZ923603 |
| Sr. no. | Sequence name | Homology | Redundancy | Seq. length | Min. E-value | Mean similarity |
|---|---|---|---|---|---|---|
| 1 | Zm_RL202 | Metal transporter Nramp5 | 130 | 669 | 9.50 × 10−47 | 84.65% |
| 2 | Zm_RL143 | Unknown | 37 | 120 | 6.20 × 10−11 | 92.78% |
| 3 | Zm_RL365 | Hypothetical protein VIGAN_UM160600, partial | 35 | 216 | 5.40 × 10−23 | 88.45% |
| 4 | Zm_RL303 | Signal anchor, putative | 2 | 219 | 1.70 × 10−23 | 85.95% |
Gene ontology annotation categorization and analysis of ESTs
All the ESTs of both libraries were carefully analysed in terms of their major gene ontology (GO) categories: biological process, molecular function and cellular component. To compare specific ESTs of both library, functional annotation were performed separately. In ZmSF, maximum numbers of annotated ESTs were grouped under GO category of biological process whereas in ZmSR highest numbers of ESTs were grouped under GO category of molecular function. Based on biological process, most of the transcripts in ZmSF were assigned to DNA repair, single organism signalling, L-ascorbic acid biosynthetic process, inositol biosynthetic process, response to cold, oxidative and salt stress, xanthophylls and fatty acid metabolic process, regulation of defense response and vitamin E biosynthetic process (Fig. 2A; ESI Table 3†). ESTs of ZmSR were categorized to transport (Fig. 3A; ESI Table 4†). Based on cellular component category, a large number of ESTs of ZmSF was assigned to the nucleus, extracellular region, mitochondrion, cytosol, organelle envelope and plasma membrane (Fig. 2B; ESI Table 5†). Interestingly, maximum ESTs of ZmSR were assigned to mitochondrion, integral component of membrane and chloroplast (Fig. 3B; ESI Table 6†). Under molecular function category, most transcripts of ZmSF were pertained to ion and nucleic acid binding, hydrolase activity, phosphatase and cyclase activity (Fig. 2C; ESI Table 9†). In contrast, ZmSR had maximum representation associated with genes involved in ion binding and transporter activity (Fig. 3C; ESI Table 10†). | ||
| Fig. 3 Gene ontology (GO)-based functional categorization of control leaf ESTs, obtained by SSH. ESTs were categorized into biological process (A), cellular component (B), and molecular function (C). | ||
InterPro scan and KEGG analysis of ESTs
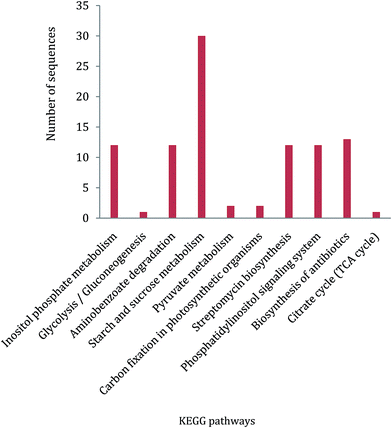
In ZmSF, 143 ESTs were without IPS domains, 49 were with IPS domains and 35 were with GO annotations (Fig. 4; ESI Table 11†). Similarly, 204 ESTs were without IPS domains and none of the EST was found with IPS domains and GO annotations (Fig. 4; ESI Table 11†) in the ZmSR. All the InterPro protein domains present in the ESTs of ZmSF were described in ESI Table 12.† In ZmSF, eight protein domains (IPR013780, IPR012850, IPR033121, IPR021109, IPR012301, IPR016040, IPR012302 and IPR008210) were detected which are related to glycosyl hydrolase (all beta), alpha-amylase (C-terminal beta-sheet), peptidase family A1 domain, aspartic peptidase domain, malic enzyme (N-terminal domain), NAD (P)-binding domain, malic enzyme (NAD-binding) and phosphoenolpyruvate carboxykinase (N-terminal) respectively (Fig. 5). Interestingly, no protein domain was detected in ESTs of ZmSR after InterPro scan annotation. KEGG pathway analysis was done to assign metabolic pathways to ESTs. In ZmSF, ESTs were assigned for 10 KEGG pathways involved in inositol phosphate metabolism, glycolysis/gluconeogenesis, aminobenzoate degradation, starch and sucrose metabolism, pyruvate metabolism, carbon fixation in photosynthetic organisms, streptomycin biosynthesis, phosphatidylinositol signaling system, biosynthesis of antibiotics and citrate cycle (Fig. 6; ESI Table 13†) but no EST was annotated to any KEGG metabolic pathway in ZmSR.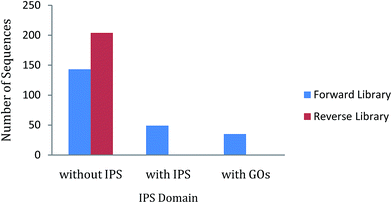 | ||
| Fig. 4 InterPro Scan (IPS) analysis showing the number of ESTs without IPS, with IPS, and GO domains in forward and reverse libraries. | ||
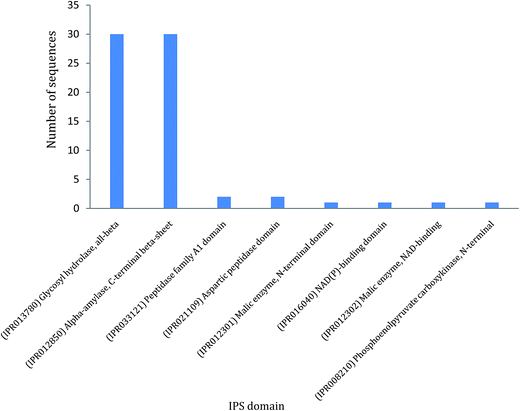 | ||
| Fig. 5 IPS domain analysis of functionally annotated ESTs from forward SSH library. Data distribution summary from BLAST2GO shows blast hits. | ||
Differential expression analysis of selected genes using semi-quantitative RT-PCR
