Open Access Article
Open Access ArticleBis-isoquinolinium and bis-pyridinium acetylcholinesterase inhibitors: in vitro screening of probes for novel selective insecticides†
Veronika Hrabcovaab,
Jan Korabecnybc,
Brigita Manyovabd,
Lenka Matouskovabd,
Tomas Kucerab,
Rafael Dolezalac,
Kamil Musilekabc,
Lukas Goreckibc,
Eugenie Nepovimovabc,
Kamil Kucaabc and
Daniel Jun *bc
*bc
aUniversity Hradec Kralove, Faculty of Science, Department of Biology, Rokitanskeho 62, 50003 Hradec Kralove, Czech Republic
bUniversity of Defence, Faculty of Military Health Sciences, Department of Toxicology and Military Pharmacy, Trebesska 1575, 500 01 Hradec Kralove, Czech Republic. E-mail: daniel.jun@unob.cz; Tel: +420 973 255 193
cUniversity Hospital, Biomedical Research Centre, Sokolska 581, 500 05 Hradec Kralove, Czech Republic
dCharles University, Faculty of Pharmacy, Department of Pharmacology and Toxicology, Akademika Heyrovskeho 1203, 500 05 Hradec Kralove, Czech Republic
First published on 10th August 2017
Abstract
Insects have a huge impact on quality of life around the world. They play roles in transmitting diseases, crop-destruction, and as residential pests. Their increased resistance to the existing pesticides and the toxicity of carbamates (CA) and organophosphates (OP) has led to the development of new environmentally safe insecticides. In our study, thirty different bis-isoquinolinium and bis-pyridinium acetylcholinesterase inhibitors were tested as candidates for potential new selective pesticides. Our compounds were evaluated in vitro on insect acetylcholinesterase (AChE) from Musca domestica and human erythrocyte AChE using the modified Ellman's method. The values of IC50 were compared and expressed via a selectivity index (SI) towards the insect enzyme. K299, K416, and K423 provided a high SI and seem to be suitable as new lead structures of novel selective anticholinesterase insecticides. Docking studies further provided the rational background uncovering the disparities in the ligand–enzyme binding modes for each AChE enzyme. In vitro assessments as well as docking studies highlighted K299 and K416 as suitable candidates for lead structures of novel pesticides. However, further evaluation is needed to confirm this statement.
Introduction
Vector-borne diseases are among the major causes of illness and death in many tropical and subtropical countries.1 In 2015, 214 million new cases of malaria, transmitted by Anopheles gambiae, globally occurred, and the disease led to 438, 000 deaths.2 Dengue fever as well as Zika virus disease is transmitted by another mosquito species, Aedes aegypti. Currently, Zika virus infections indicate rapid geographic expansion.3 Worldwide present bed bugs (Cimex lectularius) feed on human blood and cause mental stress, acute discomfort, and social stigma.4 Finally, soybean aphids (Aphis glycines) cost US farmers more than 1 billion dollars in yield losses and the purchase and application of insecticides, hampering the production of biodiesel from soybean oil.5 All the problems caused by crop-destroying, disease-carrying, and residential insects require urgent solutions.6Carbamates (CA) and organophosphates (OP) are examples of pesticides that inhibit cholinesterases, which are commonly used for insect control.7–9 Acetylcholinesterase (AChE; EC 3.1.1.7) is a serine esterase in the α/β hydrolase-fold enzyme family.10 The function of AChE is to terminate neurotransmission by rapidly hydrolyzing acetylcholine (ACh) at the cholinergic synapses and neuromuscular junctions of most invertebrates and vertebrates.7,11–13 The active site of the enzyme is located at the bottom of a deep and narrow gorge, containing a serine residue in a catalytic triad.7,13 Current anticholinesterase insecticides work through covalent modification of serine at the active site of an enzyme, thus disabling its catalytic function and incapacitating insects.7–9
Due to the long-lasting consumption of pesticides, more and more insect species have developed resistance.14–16 This resistance has been claimed to be due to three different mechanisms: reduction in insecticide sensitivity of target enzymes, decrease in insecticide penetration into the insect body, and increase in detoxification activities.17 On the other hand, acute pesticide poisonings (APPs) are responsible for significant morbidity and mortality worldwide, particularly in developing countries.18 Estimates of the total number of poisonings are quite variable, depending on the period and author. Despite some limitations, a total number of APPs in the range from 250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 500
000 to 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 were estimated yearly with 3000–30
000 were estimated yearly with 3000–30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 fatalities.19 Due to the increasing insect resistance to pesticides, off-target toxicity, and several epidemic situations of insect-transmitted diseases, the development of new selective insecticides is needed.
000 fatalities.19 Due to the increasing insect resistance to pesticides, off-target toxicity, and several epidemic situations of insect-transmitted diseases, the development of new selective insecticides is needed.
Our aim was to screen selected recently reported AChE inhibitors from our in-house library20–22 and find out if any of them provided selectivity to insect AChE. These selective inhibitors could be used as new structure leads for further research. Approximately thirty different symmetrical bis-quaternary AChE inhibitors, namely bis-isoquinolinium and bis-pyridinium salts, were evaluated as new pesticide candidates, and the selectivity of these compounds was determined. Due to the fact that Musca domestica has just one type of AChE responsible for cholinergic function,11 it has been chosen as a suitable source of insect enzyme. Finally, in silico studies were carried out to justify the results obtained from the in vitro assessment. Predictive binding poses of the selected ligands are shown in the cavity of greenbug AChE (Schizaphis graminum, SgAChE; enzyme structurally related to Anopheles gambiae and/or Aedes aegypti AChE) to validate our hypothesis for the compounds under study.
Results and discussion
Biological assay
Herein, thirty different symmetrical bis-quaternary AChE inhibitors were assayed for their selectivity using the modified spectrophotometric method described by Ellman et al.23–25 The validity of our protocol has been verified on the anti-Alzheimer's disease drug tacrine, a well-known AChE inhibitor (data not shown in the table).26,27 Musca domestica was used as a suitable source of the insect AChE. Anopheles gambiae, a mosquito that transmits malaria, has two types of AChE: paralogous AChE (AP-AChE) to the Drosophila melanogaster gene, which is responsible for cholinergic function in mosquito, and orthologous AChE (AO-AChE), which plays a crucial role in the process of reproduction. On the other hand, Musca domestica, which belongs to the true fly species, has only one type of AChE, AO-AChE, in particular.6The compounds chosen for selectivity screening can be divided into two groups based on their structural features. All these structures have been recently reported as potential candidates for the treatment for AD or myasthenia gravis.20–22 The first group consisted of symmetrical bis-pyridinium compounds (see Fig. 1), which was selected based on the linker length and its previously reported inhibition effect. The IC50 values are listed in Table 1.
