Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Selection of affinity peptides for the purification potential of porcine circovirus type 2 (PCV2) Cap virus-like particles (VLPs)†
Guanmin Zheng ab,
Qingxia Lu*b,
Fangyu Wangb,
Qianyue Jinb,
Man Tengb,
Nana Zhangb,
Tingting Renab,
Peiyang Dingb and
Gaiping Zhang*ab
ab,
Qingxia Lu*b,
Fangyu Wangb,
Qianyue Jinb,
Man Tengb,
Nana Zhangb,
Tingting Renab,
Peiyang Dingb and
Gaiping Zhang*ab
aCollege of Animal Husbandry and Veterinary Science, Henan Agricultural University, Zhengzhou 450002, People's Republic of China. E-mail: zhanggaiping2003@163.com
bKey Laboratory of Animal Immunology of the Ministry of Agriculture, Henan Provincial Key Laboratory of Animal Immunology, Henan Academy of Agricultural Sciences, Huayuan Road No. 116, Zhengzhou 450002, People's Republic of China. E-mail: luqingxia82@163.com
First published on 8th August 2017
Abstract
In this study, we describe the use of a M13 phage-displayed random peptide library for screening novel peptide motifs that specifically recognize recombinant PCV2 Cap protein for the first time. The peptide had a sequence of YHDCFSAGFCIG, and the binding affinity was observed in the nanomolar range. In the serum blocking test, the ELISA signal of the peptide was rapidly decreased when the concentration of the PCV2 positive serum was increased.
Porcine circovirus type 2 (PCV2) is a circular, single-stranded DNA virus. It is considered as the major pathogen of porcine circovirus associated-diseases (PCVAD) that causes huge economic losses for the swine industry around the world every year.1,2 Apart from post-weaning multi-systemic wasting syndrome (PMWS), PCVAD also include porcine dermatitis and nephropathy syndrome (PDNS),3 reproductive disorders, porcine respiratory disease complex (PRDC),4 nervous system lesions,5 enteritis,6,7 and proliferative and necrotizing pneumonia (PNP).8 There are three major open reading frames (ORFs) in the PCV2 genome. First, ORF1 encodes a replication-associated protein (Rep and Rep′), which is essential for the replication of viral DNA in host cells.9,10 Second, ORF2 encodes the capsid, which is a unique structure protein (Cap) and main antigenic determinant of PCV2.11 The canonical viral jelly-roll structure of PCV2 Cap was identified via its crystal structure and cryo-electron microscopy (cryo-EM) image.12,13 Finally, ORF3 encodes a protein that is not essential for PCV2 replication, but may have an apoptotic effect on PCV2-infected cells.14,15 To date, safe and effective vaccines are considered as the best control measure for PCVAD. Virus-like particles (VLPs) formed by recombinant ORF2 expressed in prokaryotic as well as eukaryotic systems are similar to the intact PCV2 particles and have been utilized as a key constituent in vaccine formulation and have protected pigs against the infection of PCV2. Currently, the common purification process for PCV2 Cap VLPs includes ultracentrifugation in CsCl or sucrose gradients and ion-exchange chromatography;16–19 however, these methods are uneconomic and waste time and energy.
A high antigen purity is an indicator of good vaccines; however, the purification takes a most part of the vaccine production cost. To improve the purification method and reduce the cost of PCV2 Cap VLPs, a simple, rapid, and effective purification method should be established. Recently, magnetic nanoparticles (MNPs) have been used as a carrier for purification due to their rapid and convenient magnetic control effects, intrinsically large loading capacity, and ease for functional modification.20,21 The surfaces of MNPs are usually modified with biological affinity ligands for targeting. Antibodies show high specificity and target diversity, but their complex structure results in high production costs, low shelf-life, and problems related to degradation, modification, aggregation and/or denaturation.22 Their applicability in purification is limited by these shortcomings. On the other hand, other biological affinity ligands such as nucleic acids,23 carbohydrates,24 chemical molecules,25 and linear, branched, and cyclic peptides,26 have low molecular weights, enhanced stability and more efficient screening strategies. Unique and short linear peptides have emerged with specific advantages, such as smaller size, lower cost, ease of synthesis, more amenable to be engineered at the molecular level, lower immunogenicity, and higher surface density, over antibodies.
The M13 phage-displayed random peptide library (M13 Ph.D. peptide library) provides a robust platform for seeking specific peptides with different molecular properties that bind to diverse targets. Nowadays, it is used in B-cell and T-cell epitope mapping,27 disease diagnosis,28 antiviral research,29 vaccine development,30 and peptide-mediated drug delivery system development. Furthermore, the target peptides identified using the M13 Ph.D. peptide library have potential use in biological affinity ligands, which play a key role in the purification of VLPs.
