Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Brazilin inhibits fibrillogenesis of human islet amyloid polypeptide, disassembles mature fibrils, and alleviates cytotoxicity†
Jingjing Guo‡
a,
Wanqi Sun‡b,
Li Lic,
Fufeng Liu *ad and
Wenyu Lu*a
*ad and
Wenyu Lu*a
aDepartment of Biochemical Engineering, Key Laboratory of Systems Bioengineering of Ministry of Education, School of Chemical Engineering and Technology, Tianjin University, Tianjin 300072, P. R. China. E-mail: fufengliu@tust.edu.cn; wenyulu@tju.edu.cn; Fax: +86-022-60602298; Tel: +86-022-60602717
bDepartment of Chemical and Biological Engineering, The University of Alabama, Tuscaloosa, AL, USA
cCollege of Marine and Environmental Sciences, Tianjin University of Science & Technology, Tianjin 300457, P. R. China
dKey Laboratory of Industrial Fermentation Microbiology of Ministry of Education, Tianjin Key Laboratory of Industrial Microbiology, National Engineering Laboratory for Industrial Enzymes, National and Local United Engineering Lab of Metabolic Control Fermentation Technology, College of Biotechnology, Tianjin University of Science & Technology, Tianjin 300457, P. R. China
First published on 8th September 2017
Abstract
Fibrillogenesis of human islet amyloid polypeptide (hIAPP) is a pathological hallmark of type II diabetes mellitus (T2DM), and the inhibition of hIAPP fibrillogenesis is an important strategy for the prevention and treatment of T2DM. In this study, the inhibitory effects of brazilin on the fibrillization and cytotoxicity of hIAPP were examined using the thioflavin T fluorescence (ThT) assay, transmission electron microscopy (TEM), circular dichroism (CD) spectroscopy, cytotoxicity assays, and molecular dynamics simulations. Both the ThT and TEM results have shown that brazilin inhibits hIAPP fibrillogenesis in a dose-dependent manner. CD studies revealed that brazilin delays the conformational transition of hIAPP from its initial α-helical to the β-sheet form. As a result, brazilin greatly alleviates hIAPP-induced cytotoxicity. Moreover, we also found that brazilin disassembles preexisting hIAPP fibrils, and alleviates the cytotoxicity of hIAPP aggregates. The results of free energy decomposition studies calculated using molecular mechanics-Poisson–Boltzmann surface area analysis revealed that hydrophobic interactions contribute more than 75% of the free energy of binding in the brazilin–hIAPP complex, while electrostatic interactions (i.e., hydrogen bonds) play a secondary role (<25%). Two binding sites of brazilin on the hIAPP pentamer were identified, encompassing the N-terminal region and the turn region. There are 11 important residues of hIAPP that strongly interact with brazilin – Asn3, Thr4, Thr9, Arg11, Asn14, Phe15, His18, Ser19, Ser20, Asn21 and Phe23. The findings presented here will contribute to a comprehensive understanding of the inhibitory effect of brazilin on the fibrillogenesis of hIAPP, which is critical for the search for more effective agents that can inhibit hIAPP fibrillogenesis.