Expression of putative candidate genes were checked by semi-quantitative RT-PCR (Fig. 7). These genes were selected on the basis of their abundance and their differential expression in both libraries. The genes selected for expression analysis are alpha-amylase (AA), inositol monophosphatase 3 (IMP), tocopherol cyclase (TC), aspartic proteinase (AP), NADP-dependent malic enzyme (NADP-ME), phosphoenolpyruvate carboxykinase (PEPCK), Zm_FL51 (FL51), Zm_FL300 (FL300), metal transporter Nramp5 (Nramp5), Zm_RL303 (RL303), Zm_RL143 (RL143) and Zm_RL365 (RL365). Semi-quantitative RT-PCR analysis showed that the expression pattern of all the studied genes largely corroborates with the SSH results. The mRNA expressions of genes AA, AP, IMP, NADP-ME, PEPCK, TC, FL51 and FL300 were found to be upregulated in BABA primed leaf (Fig. 7A). In contrast, high expression of Nramp5, RL303, RL143 and RL365 were evident in control leaf (Fig. 7B).qRT-PCR validation of candidate genes
To validate the SSH results, relative expression analyses of candidate genes from subtracted libraries were performed using qRT-PCR. Expression pattern of all the five studied genes (IMP, NADP-ME, PEPCK, AP and Trx-h) fully corroborates with the SSH findings (Fig. 8A–E). Enhanced mRNA expressions (1.4-fold to 4.8-fold) of all genes were recorded in leaves of BABA-primed plants as compared to control. | ||
| Fig. 8 Relative expression analysis of IMP (A), NADP-ME (B), PEPCK (C), AP (D) and Trx-h (E) genes in leaves of the control and BABA-primed plants. | ||
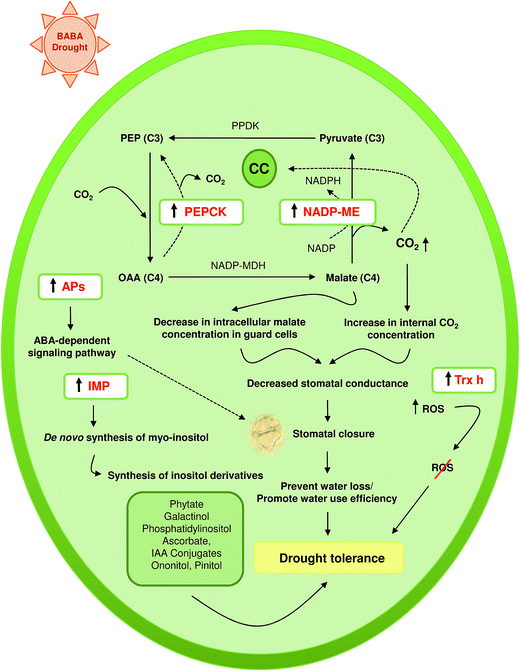
Moreover, drought stress had significant effect on the expression of BABA-responsive genes (Fig. 9). As compared to control plants, enhanced expressions of IMP, NADP-ME and Trx-h were recorded in drought challenged leaves. However, magnitude of the increase was more in BABA-primed leaves in respect of non-primed plants (Fig. 9A–C). In contrast, expression of PEPCK was down-regulated under stress (Fig. 9D). Nevertheless, magnitude of the decrease was more in non-primed leaves (0.9-fold as compared to control) than the BABA-primed plants (0.6-fold as compared to control). In addition, BABA-primed plants showed higher AP transcriptional abundance under limited water condition (Fig. 9E).
 | ||
| Fig. 9 qRT-PCR analysis of relative expression of IMP (A), NADP-ME (B), Trx-h (C), PEPCK (D) and AP (E) genes in leaves of the control, non-primed and BABA-primed plants under drought stress. | ||
Discussion
The β-aminobutyric acid (BABA) is a known potent priming agent which confers tolerance against a wide array of biotic and abiotic stresses.1,6–8,10–15 To better understand the genes which are differentially expressed upon BABA exposure, forward (BABA primed) and reverse (control) SSH libraries were constructed. In total 192 and 204 high quality expressed sequence tags (ESTs) were generated from forward and reverse SSH library respectively.Expressed sequence tags (ESTs) provide a bird's eye view of genes expressed under certain environmental cue or at a specific plant developmental stage. Suppression subtractive hybridization (SSH) is an efficient technique to categorize differentially expressed genes involved in different stress mitigation pathways and specific tissues/organs.20–23 It is a cost-effective approach for fast detection of novel genes, since SSH eliminates equally expressed and common housekeeping genes. In plants, various strategies based on subtractive libraries have been exploited to elucidate biological process specific gene complements such as response to biotic or abiotic stressors and different growth and developmental stages. Since, SSH has the advantage to reduce the representation of common transcripts and to facilitate enrichment of the differentially expressed sequences by more than 1000-fold in a single round of subtractive hybridization, it has wide application in identifying genes associated with complex organ-/tissue-specific biochemical pathways responsible for the biosynthesis of pharmaceutically important secondary metabolic compounds in specific tissues/organs of medicinal plants.21,23,45,46 To date, only a few reports are available on exploitation of SSH technique to identify and characterize genes responsive to salinity,47 drought,48 waterlogging,33 and cadmium stress.27
In this study, several differentially expressed ESTs were generated from the forward (ZmSF, BABA primed) and reverse (ZmSR, control) SSH libraries. Approx. 50% of the ESTs in both libraries did not show any significant homology to any proteins in the NCBI database, indicating a significant fraction of novel and rare transcripts (Table 1). In forward SSHL (ZmSF), alpha amylase (30) was found to be the most abundant EST followed by inositol monophosphatase 3 (12) and tocopherol cyclase (11). Other abundant ESTs of ZmSF involved in plant oxidative stress responses include thioredoxin H-type, aspartic proteinase, NADP-dependent malic enzyme and phosphoenolpyruvate carboxykinase.