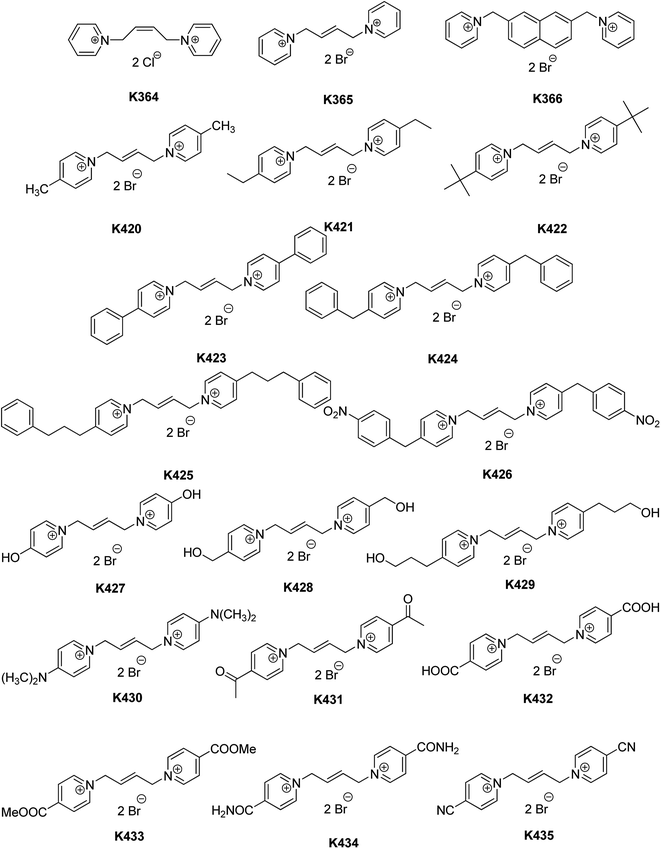 | ||
| Fig. 1 Structures of the evaluated standards and bis-pyridinium compounds (non-substituted and p-substituted). | ||
| Compound | IC50 hAChE ± SEMa (μM) | IC50 MdAChE ± SEMa (μM) | SI for MdAChE |
|---|---|---|---|
| a The IC50 values are derived from triplicate and selectivity indexes (SI) were calculated as a ratio of IC50 value for hAChE/IC50 value for MdAChE.b Data were obtained from ref. 30.c Data were obtained from ref. 31. | |||
| Propoxur | 2.094 ± 0.040b | 0.152c | 13.78 |
| Bendiocarb | 0.359 ± 0.016b | 0.058c | 6.19 |
| Compound 1 | 0.201 ± 0.019b | 0.181c | 1.11 |
| Compound 7 | 20.68 ± 9.509b | 0.051c | 405 |
| Compound 8 | 247.4 ± 106.8b | 0.654c | 378 |
| K291 | >1000 | >1000 | — |
| K293 | >1000 | >1000 | — |
| K296 | 0.881 ± 0.322 | 2.77 ± 0.08 | 0.318 |
| K299 | 0.138 ± 0.008 | 0.0226 ± 0.0006 | 6.093 |
| K302 | 0.0697 ± 0.0032 | 0.036 ± 0.001 | 1.936 |
| K364 | >1000 | >1000 | — |
| K365 | >1000 | >1000 | — |
| K366 | 8.01 ± 1.30 | 14.1 ± 1.5 | 0.568 |
| K412 | 5.10 ± 0.39 | 7.40 ± 0.39 | 0.690 |
| K413 | >1000 | 693 ± 59 | — |
| K414 | 15.78 ± 1.12 | 17.6 ± 2.1 | 0.897 |
| K415 | 38.7 ± 4.4 | 112 ± 11 | 0.346 |
| K416 | 267 ± 11 | 21.9 ± 4.7 | 12.185 |
| K417 | 8.59 ± 0.86 | 1.79 ± 0.12 | 4.789 |
| K418 | 5.87 ± 0.49 | 5.56 ± 0.49 | 1.056 |
| K419 | 0.214 ± 0.011 | 0.228 ± 0.009 | 0.936 |
| K420 | >1000 | >1000 | — |
| K421 | >1000 | >1000 | — |
| K422 | >1000 | >1000 | — |
| K423 | 4.93 ± 3.61 | 0.699 ± 0.089 | 7.058 |
| K424 | 4.09 ± 1.01 | >1000 | — |
| K425 | 0.0554 ± 0.0029 | >1000 | — |
| K426 | 5.19 ± 0.56 | 20.7 ± 2.8 | 0.251 |
| K427 | >1000 | >1000 | — |
| K428 | >1000 | >1000 | — |
| K429 | 31.5 ± 2.0 | 107 ± 15 | 0.294 |
| K430 | 1.05 ± 0.06 | 4.14 ± 0.23 | 0.253 |
| K431 | >1000 | >1000 | — |
| K432 | >1000 | >1000 | — |
| K433 | >1000 | >1000 | — |
| K434 | >1000 | >1000 | — |
| K435 | 37.3 ± 3.3 | 103 ± 11 | 0.362 |
Initially, K366 was the non-substituted bis-pyridinium salt connected with a naphthylene linkage. The IC50 value of K366 was in the micromolar range for both types of AChE. However, selectivity towards MdAChE was very low. K364 and K365 are (E) and (Z) non-substituted bispyridinium isomers with a but-(2E)-en-1,4-diyl linker, respectively. They displayed no inhibition effect in the tested concentration scale. The aromatic linkage displayed a promising inhibitory effect, opposite to the linker with a double bond. On the other hand, the substitution at the para position of bis-pyridinium inhibitors with the but-(2E)-en-1,4-diyl linker diminished the inhibitory ability.
para-Substituted bispyridinium inhibitors (K420–K435) provided a very wide inhibition effect based on the used substituent. Lipophilic moieties, such as aliphatic functional groups (K420–K422), resulted in poor inhibition as compared to the compounds with an aromatic ring. Among the aromatic functional groups (K423–K426), a phenyl-bearing compound (K423) was found to be the best inhibitor of MdAChE, having an IC50 in the nanomolar range with a SI value of 7.058, which was superior to that of the benzyl compound (K424) or the 3-phenylpropyl compound (K425). The latter compound exerted no or very low inhibition potency to MdAChE in the given concentration scale. The results showed that only K423, bearing a phenyl ring directly attached to the pyridinium moiety, was able to inhibit MdAChE, whereas chain elongation between these two moieties (K424 and K425) resulted in the loss of activity. On the other hand, a 4-nitrobenzyl compound (K426) showed inhibition in the micromolar scale although its selectivity towards MdAChE was very low. Compounds (K427–K435) containing hydrophilic functional groups also showed apparent differences in their inhibitory ability. Among them, 3-hydroxypropyl (K429), N,N-dimethylamino (K430), and 4-carbonitrile compound (K435) were superior to other hydroxyl- (K427, K428) or carbonyl-containing compounds (K431–K434), possessing IC50 values in the micromolar range. Among the hydroxyl-containing compounds (K427–K429), no clear SAR can be drawn since only K429 was able to inhibit MdAChE. However, the SI values towards MdAChE were negligible for all of them. Although K429, K430, and K435 were the best hydrophilic compounds, their inhibitory ability towards MdAChE still decreased by two orders of magnitude as compared to that of the phenyl analogue K423.