Introduction
The formation of amyloid plaques comprising different proteins has been implicated in the pathology of many diseases, including Alzheimer's disease (AD) and type II diabetes mellitus (T2DM).1 For example, the fibrillization and deposition of human islet amyloid polypeptide (hIAPP) is a pathological feature of T2DM.2,3 hIAPP is a 37-amino acid peptide, which is produced by the pancreatic islet β-cells and is co-secreted with insulin under normal physiological conditions.4,5 hIAPP is believed to play a key role in the control of gastric emptying, glucose homeostasis and suppression of glucagon release.6 It has been found that the oligomeric and/or fibrillar amyloid species of hIAPP leads to the death of β-cells and pancreatic dysfunction, contributing to islet transplant failure and T2DM. Although the fibrillization mechanism and cytotoxicity of different aggregation states of hIAPP is still under debate, it is widely accepted that inhibiting the aggregation of hIAPP might be an effective method for the treatment of T2DM.Recently, many kinds of inhibitors have been developed to prevent fibrillization of hIAPP, including small organic molecules,7–13 peptides,14–16 proteins17 and nanoparticles.18,19 Among these, small organic molecules have received special interest because of their high permeability through the blood–brain barrier and potentially low cytotoxicity.20 Several natural compounds have been found to prevent hIAPP fibrillization or to transform the mature fibrils into unstructured, off-pathway aggregates.8,21,22 For example, Chen et al.23 have found that the fibrillation and aggregation of hIAPP is affected by trehalose in a dose-dependent manner. A low dose of trehalose inhibited the conformational transition of hIAPP. Conversely, a high dose trehalose promoted the fibrillation of hIAPP. Meng et al.24 revealed that (−)-epigallocatechin gallate (EGCG) inhibits hIAPP fibrillization, produces unstructured, off-pathway aggregates, and reduces the cytotoxicity of hIAPP. Moreover, Wang, et al.25 used MD simulations to identify the possible binding sites of EGCG on the hIAPP oligomers. Components found in coffee have also been found to inhibit hIAPP amyloid formation.26 However, none of these compounds has been used for the clinical treatment of T2DM to date. Therefore, more effective natural molecules with a strong inhibitory effect on hIAPP fibrillation are urgently needed.
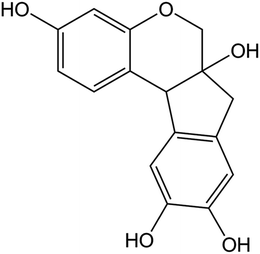
Caesalpinia sappan, a famous herb of traditional Chinese medicine, possesses antibacterial and anticoagulant properties, and is used to produce a valuable red dye, which was traditionally used for making herbal drinking water.27 The main active ingredient of Caesalpinia sappan, brazilin has received increasing attention due to its varied biological effects, including anti-cancer activity,28 induction of immunological tolerance,29 and anti-inflammatory activity.30 The chemical structure of brazilin is shown in Fig. 1. In our previous studies, brazilin was demonstrated to be an efficient inhibitor of amyloid β protein (Aβ) fibrillogenesis, as well as to be able of remodeling mature Aβ fibrils, which are associated with AD.31
Significantly, hIAPP shares a 25% degree of identity and 50% degree of similarity in sequence with Aβ,32,33 and growing evidence shows that there are common pathological processes and transcriptional pathways in T2DM and AD.34,35 Moreover, it was found that people suffering from T2DM are more likely to develop AD, and vice versa.36,37 Aggregates and depositions of hIAPP were found in AD brains,38 while Aβ and hyperphosphorylated tau deposits were also detected in the pancreases of T2DM patients,39 indicating that there is a correlation between the aggregation of Aβ and aggregation of hIAPP. In vitro studies have also revealed that Aβ and hIAPP can co-aggregate and cross-seed.40–43 For example, Zhang, et al.44 have revealed the atomic-level details of the cross-seeding interactions between Aβ and hIAPP. Since hIAPP and Aβ share a similar aggregation mechanism, and have been proven to co-aggregate with each other, efforts to find compounds that can simultaneously inhibit the aggregation of both Aβ and hIAPP might yield promising new candidates for therapeutics. We therefore investigated the inhibitory effect of brazilin on the fibrillization of hIAPP, as well as its effect on the disaggregation of preexisting fibrils and their cytotoxicity, using systematic biochemical, biophysical and cell biological experiments. Finally, MD simulations were used to explore the molecular interactions between brazilin and the hIAPP pentamer.