Carbohydrate metabolism plays a central role in plant growth and development. Alpha amylase is the key enzyme involved in the hydrolysis of starch. This enzyme is turned on during seed germination and sprouting. Treatment of seeds with 50 mM BABA was found to increase seed germination with reduced incidence of downy mildew in the sunflower plants.49 Moreover, enhanced expression of an alpha amylase gene was evident in water-stressed barley leaves.50 Study by Sato et al.51 revealed dramatic increase in alpha amylase activity and gene expression in hyacinth in response to low temperature. Elevated alpha amylase transcript (AMY1) level was reported in transgenic Arabidopsis plants conferring tolerance against salinity and drought.52 Such increased expression of amylase gene AMY3D was also documented in rice in response to osmotic stress.53 This enhanced amylase activity under stress condition is supposed to play vital role in the recovery of carbohydrates from damaged plant tissue to healthy tissues. In present study, BABA induced overexpression of alpha amylase in maize leaves might provide the energy by hydrolyzing starch into metabolizable sugars to overcome a subsequent stressful condition.
Myo-inositol monophosphatase (IMP) is the key enzyme responsible for both de novo synthesis of myo-inositol as well as breakdown of inositol (1,4,5)-trisphosphate, a second messenger involved in Ca2+ signalling. In plants, IMP is also known to catalyse dephosphorylation of L-galactose 1-phosphate thus participates in ascorbate biosynthesis as well.54,55 The cellular inositol pool plays important role in many metabolic and signalling pathways in plants.56 In plants, myo-inositol serves as the precursor and substrate for many metabolites, like phytate, galactinol, phosphatidylinositol, ascorbate, indole acetic acid conjugate, ononitol, and pinitol. These inositol derivatives are involved in plant stress adaptation in a number of species.57,58 In chickpea, CaIMP plays an essential role in improving seed germination and seedling growth under stressful conditions.59 Moreover, transgenic Arabidopsis over-expressing CaIMP displayed improved tolerance to stress during seed germination and seedling growth. Higher expressing of CaIMP gene has been shown to improve tolerance to drought.60 In our previous study, we have shown that BABA pretreatment could significantly alleviate drought stress impact on maize seedlings.9 Overexpression of inositol monophosphatase 3 in BABA-primed leaves (ZmSF) as revealed in SSH forward library and validated through qRT-PCR might contribute to drought stress acclimation.
Tocopherol cyclase (TC) catalyzes the penultimate step of tocopherol biosynthesis pathway, converting the substrate 2,3-dimethyl-5-phytyl-1,4-benzoquinone to γ-tocopherol.61 In plants, tocopherol is synthesized in the inner envelope of chloroplasts62 and gives protection to the pigments and proteins of the photosynthetic apparatus and of thylakoids lipids against oxidative degradation.63 Being an effective quencher of reactive oxygen species, it protects polyunsaturated fatty acids from lipid peroxidation.64 Enhanced accumulation of tocopherol has been reported in wheat under drought stress.65 Moreover, abiotic stress tolerance and the concentration of α-tocopherol precursor are correlated.66 Overexpression of TC (VTE1) showed enhanced tolerance to drought stress in tobacco67 and to salinity stress in rice.68 Here, tocopherol cyclase (TC) was found to be an abundant EST in forward SSHL (ZmSF), which indicates overexpression of TC in maize leaf under BABA treatment. The higher abundance of this EST is also supported by the observed upregulation of TC gene expression in semi-quantitative RT-PCR (Fig. 7A). Enhanced expression of TC might help the BABA-exposed plant to overcome a succeeding harsh condition possibly by protecting cell membranes from lipid peroxidation.
Among the abundant ESTs associated with plant defense response, thioredoxin H-type (Trx-h) was found to be overexpressed in BABA-primed leaves (ZmSF). qRT-PCR analysis also confirmed this enhanced expression of Trx-h in BABA-primed leaves (Fig. 8E). Thioredoxin family protein is a small redox protein and a key molecule participating in conformation changes of NPR1 (non-expressor of PR1). The shift from NPR1 oligomer to monomer is a requisite to activate a series of PR genes conferring tolerance against a wide range of biotic and abiotic stresses.69 Moreover, the NPR1 is a known master regulator of SA-mediated defense responses.70 Proteomic study by Liu et al.15 also revealed increased thioredoxin activity in the leaves of BABA-treated Arabidopsis thaliana. They suggest that BABA-mediated up-regulation of thioredoxin activates SA responsive genes to mitigate the simulated acid rain stress damages. In present experiment, BABA-potentiated overexpression of thioredoxin might make the maize plants more suitable to handle a subsequent stressful condition.
The aspartic proteases (APs) constitute a large family of proteolytic enzymes and play versatile roles in diverse biological processes including plant growth, development, reproduction, senescence, programmed cell death, as well as plat stress responses.71 Study by Yao et al.72 revealed that overexpression of ASPG1 (ASPARTIC PROTEASE IN GUARD CELL 1) could confer drought avoidance in Arabidopsis via ABA-dependent signaling pathway. Moreover, rice AP gene (OsAP77) has been reported to be involved in defense responses against fungal, bacterial and viral infections.73 A separate study by Timotijević et al.74 demonstrated that aspartic protease (FeAP9) gene from buckwheat resembles the overall exon–intron structure of typical aspartic proteinases. Findings revealed higher expression of FeAP9 gene in buckwheat leaves under the influence of various abiotic stresses like dark, drought and UV-B light, as well as wounding and salicylic acid. Thus, we can also presume that better performance of BABA-primed plants under drought condition9 might be due to enhanced expression of aspartic proteinase in maize leaves under BABA exposure.