The second subset of compounds consisted of bis-isoquinolinium derivatives bearing various linkers (see Fig. 2). The bis-isoquinolinium derivatives (K291–K299, K302, and K412–K419) were the most potent inhibitors in our study; only three of them (K291, K293, and K413) did not inhibit human and insect AChEs in the tested concentration range. This is probably caused by the presence of the isoquinoline heterocycle in them, which exhibits stronger stabilization via π–π or cation–π interaction to the catalytic active site (CAS) and peripheral anionic site (PAS) as compared to the bis-pyridinium compounds.21 Moreover, the used linkage and its length markedly influenced the inhibitory ability of bis-isoquinolinium compounds.21,28 Among the compounds with an aliphatic linker (K291–K299, K302, and K412–K415), those with nine and twelve methylene units (K299 and K302) were found to be the best inhibitors of MdAChE, achieving an IC50 value in the nanomolar range. K296, bearing six methylene linkers, inhibited MdAChE in the micromolar range with a low selectivity profile towards both AChEs. Furthermore, compounds bearing shorter linkers (one and three methylenes) were found to be inefficient for both enzymes, which was in agreement with previously obtained results.22,28 The presence of an oxygen heteroatom (K412 and K413) or a double bond (K414 and K415) in the linker did not yield higher affinity. In this case, the IC50 values lay in the micromolar range, and low selectivity towards MdAChE was observed. The linkage length of these compounds was four methylene units, which correlated well with the result obtained for K296.
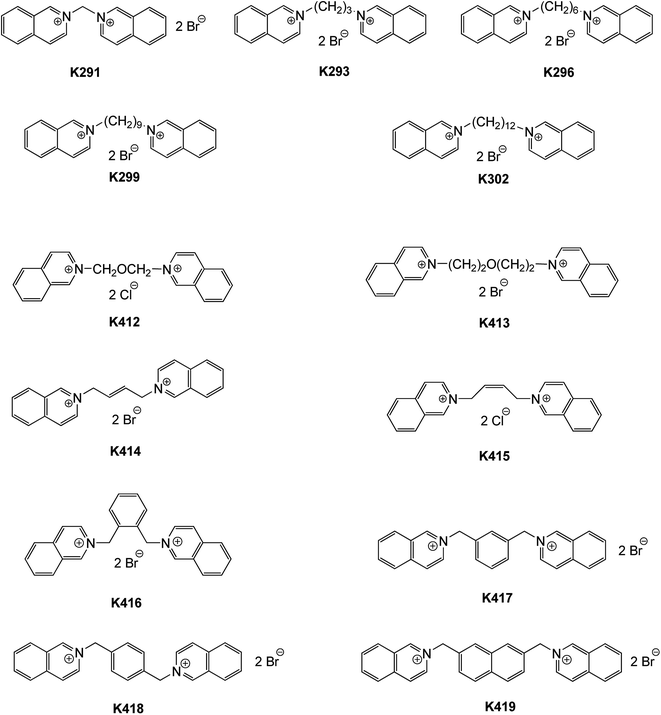 | ||
| Fig. 2 Structures of the evaluated bis-isoquinolinium compounds with aliphatic and aromatic linkers. | ||
Among bis-isoquinolinium salts bearing aromatic linkers (K416–K419), naphthylene-linked dimers (K419) were more potent as compared to xylene-linked dimers (K416–K419). K419 inhibited MdAChE in the nanomolar range, but with low selectivity. On the other hand, K416–K418 revealed high selectivity for MdAChE over hAChE. In this regard, K416 possessed the highest SI value of 12.185, and the second top-ranked compound K417 had a three-fold lower SI value of 4.789. However, the most promising compounds in terms of the IC50 values were K299, K302, and K419, achieving IC50 values in the nanomolar range.
A group of four insecticides (pyrethroids (PY), carbamates (CA), organophosphates (OP), and organochlorines (OC)) has currently been recommended by WHO pesticide evaluation scheme (WHOPES) for controlling vector-borne disease such as malaria.29 Compared to organophosphates, the carbamate insecticides probably represent a better option because they act as pseudo-irreversible AChE inhibitors with lower dermal toxicity. Moreover, two approved CA for insecticide-treated nets (ITN) are propoxur and bendiocarb, usually combined with PY. The IC50 value of hAChE for bendiocarb was found to be in the submicromolar range (IC50 0.359 μM), which was one order of magnitude lower than that of propoxur (2.094 μM).30 Based on the reported data, the SI for both of these compounds were calculated (propoxur SI 13.78 and bendiocarb 6.19).30,31 The highlighted compounds in our screening study in terms of SI are K299, K416, and K423. All these analogues resemble the SI values of the approved CA. K416 with a SI of 12.19 is superior to K423 (SI 7.06) and to K299 (SI 6.09). In terms of MdAChE inhibition potency, K299 was able to block the enzyme activity in the same range as bendiocarb. K423 had a comparable effect with propoxur. However, these data need to be taken with precaution since bendiocarb and propoxur presumably exert different modes of AChE inhibition.
Due to the fact that increase in resistance affects all major malaria vector species over all four recommended classes of insecticides,2,31 the development of new insecticides is currently a hot issue. Among the insecticides targeting AChE, a novel class of methylcarbamate hybrids has been developed.32,33 In this family, the most potent inhibitors were meta-substituted to the phenyl ring (compound 1) and possessed an IC50 value in the nanomolar range with low selectivity towards MdAChE.30,33 Compared to those of our highlighted inhibitors (K299, K302, and K419), their inhibitory effect and selectivity index were very similar. On the other hand, the ortho-substituted carbamate derivatives (compounds 7 and 8), which possessed higher values of IC50, hAChE, exhibited up to 350-fold selectivity, which significantly exceeded that of our most of the selective compounds (K299, K416, and K423) in this study. Recently, Pang et al. have described thiol-reactive compounds, which interfere with free cysteine in PAS of AgAChE; hAChE has no such free cysteine.9 These experimental thiol-compounds inhibited >95% of AgAChE activity after a 1 h exposure at 6 μM. Under this condition, hAChE activity remained unchanged.8 This suggested that inhibitors that bind to CAS and concomitantly target free cysteine may lead to highly selective insecticides. In this regard, our screened compounds are dual-binding site inhibitors; thus, in some ways, they follow the innovative Pang's dual binding site strategy.