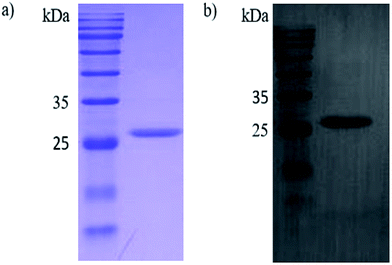
In this study, the Ph.D.-12 peptide library (New England Biolabs) was applied for the discovery of novel peptide ligands targeting the recombinant PCV2 Cap protein (r-Cap). Thus, a pET-28a-cap fusion proteins expression vector was built. The r-Cap (26 kDa) and amino acids 41–233 were expressed as their soluble form in E. coli BL21 (DE3) and purified using Ni-NTA column chromatography (Fig. 1a). To verify the reactogenicity of r-Cap, a western blot was performed using an anti-PCV2 Cap antibody (Fig. 1b). It was shown that the purified r-Cap had reactogenicity; thus, it was used as the target protein for further study.
We have biopanned a Ph.D.-12 peptide library against r-Cap to identify the linear peptide motifs that specifically bind to it. In brief, the r-Cap was immobilized onto 60 × 15 mm Petri dishes and washed with TBST after blocking. The Ph.D.-12 peptide library (1.5 × 1011 pfu) was added to the pre-coated Petri dishes, and the bound phage was eluted. After 3 rounds of biopanning, the peptides specific for r-Cap were identified. The operating process and statistical result after 3 rounds of biopanning are shown in Table S1.† With an increase in the number of rounds of biopanning, stricter conditions for selection and elution were employed. As expected, the amount of enrichment of the eluted phage clones remained the same with the increase in the number of rounds of biopanning. Samples of the eluted phage from the third round of biopanning were used to purify the plaques of the individual phage clones and analyse the DNA sequence. In this case, 23 clones were DNA-sequenced and 14 readable sequences were obtained. The sequencing results showed that 9 of the 14 sequences were identical with the amino acid sequence of YHDCFSAGFCIG. Thus, from these 9 phage clones, the one named as 3R#2 was selected for further characterization.
Indirect ELISA was performed to determine whether the 3R#2 phage-displayed peptides had specifically bound to r-Cap. Thus, the 3R#2 clones with different concentrations ranging from 106 to 1011 pfu were incubated with 10 μg mL−1 of r-Cap. The results indicated that the highest ELISA signal was found in the presence of 1011 pfu mL−1 of the 3R#2 clones, and the ELISA signal was reduced with a decrease in the amount of 3R#2 clones (Fig. 2a). In addition, the ELISA signal sharply decreased at a concentration of 1010 pfu mL−1. To further study the effects of the r-Cap concentration on the ELISA signal, 1010 pfu mL−1 of the 3R#2 phage clones was added to a pre-immobilized well with r-Cap at a variety of concentrations from 1.25 μg mL−1 to 80 μg mL−1 and measured using ELISA. The results showed that the binding ratio between the phage clones and r-Cap was highest at 40 μg mL−1 of r-Cap, and the ELISA signal increased with the increasing r-Cap concentration (Fig. 2b). As a negative control, wild type M13 phage did not bind to r-Cap (data not shown). From the above mentioned description, we found that the 3R#2 phage-displayed peptides had specifically bound to r-Cap. Interestingly, this peptide was rich in hydrophobic amino acid residues such as Ile, Phe, Cys, and Ala, and the hydrophobicity reached up to 50% in total; this suggested that hydrophobic interactions were exploited in the binding of r-Cap. In addition, two cysteine residues at 4 and 10 were also found in the 3R#2 sequence. Since 3R#2 contains an even number of Cys, it may form a disulfide-bond linked beta structure, helical structures containing S–S bonds or multiple thioether bonds. Finally, the sequence displayed on the 3R#2 clones contains two Glys, which may stabilize the peptide conformation or flexibility.