The NADP-malic enzyme (NADP-ME) converts L-malate into pyruvate, NADPH and CO2 through oxidative decarboxylation using NADP+ as coenzyme in presence of divalent metal ions.75 It provides the source of CO2 for photosynthesis in the bundle sheath chloroplasts of C4 plants and in the cytosol of CAM plants.76 In addition, it serves as a source of NADPH and pyruvate in the cytosol of various plant tissues. Apart from the diverse housekeeping functions, NADP-ME is known to play crucial role in plant defense by providing building blocks and energy for the biosynthesis of defense compounds.77 Over-expression of NADP-ME2 was reported in transgenic Arabidopsis plants conferring tolerance against salt and osmotic stress.78 In addition, rice NADP-ME genes have been reported to be up-regulated by NaCl stress.79 Decreased stomatal conductance leading to drought avoidance was reported in ME-transformed tobacco.80 Like NADP-ME, phosphoenolpyruvate carboxykinase (PEPCK) is also an important decarboxylation enzyme, which actively participates in photosynthetic CO2-concentrating mechanism in C4 and CAM plants. In bundle sheath cells, PEPCK decarboxylates oxaloacetate releasing carbon dioxide, which is finally fixed by the enzyme Rubisco. In addition, PEPCK has been reported to be involved in stomatal closure through malate metabolism.81 As compared to control, the pck1 mutant Arabidopsis plants with substantially reduced amounts of PEPCK activity,82 showed increased stomatal conductance and wider stomatal apertures resulting reduced drought tolerance. Here, the BABA-primed leaf (ZmSF) showed overexpression of NADP-ME and PEPCK. This enhanced trend is consistent with the qRT-PCR expression data for NADP-ME and PEPCK genes (Fig. 8B and C). The enhanced expression of NADP-ME and PEPCK in BABA-primed maize leaves could have been a defense strategy to overcome a subsequent osmotic stress.
In control leaf SSH library (ZmSR), metal transporter Nramp5 was found to be the most abundant EST (Table 3). This protein plays an essential role in metal homeostasis. In rice, OsNRAMP5 has been identified as Mn, Fe and Cd transporter.83 Suppression of OsNRAMP5 expression through RNAi showed high Cd translocation from root to shoot. OsNRAMP5 knockdown rice plants accumulated more Cd in shoots, which render it a promising candidate protein for Cd phytoremediation.83 BABA has been reported to enhance Cd tolerance in Arabidopsis14 and soybean.13 In present experiment, BABA pre-exposure found to down regulate the Nramp5 (ZmSF). Semi-quantitative PCR analysis showed that expression pattern of Nramp5 largely corroborates with the SSH result. The expression of Nramp5 was high in control leaf but relatively low in BABA-primed leaves (Fig. 7B). BABA-mediated down regulation of Nramp5 might provide tolerance to heavy metal stress particularly Cd. Unknown protein, hypothetical protein, signal anchored and novel proteins were also abundant ESTs in control leaf SSH library (ZmSR). Hypothetical protein exhibited similarity with the dehydration responsive protein which might render control plants drought susceptible.
Gene ontology (GO) annotations of all the subtractive ESTs were performed using the Blast2GO program.43 In ZmSF, functional classification of ESTs in biological process categories revealed that metabolic process (GO:0008152), cellular process (GO:0009987), response to temperature stimulus (GO:0009266), response to stress (GO:0006950), biosynthetic process (GO:0009058), response to abiotic stimulus (GO:0009628), signal transduction (GO:0007165), inositol phosphate dephosphorylation (GO:0046855) and phloem sucrose loading (GO:0009915) were the highly represented groups (ESI Table 14†). Presence of these well-represented categories indicate that BABA-primed plants is getting ready to tackle stressful condition. In molecular function, sequences related to cation binding (GO:0043169), nucleic acid binding (GO:0003676), hydrolase activity, acting on acid anhydrides (GO:0016817), amylase activity (GO:0016160), hydrolase activity, acting on ester bonds (GO:0016788), tocopherol cyclase activity (GO:0009976), hydrolase activity, acting on glycosyl bonds (GO:0016798) and phosphatase activity (GO:0016791) were the abundant categories (ESI Table 15†). In ZmSF, ESTs belonging to major subgroups of cellular components included sequences related to nucleus (GO:0005634), extracellular region (GO:0005576), mitochondrion (GO:0005739), plasma membrane (GO:0005886), cytosol (GO:0005829), plastoglobule (GO:0010287), chloroplast thylakoid (GO:0009534) and chloroplast envelope (GO:0009941) (ESI Table 16†). In ZmSR, functionally annotated ESTs were assigned to transport (GO:0006810) in terms of biological process (ESI Table 17†). In molecular function, transcripts belonged to transporter activity (GO:0005215) and zinc ion binding (GO:0008270) were major categories (ESI Table 18†). In ZmSR, ESTs related to mitochondrion (GO:0005739), integral component of membrane (GO:0016021) and chloroplast (GO:0009507) were major subgroups of cellular components (ESI Table 19†).
In ZmSF, functionally annotated ESTs were found to be involved in ten pathways based on KEGG analysis including starch and sucrose metabolism, inositol phosphate metabolism, phosphatidylinositol signaling system, aminobenzoate degradation, streptomycin biosynthesis, biosynthesis of antibiotics, pyruvate metabolism, glycolysis/gluconeogenesis, carbon fixation in photosynthetic organisms, and citrate cycle. In addition, it had been observed that maximum numbers of ESTs were associated with starch and sucrose metabolism followed by inositol metabolism, biosynthesis of antibiotics, pyruvate metabolism, glycolysis/gluconeogenesis, carbon fixation in photosynthetic organisms, and citrate cycle pathway (Fig. 6; ESI Table 13†). Interestingly, functionally annotated ESTs of ZmSR was not assigned with any KEGG pathway. Presence of ESTs linked with diverse pathways related to carbohydrate metabolism in the BABA primed SSH library, might help in plant growth and development under subsequent adverse conditions.