Materials and methods
Materials
Lyophilized hIAPP powder (>95%) was obtained from GL Biochem (Shanghai, China). Brazilin (HPLC ≥98%) was received from Yuanye Bio-Tech (Shanghai, China). 1,1,1,3,3,3-Hexafluoro-2-propanol (HFIP), dimethyl sulfoxide (DMSO), thioflavin T (ThT), and 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) were purchased from Sigma (Sigma-aldrich, St. Louis, MO, USA). The rat insulinoma cell line INS-1 was obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Fetal bovine serum (FBS) and Roswell Park Memorial Institute 1640 medium were purchased from Invitrogen (Carlsbad, CA, USA). All other chemicals were of the highest purity available from local sources.Peptide preparation
The lyophilized hIAPP powder was stored at −80 °C. It was first dissolved in HFIP to a concentration of 1.0 mg mL−1, and then incubated at 4 °C for 2 h. In order to remove any pre-existing aggregates, the solutions were centrifuged at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 20 min at 4 °C, following sonication in an ice bath for 2 min. The top 75% portions of the supernatants were carefully collected and lyophilized. The resulting aggregate-free freeze-dried hIAPP was stored at −20 °C until final use.
000g for 20 min at 4 °C, following sonication in an ice bath for 2 min. The top 75% portions of the supernatants were carefully collected and lyophilized. The resulting aggregate-free freeze-dried hIAPP was stored at −20 °C until final use.
Aggregation and disaggregation experiments
Lyophilized hIAPP was dissolved in HFIP to a final concentration of 1.58 mM, and centrifuged at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 20 min at 4 °C after sonication in an ice bath. After this, the resulting stock solution of hIAPP was diluted with Tris–HCl (20 mM, pH 7.4) buffer solutions, containing different concentrations of brazilin. The final concentration of hIAPP was 25 μM. The resulting solutions were incubated under constant shaking at 25 °C. For the disaggregation experiments, hIAPP monomers were first incubated as described above for 24 h, which is sufficiently long for hIAPP to grow into mature fibrils.45 Subsequently, equal volumes of brazilin solutions with different concentrations were added into the fibril solutions. The resulting disaggregation solutions were incubated at 25 °C under constant shaking.
000g for 20 min at 4 °C after sonication in an ice bath. After this, the resulting stock solution of hIAPP was diluted with Tris–HCl (20 mM, pH 7.4) buffer solutions, containing different concentrations of brazilin. The final concentration of hIAPP was 25 μM. The resulting solutions were incubated under constant shaking at 25 °C. For the disaggregation experiments, hIAPP monomers were first incubated as described above for 24 h, which is sufficiently long for hIAPP to grow into mature fibrils.45 Subsequently, equal volumes of brazilin solutions with different concentrations were added into the fibril solutions. The resulting disaggregation solutions were incubated at 25 °C under constant shaking.
Thioflavin T (ThT) fluorescence assay
Fibrillization of hIAPP in the presence of different concentrations of brazilin was monitored using the ThT fluorescence assay. A 2 mM ThT stock was prepared by adding 32.8 mg of ThT powder into 50 mL of deionized water. The resulting ThT stock was further diluted using Tris buffer (20 mM, pH 7.4) to reach a final concentration of 25 μM. At different time points, 200 μL sample aliquots were added into 2 mL of ThT solution (25 μM, pH 7.4). An LS55 fluorescence spectrometer (Perkin-Elmer, MA, USA) was used to measure the fluorescence intensity, with the excitation and emission wavelengths set to 440 nm and 480 nm, respectively. Both the excitation and emission slit widths were 5 nm. The fluorescence intensity of an otherwise equivalent solution without hIAPP was subtracted as background from each read with the target protein. Each measurement was done in triplicate.Transmission electron microscopy (TEM)
After 24 h of incubation, 10 μL samples from the aggregation and disaggregation assays were deposited onto 300-mesh carbon-coated copper grids and negatively stained with 2% phosphotungstic acid. The resulting samples were inspected under a JEM 100-CXII transmission electron microscope (JEOL, Tokyo, Japan) at 100 kV.Circular dichroism (CD) spectroscopy
Far-UV (200–260 nm) CD spectra of 25 μM hIAPP samples with or without brazilin were recorded in a 1 mm path length quartz cell on a J-810 CD spectrometer (Jasco, Japan) at 25 °C with a scan speed of 20 nm min−1 and a 1 nm bandwidth. The CD spectra of samples otherwise equivalent samples without hIAPP were subtracted as backgrounds. All spectra were recorded as the averages of 3 consecutive scans.Cytotoxicity assay