Moreover, it was described that bis-pyridinium compounds bearing a polymethylene linker of up to four carbons did not inhibit the response of the nicotinic ACh receptor.34 Furthermore, a representative compound of bis-isoquinolinium salts was also evaluated for its nicotinic effect, and no inhibitory ability was confirmed.35 Therefore, we do not speculate any inhibitory effect of our most promising compounds (K299, K416, and K423) on the nicotinic ACh receptors.
Docking studies
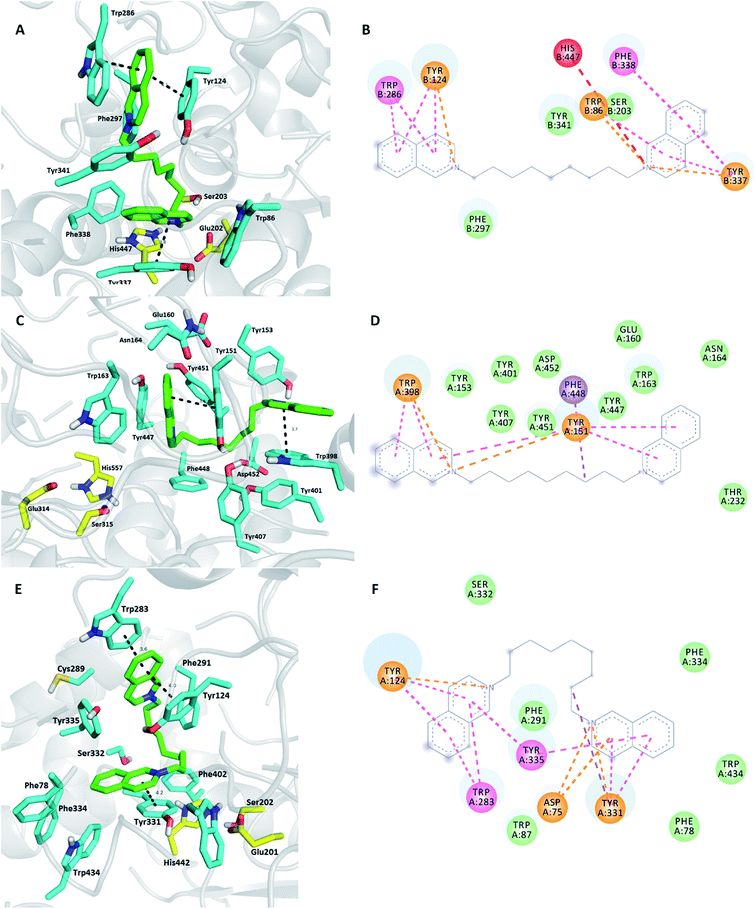
To explain the reasons for varied potencies between hAChE and MdAChE inhibition, several ligands were docked into their active sites as these enzymes were involved in the biological assay. In this context, we followed our previously described protocol.36–39 Moreover, the prediction of the ligand anchored onto the cavity of greenbug AChE (Schizaphis graminum, SgAChE) was carried out. The rationale for selection of this enzyme is structural similarity to Anopheles gambiae AChE.13 Moreover, the structure of SgAChE is freely available from the RCSB Protein Data Bank as a 3D theoretical model (PDB ID: 2HCP). The crystal structure of hAChE (PDB ID: 4EY7) with the well-known Alzheimer's disease drug donepezil was used as the template receptor for docking.32,40,41 Donepezil, likewise the intended ligands used in this study, is a dual-binder, i.e. a compound capable of simultaneously interacting with both anionic sites of AChE. MdAChE enzyme is not freely available from the RCSB Protein Data Bank; hence, this enzyme has been created based on the homology modeling technique and further validated before performing the in silico calculations (see the Experimental section). Based on the selectivity and activity profiles, we have chosen K299, being the most potent inhibitor of MdAChE from the family of bis-isoquinolinium salts, K416, having the highest selectivity ratio index for MdAChE under this study, K423, the highlighted MdAChE inhibitor from bis-pyridinium salts, and K425, as the highly effective hAChE binder with no activity towards MdAChE in the tested concentration scale.Initially, K299 revealed a dual-binding site character in all three models of AChE (Fig. 3), but with several divergences that might explain different enzyme affinity.22 Regarding the hAChE enzyme (Fig. 3A and B), the proximal isoquinoline heterocyclic core is engaged in parallel π–π/cation–π interactions with Tyr337 (3.8 Å), T-shaped π–π/cation–π interaction with Trp86 (3.5 Å), and π–π stacking with Phe338 (3.7 Å), whereas the distal isoquinoline moiety is sandwiched between Trp286 (3.6 Å) and Tyr124 (4.1 Å) via π–π and/or cation–π stacking. The linker combining both isoquinolines is responsible for weak van der Waals contact to Phe297 and Tyr341. Ser203 from the catalytic triad displayed a hydrophobic interaction to K299. On the contrary, His447 demonstrated unfavorable positive–positive repulsion to charged nitrogen from the proximal isoquinoline moiety. The K299-MdAChE (Fig. 3C and D) complex is somewhat different from the K299-hAChE ternary complex.
The overall arrangement in the K299-MdAChE complex placed the ligand in proximity of Tyr151, making this amino acid a cornerstone for contacting both isoquinolines as well as nine-methylene linker via either π–π/cation–π or hydrophobic interactions. Several other important amino acid residues provided contact to K299. In this case, no catalytic triad residue showed contact to K299. The last model K299-SgAChE in some way combines positive features of the previously mentioned K299-hAChE and K299-MdAChE ternary complexes. Tyr335 (SgAChE) is equal to Tyr151 (MdAChE) and hence fulfils the role of the central amino acid in which proximity K299 lies. The distal isoquinoline core is sandwiched between Trp283 (3.6 Å) and Tyr124 (4.0 Å).
In terms of the balanced selectivity and potency prediction using docking studies, all the abovementioned observations indicate that K299 can be the lead candidate in the search for a novel pesticide. However, further evaluation is necessarily needed to confirm this statement.