To further elucidate the functionally annotated ESTs, InterPro scan analysis for protein domain detection was performed. In ZmSF, total eight protein domains were detected (ESI Table 12†), whereas in ZmSR no protein domain was identified. In ZmSF, maximum protein domains are associated with the glycosyl hydrolase (all-beta) and alpha-amylase (C-terminal beta-sheet) domain (ESI Table 12†). These domains are found in several members of the glycosyl hydrolase family 13 that hydrolyse the glycosidic bonds of glycogen, starch and related polysaccharides. Peptidase family A1 domain and Aspartic peptidase domain protein belongs to aspartic peptidase A1 family members, also known as aspartyl proteases. BABA pre-treatment was shown to induce resistance against Phytophthora infestans and Fusarium solani in potato through higher accumulation of StAP1 (aspartyl protease) protein.84 Other protein domains were associated with NAD(P) malic enzyme. These domains represent NAD- and NADP-binding domains with a core Rossmann-type fold consisting of 3-layers alpha/beta/alpha. Malic enzymes (malate oxidoreductases) accelerate the oxidative decarboxylation of malate to form pyruvate. This reaction is important in a number of metabolic pathways. The carbon dioxide released from the reaction may be used in sugar production in Calvin cycle.85 NADP-ME transcripts accumulation has been found to be increased during salt stress in Mesembryanthemum crystallinum.86 Phosphoenolpyruvate carboxykinase, N-terminal was associated with the N-terminal domain found in both GTP-utilising and ATP-utilising phosphoenolpyruvate carboxykinase enzymes. It has an alpha/beta topology. Phosphoenolpyruvate carboxykinase (PEPCK) catalyzes the decarboxylation of oxaloacetate to phosphoenolpyruvate and carbon dioxide by using either ATP or GTP as a source of phosphate in gluconeogenesis. In interpreting our SSH results, we have proposed a comprehensive hypothetical model about the possible roles of BABA responsive genes in regulating BABA potentiated defense to confer tolerance against various environmental challenges (Fig. 10).
 | ||
| Fig. 10 Schematic illustration summarizing significantly changed BABA-responsive genes and their involvement in stress mitigation. | ||
Plant responds to drought by controlling the rate of water loss from leaves through modulating stomatal aperture size. Potassium plays key role during stomatal opening. The entry of potassium via specific potassium channel in guard cells is followed by accumulation of malate and chloride, resulting opening of stomata.87 In C4 plants like maize, the NADP-ME converts malate into pyruvate, NADPH and CO2 via oxidative decarboxylation reaction. Growing evidence indicates that such NADP-ME mediated malate degradation in guard cells may lead to stomatal closure.88 Transgenic tobacco plants overexpressing maize NADP-ME showed drought tolerance through altered malate metabolism in guard cells.89 Findings reveal that ME-regulated stomatal closure and altered plant water-use efficiency might be due to sudden decline in the intracellular malate concentration in guard cells rendering them unavailable for proper stomatal opening or due to elevated CO2 level in the whole leaves as result of ME expression in mesophyll cells. Our previous study revealed that BABA pre-exposed maize plants (BABA-primed) performed better under drought stress than non-primed plants.9 In the present drought experiment, elevated NADP-ME transcript level in BABA primed plants (Fig. 9B) might confer tolerance against limited water condition through NADP-ME mediated altered malate metabolism. Moreover, the CO2 generated during conversion of malate to pyruvate might contribute to increase in internal CO2 level, which renders BABA primed plants more drought tolerant by decreasing stomatal conductance. In addition, stomatal closure under reduced water availability is driven by ABA hormone. Recent study by Yao et al.72 suggests that overexpression of APs confer drought avoidance in Arabidopsis through ABA signalling in guard cells. Comparatively, high APs expression might also help the BABA-primed plants (Fig. 9E) to mitigate drought stress by reducing water loss through stomatal closure. Higher abundance of Trx-h transcript (Fig. 9C) might be another determining factor for better performance of BABA-primed maize plants in combating drought induced oxidative stress damages. In interpreting our data, we have proposed a hypothetical model for drought tolerance mechanism in maize by taking into account the possible roles of BABA-responsive genes in different pathways leading to stress tolerance under reduced availability of water (Fig. 11).
Conclusion
The present study was conducted with the aim to identify rare and differentially expressed transcripts in BABA-primed maize leaf. Findings revealed that a number of stress-related genes were upregulated by BABA. A total 204 ESTs from reverse and 192 ESTs from forward libraries were identified and analyzed. BLASTX data revealed that 130 ESTs (63.72%) from ZmSR and 62 ESTs (32.29%) from ZmSF encoded for polypeptides/proteins (known or putative proteins). Contigs assembly provided 17 unique clones from ZmSF and 4 unique clones from ZmSR library. A total of 31 unigenes were identified to be up- or down-regulated by BABA. Of these 31 genes, 9 genes could be classified into different functional categories based on their putative roles. It is likely that those genes, which were up- or down-regulated by exogenous BABA treatment, might have relevance, since BABA plays an essential role in adaptive responses to environmental stimuli and many developmental processes in plants. Functional annotation identified some secondary metabolic pathway genes, which might have a regulatory role in the inositol phosphate metabolism, glycolysis/gluconeogenesis, aminobenzoate degradation, starch and sucrose metabolism, pyruvate metabolism, carbon fixation, phosphatidylinositol signaling system and citrate cycle. Some ESTs did not exhibit any significant similarity to any proteins in the NCBI database, indicating a significant fraction of novel and rare expressed transcripts. Our study revealed that BABA responsive genes were involved in diverse biological and metabolic processes such as seed germination, seedling growth, phosphatidylinositol signalling, sucrose metabolism, pyruvate metabolism, glycolysis/gluconeogenesis, photosynthesis and citrate cycle. Moreover, the functionally annotated ESTs were found to be involved in stress mitigation e.g. drought (alpha amylase, inositol monophosphatase, tocopherol cyclase, aspartic proteinase, NADP-malic enzyme, phosphoenolpyruvate carboxykinase), salinity (alpha amylase, NADP-malic enzyme, tocopherol cyclase), osmotic stress (NADP-malic enzyme), disease resistance (aspartic proteinase). All these valuable information provide a first step for further characterization of BABA-mediated candidate genes involved in various stress tolerance. Moreover, further research needs to be done for novel and rare expressed transcripts to elucidate their function in BABA-potentiated stress adaptation mechanism. All the information would not only enrich us in deeper understanding of BABA-mediated plant response to environmental stressors but would also enable us to design genetically engineered stress tolerant crops. Overall, our study adds new insights into the BABA potentiated defense mechanisms in plants.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This research was supported by grant from the Council of Scientific & Industrial Research (CSIR), New Delhi, India through R&D project 38(1309)/11/EMR-II. Part of the work was done in West Bengal State University, when the corresponding author (ZH) was a faculty there. We are thankful to the Director, Indian Institute of Maize Research (IIMR), Pusa Campus, New Delhi, India for providing maize seeds. Financial supports from DST-FIST, Department of Botany and DST-PURSE Programme of University of Kalyani are also duly acknowledged. The authors A. K. S., I. A., S. G., S. A., A. A. thankfully acknowledge the financial support provided by the Department of Science & Technology, New Delhi as DST INSPIRE SRF, Council of Scientific and Industrial Research (CSIR), New Delhi as CSIR-JRF and SRF, Science and Engineering Research Board (SERB), New Delhi as N-PDF and University of Kalyani as University Research Scholar respectively.References
- D. Macarisin, M. E. Wisniewski, C. Bassett and T. W. Thannhauser, Plant, Cell Environ., 2009, 32, 1612–1631 CrossRef CAS.