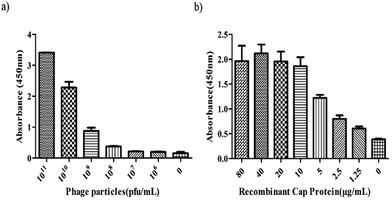 | ||
| Fig. 2 The indirect ELISA assays. The effects of the 3R#2 phage concentration (0–1011 pfu mL−1) (a) and r-Cap (0–80 μg mL−1) (b) on the ELISA signal. | ||
A further surface plasmon resonance (SPR) experiment was performed with r-Cap and the 3R#2 phage clones to measure their equilibrium dissociation binding affinity. Fig. 3a shows the equilibrium dissociation binding constant (Kd) of the 3R#2 phage clones. In detail, the Kd value of 3R#2 was 98.5 nM. The blocking effect of PCV2 positive serum (prepared by the Henan Provincial Key Laboratory of Animal Immunology) on the binding event between the 3R#2 phage clones and r-Cap was also investigated. First, the PCV2 positive serum ranging from 0.1% to 1% was incubated at 10 μg mL−1 and 20 μg mL−1 of r-Cap for 1 h at 37 °C. Second, 1011 pfu mL−1 of the 3R#2 clones was added to the wells for 1 h at 37 °C. Third, the HRP-conjugated anti-M13 monoclonal antibody was added to the wells for 1 h at 37 °C. Finally, the ELISA signals were measured at a wavelength of 450 nm. Interestingly, the ELISA signal of the 3R#2 clones was rapidly decreased when the concentration of PCV2 positive serum was increased (Fig. 3b). As a consequence, we concluded that the 3R#2 phage-displayed peptide screened using the Ph.D.-12 peptide library was found to be a potential affinity ligand for the purification of r-Cap; this suggested that it might have potential for PCV2 VLPs purification.
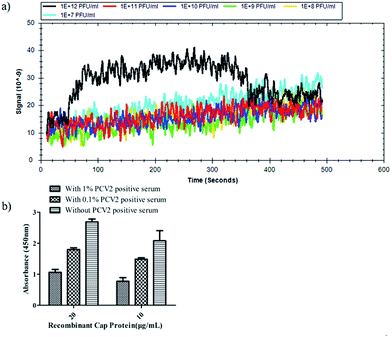 | ||
| Fig. 3 The equilibrium dissociation binding constant (Kd) obtained by SPR (a) and serum blocking test (b). | ||
Conclusions
In this study, we have demonstrated, for the first time, the use of a phage display technique for biopanning novel peptide motifs that specifically recognize target proteins. The 3R#2 peptide specific for target proteins had a sequence of YHDCFSAGFCIG, and ELISA and SPR assays were performed to evaluate the binding behaviour of this novel peptide. The results of the SPR assays show that the equilibrium dissociation binding constant of the 3R#2 peptide was found to be in the nanomolar range for its target. In the serum blocking test, the OD value (450 nm) rapidly dropped as the concentration of the PCV2 positive serum increased. To the best of our knowledge, it may be conceivable that the newly identified recognition peptide provides new peptide-based affinity ligands for the purification of PCV2 VLPs. Because the phage used in this study can be generated using approximately 5 copies of the peptide per phage, the true Kd value of the free peptides for the target protein may be different due to the loss of the functional affinity. Further work will be required to explore the performance of synthetic peptides upon immobilization using MNPs.Acknowledgements
The present study was funded by the Key Science and Technology Program of Henan Province (162102110136) and Science-Technology Foundation for Outstanding Young Scientists of Henan Academy of Agricultural Sciences (2016YQ28).Notes and references
- Z. Afghah, B. Webb, X. J. Meng and S. Ramamoorthy, Vet. Microbiol., 2017, 206, 21–28 CrossRef CAS PubMed.
- G. Franzo, C. M. Tucciarone, M. Cecchinato and M. Drigo, Sci. Rep., 2016, 6, 39458 CrossRef CAS PubMed.
- C. Rosell, J. Segales, J. A. Ramos-Vara, J. M. Folch, G. M. Rodriguez-Arrioja, C. O. Duran, M. Balasch, J. Plana-Duran and M. Domingo, Vet. Rec., 2000, 146, 40–43 CrossRef CAS PubMed.
- J. Kim, H. K. Chung and C. Chae, Vet. J., 2003, 166, 251–256 CrossRef CAS PubMed.
- G. W. Stevenson, M. Kiupel, S. K. Mittal, J. Choi, K. S. Latimer and C. L. Kanitz, J. Vet. Diagn. Invest., 2001, 13, 57–62 CrossRef CAS PubMed.
- J. Kim, Y. Ha, K. Jung, C. Choi and C. Chae, Can. J. Vet. Res., 2004, 68, 218–221 Search PubMed.
- T. K. Jensen, H. Vigre, B. Svensmark and V. Bille-Hansen, J. Comp. Pathol., 2006, 135, 176–182 CrossRef CAS PubMed.