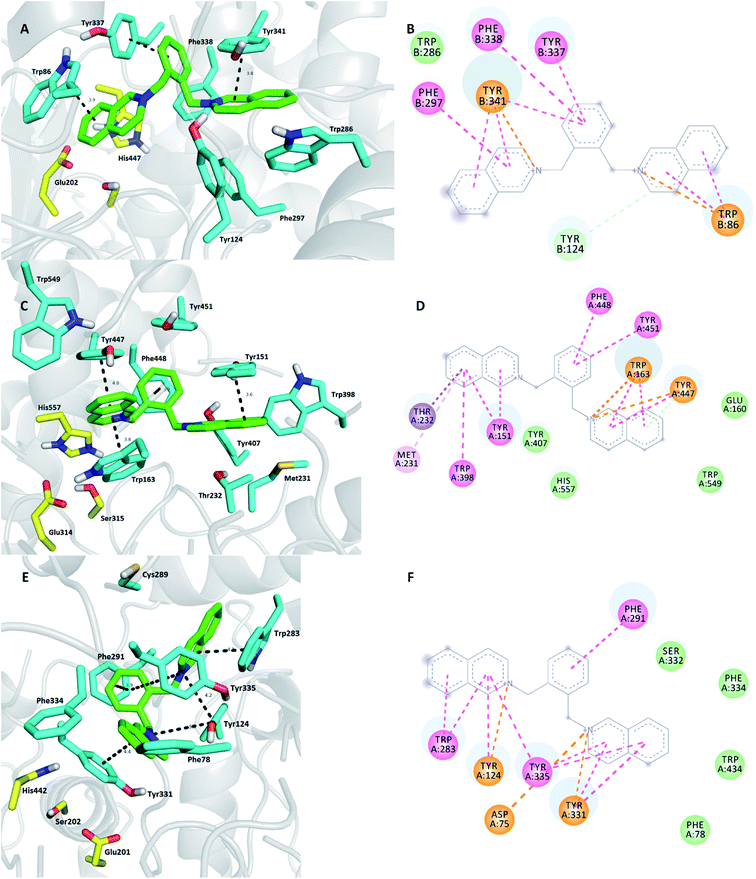
Continuing with the bis-isoquinolinium subset, the compound K416 (Fig. 4) was preferably selected for docking simulation instead of other aromatically linked bis-isoquinolinium salts (K417, K418, and K419) based on the highest selectivity profile towards MdAChE. It is well-known that introduction of a flexible linker in combination with an aromatic moiety leads to activity enhancement.32,42 This is in accordance ith the aromatic residue-abundant character of the mid-gorge region of the AChE enzyme. Indeed, this might be the case for some derivatives from the family of aromatically linked bis-isoquinolinium salts. However, hybrids used in this study combined only the short methylene chain with the aromatic part, which in general did not reach the inhibition potency of alkyl-tethered bis-isoquinolinium salts. This is presumably caused by the insufficient length of aromatically linked bis-isoquinolinium salts between the catalytic anionic site (CAS) moiety and the peripheral anionic site (PAS) binder, i.e. between both isoquinoline cores. The distance between these regions for an ideal inhibitor was found to be around 20 Å.43 Apart from all these, K416 is specific as compared to other inhibitors inspected in this study because it contains an o-disubstituted xylylene moiety. This liability is plausibly responsible for less linear properties in K416 as compared to those in other aromatically linked inhibitors (K417, K418, and K419), thus preserving steric rigidity leading to specific interactions and discrepancies in the binding sites of each AChE enzyme. Upon closer observation of the 3D complex of K416-hAChE (Fig. 4A and B), it was evident that the distal isoquinoline core was not able to establish favorable π–π/cation–π contact with Trp286, a crucial PAS amino acid residue. This amino acid residue is only involved in very weak hydrophobic contact. On the contrary, the proximal isoquinoline moiety is engaged in face-to-face π–π/cation–π interaction with Tyr341 (3.8 Å). This amino acid residue is also implicated in distorted π–π stacking with the connecting o-xylylene moiety with the distance of 3.8 Å. At the opposite site, deep in the gorge of hAChE, proximal isoquinoline can be found, forming parallel π–π contact with Trp86 (3.9 Å). No direct interaction can be observed between K416 and the catalytic triad residues of hAChE. In contrast, K416 revealed edge-to-edge ligand anchoring supported by parallel π–π interactions with Tyr151 (3.6 Å) and a sandwich orientation to Tyr447 (4.0 Å) and Trp163 (3.8 Å) in the PAS and CAS regions of MdAChE, respectively. Additionally, His557 from the catalytic triad furnished plausible hydrophobic contact with K416. Several other interactions can be found as well, e.g. σ–π contact between the distal isoquinoline heterocycle and Thr232, alkyl–π interaction with Met231, van der Waals interactions with Tyr407, Trp549, and Glu160, and many others. All the amino acid residues encompassing the K416 in the cavity of MdAChE are highlighted in Fig. 4(C and D). These findings justify the high potency and selectivity of K416 for MdAChE over hAChE. The investigation of the predictive model of K416 in SgAChE (Fig. 4E and F) is closely related to that of the K299-SgAChE complex with Tyr335 networking both isoquinolines. In this case, a plethora of attractive interactions can be found such as plausible cation–π interactions with Tyr124, Asp75, and Tyr331. The catalytic triad seems to stand aside of enzyme–ligand anchoring.