- G. J. Beckers, M. Jaskiewicz, Y. Liu, W. R. Underwood, S. Y. He, S. Zhang and U. Conrath, Plant Cell, 2009, 21, 944–953 CrossRef CAS PubMed.
- U. Conrath, G. J. M. Beckers, V. Flors, P. García-Agustín, G. Jakab, F. Mauch, M. A. Newman, C. M. Pieterse, B. Poinssot, M. J. Pozo, A. Pugin, U. Schaffrath, J. Ton, D. Wendehenne, L. Zimmerli and B. Mauch-Mani, Mol. Plant Microbe Interact., 2006, 19, 1062–1071 CrossRef CAS PubMed.
- I. Baccelli and B. Mauch-Mani, Plant Mol. Biol., 2016, 91, 703–711 CrossRef CAS PubMed.
- G. C. Papavizas, Plant Dis. Rep., 1964, 48, 537–541 CAS.
- L. Zimmerli, G. Jakab, J. P. Metraux and B. Mauch-Mani, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 12920–12925 CrossRef CAS PubMed.
- J. Ton and B. Mauch-Mani, Plant J., 2004, 38, 119–130 CrossRef CAS PubMed.
- J. Ton, G. Jakab, V. Toquin, V. Flors, A. Iavicoli, M. N. Maeder, J. P. Metraux and B. Mauch-Mani, Plant Cell, 2005, 17, 987–999 CrossRef CAS PubMed.
- A. K. Shaw, P. K. Bhardwaj, S. Ghosh, S. Roy, S. Saha, A. R. Sherpa, S. K. Saha and Z. Hossain, Environ. Sci. Pollut. Res., 2016, 23, 2437–2453 CrossRef CAS PubMed.
- A. Sos-Hegedus, Z. Juhasz, P. Poor, M. Kondrak, F. Antal, I. Tari, B. Mauch-Mani and Z. Banfalvi, PLoS One, 2014, 9, e114297 Search PubMed.
- Y. L. Du, Z. Y. Wang, J. W. Fan, N. C. Turner, T. Wang and F. M. Li, J. Exp. Bot., 2012, 63, 4849–4860 CrossRef CAS PubMed.
- G. Jakab, J. Ton, V. Flors, L. Zimmerli, J. P. Métraux and B. Mauch-Mani, Plant Physiol., 2005, 139, 267–274 CrossRef CAS PubMed.
- Z. Hossain, T. Makino and S. Komatsu, J. Proteomics, 2012, 75, 4151–4164 CrossRef CAS PubMed.
- S. Q. Cao, G. Ren, L. Jiang, H. B. Yuan and G. H. Ma, Russ. J. Plant Physiol., 2009, 56, 575–579 CrossRef CAS.
- T. Liu, X. Jiang, W. Shi, J. Chen, Z. Pei and H. Zheng, Proteomics, 2011, 11, 2079–2094 CrossRef CAS PubMed.
- D. V. Rebrikov, Cold Spring Harb Protoc., 2008, pdb.top21 Search PubMed.
- M. Clement, A. Lambert, D. Herouart and E. Boncompagni, Gene, 2008, 426, 15–22 CrossRef CAS PubMed.
- L. Diatchenko, Y. L. Lau, A. P. Campbell, A. Chenchik, F. Moqadam, B. Huang, S. Lukyanov, K. Lukyanov, N. Gurskaya, E. D. Sverdlov and P. D. Siebert, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 6025–6030 CrossRef CAS.
- B. Gurung, P. K. Bhardwaj and N. C. Talukdar, Funct. Integr. Genomics, 2016, 16, 619–639 CrossRef CAS PubMed.
- D. Divya, T. Singh, S. Nair and J. S. Bentur, Funct. Integr. Genomics, 2016, 16, 153–169 CrossRef CAS PubMed.
- L. K. Narnoliya, R. Rajakani, N. S. Sanwan, V. Gupta and R. S. Sangwan, Mol. Biol. Rep., 2014, 41, 413147–413162 CrossRef PubMed.
- P. K. Bhardwaj, R. Kapoor, D. Mala, G. Bhagwat, V. Acharya, A. K. Singh, S. K. Vats, P. S. Ahuja and S. Kumar, Sci. Rep., 2013, 3, 1022 CrossRef PubMed.
- S. Kumar, S. Kalra, S. Kumar, J. Kaur and K. Singh, Gene, 2012, 511, 79–87 CrossRef CAS PubMed.
- B. Ouyang, T. Yang, H. Li, L. Zhang, Y. Zhang, J. Zhang, Z. Fei and Z. Ye, J. Exp. Bot., 2007, 58, 507–520 CrossRef CAS PubMed.
- Y. Y. Wong, C. L. Ho, P. D. Nguyen, S. S. Teo, J. A. Harikrishna, R. A. Rahim and M. C. V. L. Wong, Aquat. Bot., 2007, 86, 117–122 CrossRef CAS.
- B. B. Sahu and B. P. Shaw, BMC Plant Biol., 2009, 9, 69 CrossRef PubMed.
- Q. Dai, B. Huang, Z. Yang, J. Yuan and J. Yang, Front. Environ. Sci. Eng., 2010, 4, 449 CrossRef CAS.