- R. Drolet, R. Larochelle, M. Morin, B. Delisle and R. Magar, Vet. Pathol., 2003, 40, 143–148 CrossRef CAS PubMed.
- A. K. Cheung, Virology, 2003, 313, 452–459 CrossRef CAS PubMed.
- T. Finsterbusch and A. Mankertz, Virus Res., 2009, 143, 177–183 CrossRef CAS PubMed.
- K. P. Horlen, S. S. Dritz, J. C. Nietfeld, S. C. Henry, R. A. Hesse, R. Oberst, M. Hays, J. Anderson and R. R. Rowland, J. Am. Vet. Med. Assoc., 2008, 232, 906–912 CrossRef PubMed.
- R. Khayat, N. Brunn, J. A. Speir, J. M. Hardham, R. G. Ankenbauer, A. Schneemann and J. E. Johnson, J. Virol., 2011, 85, 7856–7862 CrossRef CAS PubMed.
- Z. Liu, F. Guo, F. Wang, T. C. Li and W. Jiang, Structure, 2016, 24, 319–328 CrossRef CAS PubMed.
- N. M. Juhan, T. LeRoith, T. Opriessnig and X. J. Meng, Virus Res., 2010, 147, 60–66 CrossRef CAS PubMed.
- J. Liu, I. Chen and J. Kwang, J. Virol., 2005, 79, 8262–8274 CrossRef CAS PubMed.
- P. C. Wu, W. L. Lin, C. M. Wu, J. N. Chi, M. S. Chien and C. Huang, Appl. Microbiol. Biotechnol., 2012, 95, 1501–1507 CrossRef CAS PubMed.
- L. J. Liu, T. Suzuki, H. Tsunemitsu, M. Kataoka, N. Ngata, N. Takeda, T. Wakita, T. Miyamura and T. C. Li, Arch. Virol., 2008, 153, 2291–2295 CrossRef CAS PubMed.
- S. A. Bucarey, J. Noriega, P. Reyes, C. Tapia, L. Saenz, A. Zuniga and J. A. Tobar, Vaccine, 2009, 27, 5781–5790 CrossRef CAS PubMed.
- M. Zaveckas, S. Snipaitis, H. Pesliakas, J. Nainys and A. Gedvilaite, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2015, 991, 21–28 CrossRef CAS PubMed.
- S. Yang, H. Ouyang, X. Su, H. Gao, W. Kong, M. Wang, Q. Shu and Z. Fu, Biosens. Bioelectron., 2016, 78, 174–180 CrossRef CAS PubMed.
- W. Wang, X. Ji, H. B. Na, M. Safi, A. Smith, G. Palui, J. M. Perez and H. Mattoussi, Langmuir, 2014, 30, 6197–6208 CrossRef CAS PubMed.
- C. S. Fernandes, I. Barbosa, R. Castro, A. S. Pina, A. S. Coroadinha, A. Barbas and A. C. Roque, Biotechnol. J., 2016, 11, 1513–1524 CrossRef CAS PubMed.
- D. Cheng, M. Yu, F. Fu, W. Han, G. Li, J. Xie, Y. Song, M. T. Swihart and E. Song, Anal. Chem., 2016, 88, 820–825 CrossRef CAS PubMed.
- K. El-Boubbou, C. Gruden and X. Huang, J. Am. Chem. Soc., 2007, 129, 13392–13393 CrossRef CAS PubMed.
- X. Yang, X. Zhou, M. Zhu and D. Xing, Biosens. Bioelectron., 2016, 91, 238–245 CrossRef PubMed.
- F. Y. Kuo, W. L. Lin and Y. C. Chen, Nanoscale, 2016, 8, 9217–9225 RSC.
- C. H. Wu, I. J. Liu, R. M. Lu and H. C. Wu, J. Biomed. Sci., 2016, 23, 8 CrossRef PubMed.
- B. Bakhshinejad and M. Sadeghizadeh, World J. Gastroenterol., 2014, 20, 11671–11683 CrossRef PubMed.
- Y. He, X. Ye, P. Tiollais, J. Zhang, J. Zhang, J. Liu and Y. Xie, Acta Biochim. Biophys. Sin., 2014, 46, 691–698 CrossRef CAS PubMed.
- W. S. Tan and K. L. Ho, World J. Gastroenterol., 2014, 20, 11650–11670 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details including biopanning of M13 phage. See DOI: 10.1039/c7ra05790c |
| This journal is © The Royal Society of Chemistry 2017 |