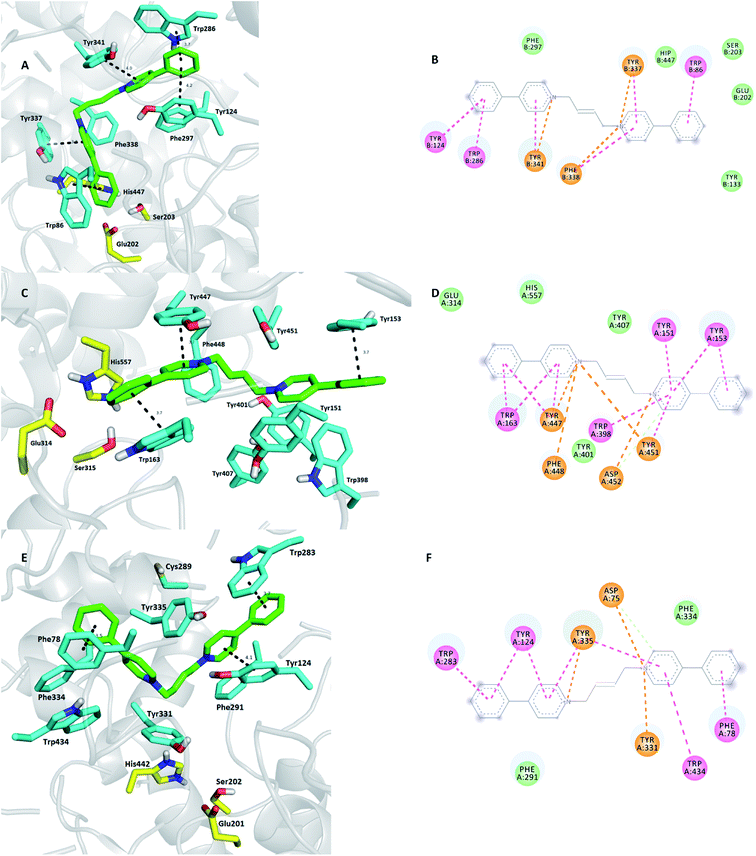
Via probing K423, the most potent and selective MdAChE binder from the bis-pyridinium family containing a but-(2E)-en-1,4-diyl linker, some structure diversity in the specific AChE active sites can be concluded. Considering the K423-hAChE complex (Fig. 5A and B), K423 furnished a typical dual-binding site pattern spanning the cavity gorge from the entrance to the bottom of the deep gorge. Every aromatic part of the ligand revealed face-to-face π–π and/or cation–π interactions with its amino acid counterpart proximally to the enzyme center in the order Trp286 (3.7 Å), Tyr341 (4.0 Å), Tyr337 (3.7 Å), and Trp86 (4.0 Å). Interestingly, all the catalytic triad residues are connected through hydrophobic interaction to K423. In contrast to hAChE, K423 mimics a similar spatial orientation in MdAChE (Fig. 5C and D), extending from the rim to underneath the gorge. Similarly, each aromatic part of the ligand displayed favorable π–π and/or cation–π contacts with aromatic amino acid residues delineating the cavity enzyme. Stronger affinity of K423 to MdAChE over hAChE might be provided by the web of several cation π interactions, as illustrated in Fig. 4D. The model of K423-SgAChE (Fig. 5E and F) gave the worst fit to the active site and thus could be considered as the potential pesticide with the lowest interest in this study. This is mainly speculated to be due to the unbeneficial occupation of the cavity of SgAChE where some face-to-face π–π stacking is missing and also the density of cation–π interactions is not so high.
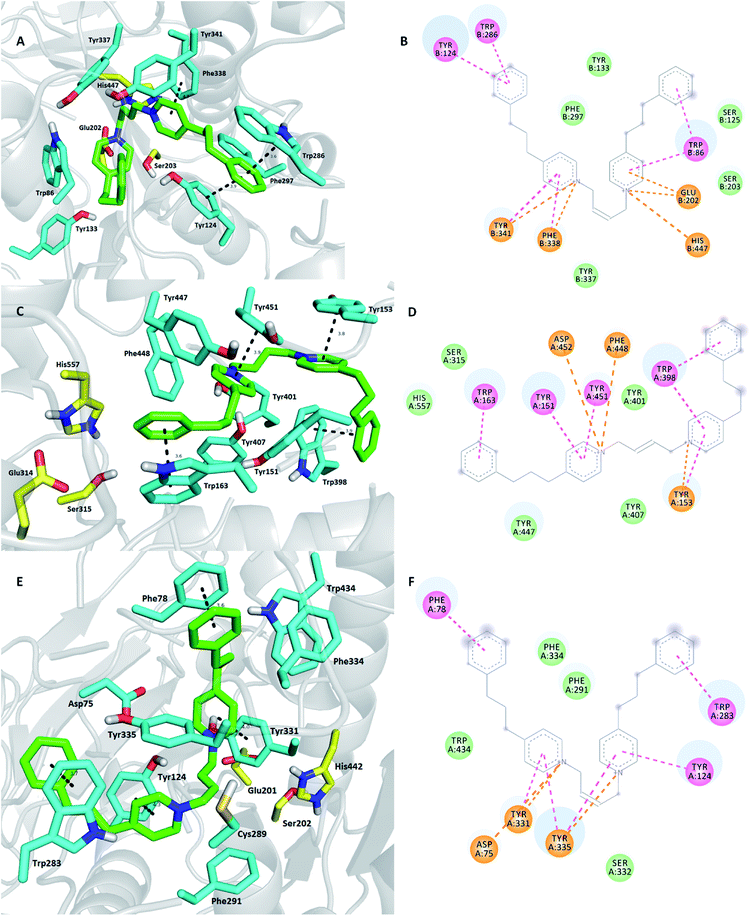
Lastly, our in silico study disclosed K425 as the ligand with no activity towards MdAChE and high affinity to the hAChE enzyme in the tested concentration scale (Fig. 6).28 Taking into account all the findings, no clear results or SAR data can be drawn based on the molecular level of the enzyme–ligand interaction. This is indeed true for all three cases of K425 hAChE, K425 MdAChE, and K425 SgAChE where no clear differences can be observed in the ligand lodging. Presumably, more definite results might be generated via molecular dynamics or Monte Carlo methods; however, these techniques are beyond the scope of this study.44,45 The docking score calculated by the Vina score (hybrid scoring function combining empirical and knowledge-based scoring) for K425 hAChE, K425 MdAChE, and K425 SgAChE complexes was −12.7 kcal mol−1, −12.4 kcal mol−1, and −13.0 kcal mol−1, respectively, which slightly correlated with the obtained in vitro results. Moreover, the energetic balance obtained for the K425 SgAChE complex does not discriminate this ligand from further development as a selective pesticide. Note that further biological assays towards SgAChE or related enzymes are necessarily needed to make any clear conclusion.
Conclusions
In summary, 34 compounds were evaluated for their inhibition ability to human and insect AChE. Their potency to inhibit hAChE and MdAChE was tested in vitro, and SI values were calculated to establish their selectivity. The most promising compounds in terms of selectivity were K299, K416, and K423, being more selective than the approved carbamates for controlling vector-borne disease. Consequently, the promising selectivity of these screened compounds was determined via docking study with hAChE, MdAChE, and SgAChE. The apparent interactions of π–π or π–cationic origin were described with several discrepancies indicating the binding differences. The docking study highlighted K299 and K416 as the candidates for lead structures in novel pesticides. However, further evaluation is needed to confirm this proof-of-concept.Acknowledgements
The authors would like to thank Jitka Turanova for her technical assistance. The work was financially supported by the by Ministry of Health of the Czech Republic (No. 16-34390A) and by Ministry of Education, Youth and Sports of the Czech Republic (No. SV/UHK2109 and No. SV/FVZ201601). Computational resources were provided by the CESNET LM2015042 and the CERIT Scientific Cloud LM2015085, provided under the programme “Projects of Large Research, Development, and Innovations Infrastructures”. The experiments were performed in compliance with the Czech legislation and institutional guidelines of the Faculty of Military Health Sciences, University of Defence. The institutional ethical committee of the Faculty of Military Health Sciences, University of Defence approved the experiment. The informed consents were obtained from all human subjects who provided the blood samples.Notes and references
- WHO/WHOPES, WHO Pesticides Evaluation Scheme, WHO, cited 2015 May 15, http://www.who.int/whopes/questions/en/.