- H. S. Aguilar-Hernández, L. Santos, F. León-Galván, A. Barrera-Pacheco, E. Espitia-Rangel, A. De León-Rodríguez, R. G. Guevara-Gonzálezc and A. P. B. de la Rosaa, J. Plant Physiol., 2011, 168, 2102–2109 CrossRef PubMed.
- H. Ding, Z. M. Zhang, F. F. Qin, L. X. Dai, C. J. Li, D. W. Ci and W. W. Song, Electron. J. Biotechnol., 2014, 7, 304–310 CrossRef.
- D. Jia, B. Zhang, P. P. Zhang, J. Y. Zhang, Y. H. Liu, J. S. Wang and R. Y. Ma, Russ. J. Plant Physiol., 2015, 62, 93 CrossRef CAS.
- H. Sävestrand, M. Brosché, M. Ängehagen and A. Strid, Plant, Cell Environ., 2000, 23, 689–700 CrossRef.
- H. T. Nguyen, J. Leipner, P. Stamp and O. Guerra-Peraza, Plant Physiol. Biochem., 2009, 47, 116–122 CrossRef CAS PubMed.
- X. Zou, Y. Jiang, L. Liu, Z. Zhang and Y. Zheng, BMC Plant Biol., 2010, 10, 189 CrossRef PubMed.
- G. F. Zhou, Y. Z. Liu, O. Sheng, Q. J. Wei, C. Q. Yang and S. A. Peng, Front. Plant Sci., 2015, 5, 795 Search PubMed.
- W. L. Guo, R. G. Chen, Z. H. Gong, Y. X. Yin and D. W. Li, PLos One, 2013, 8, e66667 CAS.
- E. Luna, M. van Hulten, Y. Zhang, O. Berkowitz, A. López, P. Pétriacq, M. A. Sellwood, B. Chen, M. Burrell, A. van de Meene, C. M. J. Pieterse, V. Flors and J. Ton, Nat. Chem. Biol., 2014, 10, 450–456 CrossRef CAS PubMed.
- P. Singh, Y. C. Kuo, S. Mishra, C. H. Tsai, C. C. Chien, C. W. Chen, M. Desclos-Theveniau, P. W. Chu, B. Schulze, D. Chinchilla, T. Boller and L. Zimmerli, Plant Cell, 2012, 24, 1256–1270 CrossRef CAS PubMed.
- C. Po-Wen, P. Singh and L. Zimmerli, Mol. Plant Pathol., 2013, 14, 58–70 CrossRef PubMed.
- L. Zimmerli, B. H. Hou, C. H. Tsai, G. Jakab, B. Mauch-Mani and S. Somerville, Plant J., 2008, 53, 144–156 CrossRef CAS PubMed.
- C. Tsai, P. Singh, C. Chen, J. Thomas, J. Weber, B. Mauch-Mani and L. Zimmerli, Plant J., 2011, 65, 469–479 CrossRef CAS PubMed.
- S. Van der Ent, M. Van Hulten, M. J. Pozo, T. Czechowski, M. K. Udvardi, C. M. Pieterse and J. Ton, New Phytol., 2009, 183, 419–431 CrossRef CAS PubMed.
- S. Ghawana, A. Paul, H. Kumar, A. Kumar, H. Singh, P. K. Bhardwaj, A. Rani, R. S. Singh, J. Raizada, K. Singh and S. Kumar, BMC Res. Notes, 2011, 4, 85 CrossRef CAS PubMed.
- A. Conesa, S. Gotz, J. Garcia-Gomez, J. Terol, M. Talon and M. Robles, Bioinformatics, 2005, 21, 3674–3676 CrossRef CAS PubMed.
- K. J. Livak and T. D. Schmittgen, Methods, 2001, 25, 402–408 CrossRef CAS PubMed.
- F. He, Y. Zhu and Y. Zhang, Plant Cell Rep., 2008, 27, 923–930 CrossRef CAS PubMed.
- R. Rajakani, L. Narnoliya, N. S. Sangwan, R. S. Sangwan and V. Gupta, Tree Genet. Genomes, 2014, 10, 1331–1351 CrossRef.
- D. Ding, Z. Xiao, H. Xiao, T. Xia, Y. L. Zheng and F. Qiu, Genes Genomics, 2012, 34, 265–273 CrossRef CAS.
- J. Zheng, J. Zhao, Y. Tao, J. Wang, Y. Liu, J. Fu, Y. Jin, P. Gao, J. Zhang, Y. Bai and G. Wang, Plant Mol. Biol., 2004, 55, 807–823 CrossRef CAS PubMed.
- P. Nandeeshkumar, B. R. Sarosh, K. R. Kini, H. S. Prakash and H. S. Shetty, Arch. Phytopathol. Plant Prot., 2009, 42, 1020–1032 CrossRef CAS.
- J. V. Jacobsen, A. D. Hanson and P. M. Chandler, Plant Physiol., 1986, 80, 350–359 CrossRef CAS PubMed.
- A. Sato, H. Okubo and K. Saitou, J. Am. Soc. Hortic. Sci., 2006, 131, 185–191 CAS.
- H. S. Ma, D. Liang, P. Shuai, X. L. Xia and W. L. Yin, J. Exp. Bot., 2010, 61, 4011–4019 CrossRef CAS PubMed.
- Y. S. Hwang, E. E. Karrer, B. R. Thomas, L. Chen and R. L. Rodriguez, Plant Mol. Biol., 1998, 36, 331–341 CrossRef CAS PubMed.
- W. A. Laing, S. Bulley, M. Wright, J. Cooney, D. Jensen, D. Barraclough and E. MacRae, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 16976–16981 CrossRef CAS PubMed.
- J. Torabinejad, J. L. Donahue, B. N. Gunesekera, M. J. Allen-Daniels and G. E. Gillaspy, Plant Physiol., 2009, 150, 951–961 CrossRef CAS PubMed.
- F. A. Loewus and M. W. Loewus, Annu. Rev. Plant Physiol., 1983, 34, 137–161 CrossRef CAS.
- R. F. Irvine and M. J. Schell, Nat. Rev. Mol. Cell Biol., 2001, 2, 327–338 CrossRef CAS PubMed.