- WHO/Fact Sheet, World Malaria Report 2015, WHO, cited 2016 Sep 21, http://www.who.int/malaria/media/world-malaria-report-2015/en/.
- WHO/Zika virus, WHO, cited 2016 Feb 3, http://www.who.int/mediacentre/factsheets/zika/en/.
- C. Boase, Bedbugs – back from the brink, Pestic. Outlook, 2001, 12(4), 159–162 RSC.
- D. W. Ragsdale, B. P. McCornack, R. C. Venette, B. D. Potter, I. V. MacRae and E. W. Hodgson, et al., Economic threshold for soybean aphid (Hemiptera: Aphididae), J. Econ. Entomol., 2007, 100(4), 1258–1267 CrossRef CAS PubMed.
- Y.-P. Pang, Chapter Six – Insect Acetylcholinesterase as a Target for Effective and Environmentally Safe Insecticides, in Advances in Insect Physiology, ed. E. Cohen, Academic Press, 2014, vol. 46, p. 435–494, available from: http://www.sciencedirect.com/science/article/pii/B9780124170100000069 Search PubMed.
- Y.-P. Pang, Novel acetylcholinesterase target site for malaria mosquito control, PLoS One, 2006, 1, e58 Search PubMed.
- Y.-P. Pang, S. K. Singh, Y. Gao, T. L. Lassiter, R. K. Mishra and K. Y. Zhu, et al., Selective and irreversible inhibitors of aphid acetylcholinesterases: steps toward human-safe insecticides, PLoS One, 2009, 4(2), e4349 Search PubMed.
- Y.-P. Pang, Species marker for developing novel and safe pesticides, Bioorg. Med. Chem. Lett., 2007, 17(1), 197–199 CrossRef CAS PubMed.
- S. B. Walsh, T. A. Dolden, G. D. Moores, M. Kristensen, T. Lewis and A. L. Devonshire, et al., Identification and characterization of mutations in housefly (Musca domestica) acetylcholinesterase involved in insecticide resistance, Biochem. J., 2001, 359(1), 175–181 CrossRef CAS PubMed.
- Y. Lu, Y. Park, X. Gao, X. Zhang, J. Yao and Y.-P. Pang, et al., Cholinergic and non-cholinergic functions of two acetylcholinesterase genes revealed by gene-silencing in Tribolium castaneum, Sci. Rep., 2012, 2, 288 CrossRef PubMed.
- Y. H. Kim, J. Y. Choi, Y. H. Je, Y. H. Koh and S. H. Lee, Functional analysis and molecular characterization of two acetylcholinesterases from the German cockroach, Blattella germanica, Insect Mol. Biol., 2010, 19(6), 765–776 CrossRef CAS PubMed.
- Y.-P. Pang, S. Brimijoin, D. W. Ragsdale, K. Y. Zhu and R. Suranyi, Novel and viable acetylcholinesterase target site for developing effective and environmentally safe insecticides, Curr. Drug Targets, 2012, 13(4), 471–482 CrossRef CAS PubMed.
- R. N'Guessan, F. Darriet, P. Guillet, P. Carnevale, M. Traore-Lamizana and V. Corbel, et al., Resistance to carbosulfan in Anopheles gambiae from Ivory Coast, based on reduced sensitivity of acetylcholinesterase, Med. Vet. Entomol., 2003, 17(1), 19–25 CrossRef.
- J. M. Mutunga, T. D. Anderson, D. T. Craft, A. D. Gross, D. R. Swale and F. Tong, et al., Carbamate and pyrethroid resistance in the akron strain of Anopheles gambiae, Pestic. Biochem. Physiol., 2015, 121, 116–121 CrossRef CAS PubMed.
- T. S. Awolola, O. A. Oduola, C. Strode, L. L. Koekemoer, B. Brooke and H. Ranson, Evidence of multiple pyrethroid resistance mechanisms in the malaria vector Anopheles gambiae sensu stricto from Nigeria, Trans. R. Soc. Trop. Med. Hyg., 2009, 103(11), 1139–1145 CrossRef CAS PubMed.
- D. J. Im, W. T. Kim and K. S. Boo, Purification and Partial cDNA Sequence of Acetylcholinesterase from a Korean Strain of the Housefly, Musca domestica, J. Asia-Pac. Entomol., 2004, 7(1), 81–87 CrossRef.
- J. Jeyaratnam, Acute pesticide poisoning: a major global health problem, World Health Stat. Q. Rapp. Trimest. Stat. Sanit. Mond., 1990, 43(3), 139–144 CAS.
- T. Satoh and R. C. Gupta, Anticholinesterase Pesticides: Metabolism, Neurotoxicity, and Epidemiology, John Wiley & Sons, 2011. p. 645 Search PubMed.
- K. Musilek, R. Pavlikova, J. Marek, M. Komloova, O. Holas and M. Hrabinova, et al., The preparation, in vitro screening and molecular docking of symmetrical bisquaternary cholinesterase inhibitors containing a but-(2E)-en-1,4-diyl connecting linkage, J. Enzyme Inhib. Med. Chem., 2011, 26(2), 245–253 CrossRef CAS PubMed.
- K. Musilek, M. Komloova, O. Holas, M. Hrabinova, M. Pohanka and V. Dohnal, et al., Preparation and in vitro screening of symmetrical bis-isoquinolinium cholinesterase inhibitors bearing various connecting linkage – Implications for early Myasthenia gravis treatment, Eur. J. Med. Chem., 2011, 46(2), 811–818 CrossRef CAS PubMed.
- K. Musilek, M. Komloova, V. Zavadova, O. Holas, M. Hrabinova and M. Pohanka, et al., Preparation and in vitro screening of symmetrical bispyridinium cholinesterase inhibitors bearing different connecting linkage—initial study for Myasthenia gravis implications, Bioorg. Med. Chem. Lett., 2010, 20(5), 1763–1766 CrossRef CAS PubMed.
- G. L. Ellman, K. D. Courtney, V. Andres and R. M. Feather-Stone, A new and rapid colorimetric determination of acetylcholinesterase activity, Biochem. Pharmacol., 1961, 7, 88–95 CrossRef CAS PubMed.
- M. Pohanka, J. Z. Karasova, K. Kuca, J. Pikula, O. Holas and J. Korabecny, et al., Colorimetric dipstick for assay of organophosphate pesticides and nerve agents represented by paraoxon, sarin and VX, Talanta, 2010, 81(1–2), 621–624 CrossRef CAS PubMed.