- E. A. Kido, J. R. Ferreira Neto, R. L. Silva, L. C. Belarmino, J. P. Bezerra Neto, N. M. Soares-Cavalcanti, V. Pandolfi, M. D. Silva, A. L. Nepomuceno and A. M. Benko-Iseppon, BMC Bioinf., 2013, 14, S7 CrossRef CAS PubMed.
- S. C. Saxena, P. Salvi, H. Kaur, P. Verma, B. P. Petla, V. Rao, N. Kamble and M. Majee, J. Exp. Bot., 2013, 64, 5623–5639 CrossRef CAS PubMed.
- A. Joshi-Saha and K. S. Reddy, J. Exp. Bot., 2015, 66, 5683–5690 CrossRef CAS PubMed.
- S. E. Sattler, E. B. Cahoon, S. J. Coughlan and D. DellaPenna, Plant Physiol., 2003, 132, 2184–2195 CrossRef CAS PubMed.
- H. Lichtenthaler, U. Prenzel, R. Douce and J. Joyard, Biochim. Biophys. Acta, 1981, 641, 99–105 CrossRef CAS.
- M. Kanwischer, S. Porfirova, E. Bergmuller and P. Dormann, Plant Physiol., 2005, 137, 713–723 CrossRef CAS PubMed.
- P. M. Bramley, I. Elmadfa, A. Kafatos, F. J. Kelly, Y. Manios, H. E. Roxborough, W. Schuch, P. J. A. Sheehy and K. H. Wagner, J. Sci. Food Agric., 2000, 80, 913–938 CrossRef CAS.
- H. B. Shao, L. Y. Chu, G. Wu, J. H. Zhang, Z. H. Lu and Y. C. Hu, Colloids Surf., B, 2007, 54, 143–149 CrossRef CAS PubMed.
- L. Pourcel, J. M. Routaboul and V. Cheynier, Trends Plant Sci., 2007, 12, 29–36 CrossRef CAS PubMed.
- X. Liu, X. Hua, J. Guo, D. Qi, L. Wang, Z. Liu, Z. Jin, S. Chen and G. Liu, Biotechnol. Lett., 2008, 30, 1275–1280 CrossRef CAS PubMed.
- S. Q. Ouyang, S. J. He, P. Liu, Z. Wanke, Z. Jinsong and C. Shouyi, Sci. China: Life Sci., 2011, 54, 181–188 CrossRef CAS PubMed.
- M. Kinkema, W. Fan and X. Dong, Plant Cell, 2000, 12, 2339–2350 CrossRef CAS.
- Z. Mou, W. Fan and X. Dong, Cell, 2003, 113, 935–944 CrossRef CAS PubMed.
- I. Simoes and C. Faro, Eur. J. Biochem., 2004, 271, 2067–2075 CrossRef CAS PubMed.
- X. Yao, W. Xiong, T. Ye and Y. Wu, J. Exp. Bot., 2012, 63, 2579–2593 CrossRef CAS PubMed.
- M. M. Alam, H. Nakamura, H. Ichikawa, A. Miyao, H. Hirochika, K. Kobayashi, N. Yamaoka and M. Nishiguchi, Rice, 2014, 7, 9 CrossRef PubMed.
- G. S. Timotijević, M. D. J. Milisavljević, S. R. Radović, M. M. Konstantinović and V. R. Maksimović, J. Plant Physiol., 2010, 167, 61–68 CrossRef PubMed.
- M. F. Drincovich, P. Casati and C. S. Andreo, FEBS Lett., 2001, 490, 1–6 CrossRef CAS PubMed.
- G. E. Edwards and C. S. Andreo, Phytochemistry, 1992, 31, 1845–1857 CrossRef CAS PubMed.
- P. Casati, M. F. Drincovich, G. E. Edwards and C. S. Andreo, Photosynth. Res., 1999, 61, 99–105 CrossRef CAS.
- S. Liu, Y. Cheng, X. Zhang, Q. Guan, S. Nishiuchi, K. Hase and T. Takano, Plant Mol. Biol., 2007, 64, 49–58 CrossRef CAS PubMed.
- W. Chi, J. H. Yang, N. H. Wu and F. Zhang, Biosci., Biotechnol., Biochem., 2004, 68, 1865–1874 CrossRef CAS PubMed.
- M. M. Laporte, B. Shen and M. C. Tarczynski, J. Exp. Bot., 2002, 53, 699–705 CrossRef CAS PubMed.
- S. Penfield, S. Clements, K. J. Bailey, A. D. Gilday, R. C. Leegood, J. E. Gray and I. A. Graham, Plant J., 2012, 69, 679–688 CrossRef CAS PubMed.
- S. Penfield, E. L. Rylott, A. D. Gilday, S. Graham, T. R. Larson and I. A. Graham, Plant Cell, 2004, 16, 2705–2718 CrossRef CAS PubMed.
- Y. Ishimaru, R. Takahashi, K. Bashir, H. Shimo, T. Senoura, K. Sugimoto, K. Ono, M. Yano, S. Ishikawa, T. Arao, H. Nakanishi and N. K. Nishizawa, Sci. Rep., 2012, 2, 286 CrossRef PubMed.
- F. P. Olivieri, M. C. Lobato, E. Gonzalez Altamiranda, G. R. Daleo, M. Huarte, M. G. Guevara and A. B. Andreu, Eur. J. Plant Pathol., 2009, 123, 47–56 CrossRef CAS.
- J. J. Long, J. L. Wangs and J. O. Berry, J. Biol. Chem., 1994, 269, 2821–2833 Search PubMed.
- J. C. Cusiiman, Eur. J. Biochem., 1992, 208, 259–266 CrossRef.
- E. Zeiger, Annu. Rev. Plant Physiol., 1983, 34, 441–475 CrossRef CAS.
- W. H. Outlaw, J. Manchester and P. H. Brown, Plant Physiol., 1981, 68(5), 1047–1051 CrossRef CAS PubMed.
- M. M. Laporte, B. Shen and M. C. Tarczynski, J. Exp. Bot., 2002, 53(369), 699–705 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra06220f |
| This journal is © The Royal Society of Chemistry 2017 |