- F. Worek, G. Reiter, P. Eyer and L. Szinicz, Reactivation kinetics of acetylcholinesterase from different species inhibited by highly toxic organophosphates, Arch. Toxicol., 2002, 76(9), 523–529 CrossRef CAS PubMed.
- O. Soukup, D. Jun, J. Zdarova-Karasova, J. Patocka, K. Musilek and J. Korabecny, et al., A resurrection of 7-MEOTA: a comparison with tacrine, Curr. Alzheimer Res., 2013, 10(8), 893–906 CrossRef CAS PubMed.
- F. Zemek, L. Drtinova, E. Nepovimova, V. Sepsova, J. Korabecny and J. Klimes, et al., Outcomes of Alzheimer's disease therapy with acetylcholinesterase inhibitors and memantine, Expert Opin. Drug Saf., 2014, 13(6), 759–774 CAS.
- M. Komloova, K. Musilek, A. Horova, O. Holas, V. Dohnal and F. Gunn-Moore, et al., Preparation, in vitro screening and molecular modelling of symmetrical bis-quinolinium cholinesterase inhibitors—implications for early Myasthenia gravis treatment, Bioorg. Med. Chem. Lett., 2011, 21(8), 2505–2509 CrossRef CAS PubMed.
- WHO/WHO Pesticide Evaluation Scheme, “WHOPES”, WHO, cited 2016 Sep 21, http://www.who.int/whopes/en/.
- Y. Jiang, D. Swale, P. R. Carlier, J. A. Hartsel, M. Ma and F. Ekström, et al., Evaluation of novel carbamate insecticides for neurotoxicity to non-target species, Pestic. Biochem. Physiol., 2013, 106(3), 156–161 CrossRef CAS.
- D. R. Swale, P. R. Carlier, J. A. Hartsel, M. Ma and J. R. Bloomquist, Mosquitocidal carbamates with low toxicity to agricultural pests: an advantageous property for insecticide resistance management, Pest Manage. Sci., 2015, 71(8), 1158–1164 CrossRef CAS PubMed.
- WHO, Insecticide resistance, WHO, 2016 Sep 20, http://www.who.int/malaria/areas/vector_control/insecticide_resistance/en/.
- J. A. Hartsel, D. M. Wong, J. M. Mutunga, M. Ma, T. D. Anderson and A. Wysinski, et al., Re-engineering aryl methylcarbamates to confer high selectivity for inhibition of Anopheles gambiae versus human acetylcholinesterase, Bioorg. Med. Chem. Lett., 2012, 22(14), 4593–4598 CrossRef CAS PubMed.
- A. Ring, B. O. Strom, S. R. Turner, C. M. Timperley, M. Bird and A. C. Green, et al., Bispyridinium Compounds Inhibit Both Muscle and Neuronal Nicotinic Acetylcholine Receptors in Human Cell Lines, PLoS One, 2015, 10(8), e0135811 Search PubMed.
- V. Sepsova, J. Krusek, J. Zdarova Karasova, F. Zemek, K. Musilek and K. Kuca, et al., The interaction of quaternary reversible acetylcholinesterase inhibitors with the nicotinic receptor, Physiol. Res., 2014, 63(6), 771–777 CAS.
- E. Nepovimova, J. Korabecny, R. Dolezal, K. Babkova, A. Ondrejicek and D. Jun, et al., Tacrine-Trolox Hybrids: A Novel Class of Centrally Active, Nonhepatotoxic Multi-Target-Directed Ligands Exerting Anticholinesterase and Antioxidant Activities with Low In Vivo Toxicity, J. Med. Chem., 2015, 58(22), 8985–9003 CrossRef CAS PubMed.
- J. Korabecny, R. Dolezal, P. Cabelova, A. Horova, E. Hruba and J. Ricny, et al., 7-MEOTA-donepezil like compounds as cholinesterase inhibitors: Synthesis, pharmacological evaluation, molecular modeling and QSAR studies, Eur. J. Med. Chem., 2014, 82, 426–438 CrossRef CAS PubMed.
- J. Korabecny, M. Andrs, E. Nepovimova, R. Dolezal, K. Babkova and A. Horova, et al., 7-Methoxytacrine-p-Anisidine Hybrids as Novel Dual Binding Site Acetylcholinesterase Inhibitors for Alzheimer's Disease Treatment, Molecules, 2015, 20(12), 22084–22101 CrossRef CAS PubMed.
- S. K. Lee, M. K. Park, H. E. Jhang, J. Yi, K. Nahm and D. W. Cho, et al., Preparation of 7-Methoxy Tacrine Dimer Analogs and Their In vitro/In silico Evaluation as Potential Cholinesterase Inhibitors, Bull. Korean Chem. Soc., 2015, 36(6), 1654–1660 CrossRef CAS.
- J. Cheung, M. J. Rudolph, F. Burshteyn, M. S. Cassidy, E. N. Gary and J. Love, et al., Structures of human acetylcholinesterase in complex with pharmacologically important ligands, J. Med. Chem., 2012, 55(22), 10282–10286 CrossRef CAS PubMed.
- M. Harel, G. Kryger, T. L. Rosenberry, W. D. Mallender, T. Lewis and R. J. Fletcher, et al., Three-dimensional structures of Drosophila melanogaster acetylcholinesterase and of its complexes with two potent inhibitors, Protein Sci., 2000, 9(6), 1063–1072 CrossRef CAS PubMed.
- J. Korabecny, L. Janovec, K. Musilek, F. Zemek, A. Horova and E. Nepovimova, et al., Comparison of Novel Tacrine and 7-MEOTA Derivatives with Aromatic and Alicyclic Residues: Synthesis, Biological Evaluation and Docking Studies, Lett. Org. Chem., 2013, 10(4), 291–297 CrossRef CAS.
- K. Musilek, O. Holas, J. Misik, M. Pohanka, L. Novotny and V. Dohnal, et al., Monooxime-monocarbamoyl Bispyridinium Xylene-Linked Reactivators of Acetylcholinesterase-Synthesis, In vitro and Toxicity Evaluation, and Docking Studies, ChemMedChem, 2010, 5(2), 247–254 CrossRef CAS PubMed.
- J. D. Durrant and J. A. McCammon, Molecular dynamics simulations and drug discovery, BMC Biol., 2011, 9, 71 CrossRef CAS PubMed.
- E. Paquet and H. L. Viktor, Molecular dynamics, monte carlo simulations, and langevin dynamics: a computational review, BioMed Res. Int., 2015, 2015, 183918 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra05838a |
| This journal is © The Royal Society of Chemistry 2017 |