The INS-1 cells were maintained in Roswell Park Memorial Institute 1640 medium with 10% FBS, 100 U mL−1 penicillin, 100 μg mL−1 streptomycin, 0.11 g L−1 L-glutamine, 0.11 g L−1 sodium pyruvate, 5.6 mM glucose and 1 mg L−1 2-mercaptoethanol, and cultured at 37 °C in a humidified atmosphere comprising 5% CO2.The MTT assay was used to measure cytotoxicity towards the INS-1 cells. Briefly, the cells were seeded into 96-well plates at a density of ∼5 × 103 cells per well. After 24 h of incubation, the prepared hIAPP species were added into the wells to a final concentration of 25 μM. Following another 24 h of incubation at 37 °C, MTT was added to a final concentration of 0.5 mg mL−1 and incubated at 37 °C for another 4 h. After this, the cells in the 96-well plates were pelleted by centrifugation at 1500 rpm for 10 min, and the supernatants were replaced with 100 μL of DMSO per well. After formazan was fully dissolved, the absorbance at 570 nm was measured. Each record represents the average of 6 replicates. Statistical significance was determined by analysis of variance using the one-tailed t-test, and differences with p < 0.05 were considered to be statistically significant.
Molecular dynamics simulations
In this study, both the pentamer and the monomer of hIAPP were used to explore the interactions between brazilin and hIAPP. The coordinates of monomeric hIAPP were taken from PDB entry 5MGQ,46 which was used to probe the inhibitory effect of brazilin on the conformational transition of hIAPP monomer. The protofibrillar pentamer of hIAPP was used to probe the molecular interactions between brazilin and hIAPP.47 The initial structure of the hIAPP pentamer was kindly provided by Prof. Tycko R. from National Institutes of Health, USA. The 3D models of the pentamer and monomer of hIAPP are shown in Fig. S1A and B,† respectively. The initial structure of brazilin was taken from the PubChem Compound Database (http://www.ncbi.nlm.nih.gov/pccompound). The GROMOS96 53a6 force–field parameters of brazilin were sourced from the Automated Topology Builder and Repository 2.0 webserver (https://atb.uq.edu.au/).48 Subsequently, the atomic charges and charge groups of brazilin were corrected to achieve better agreement with the GROMOS96 53A6 force field parameter set.49 Here, the molar ratio of brazilin to hIAPP (i.e., 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for the monomer or 2
1 for the monomer or 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for the pentamer) was slightly higher than that used in most of the biophysical and biological experiments, which was often used in many MD simulation studies.10,50–53 For example, Raman et al.54 have utilized MD simulations in addressing the inhibition effect of ibuprofen on Aβ fibrillization. The peptide/ibuprofen molar ratios are higher than that used experimentally. So we believed that our simulation results would be directly comparable to those experimental studies.
1 for the pentamer) was slightly higher than that used in most of the biophysical and biological experiments, which was often used in many MD simulation studies.10,50–53 For example, Raman et al.54 have utilized MD simulations in addressing the inhibition effect of ibuprofen on Aβ fibrillization. The peptide/ibuprofen molar ratios are higher than that used experimentally. So we believed that our simulation results would be directly comparable to those experimental studies.
The hIAPP monomer or pentamer was firstly put into a rectangular box of 7 nm × 7 nm × 9 nm. Then, 10 brazilin molecules were located randomly around the pentamer. Thereafter, water molecules were added into the box and negative ions (Cl−) were used to neutralize the simulation systems. The simple point charge (SPC) model was used to describe water. We first performed 1000 energy minimization steps to relax the simulation system, after which the thus relaxed system was equilibrated for 1 ns by successively using an isochoric–isothermal ensemble and an isothermal–isobaric ensemble, via the Berendsen weak coupling method.55 Finally, three MD simulations of 100 ns under different initial conditions were carried out by assigning different initial velocities to each atom of the simulation system. All of the MD simulations were performed close to physiological temperature (i.e., 310 K) and a pressure of 1 bar.
We performed all-atom MD simulations using the GROMACS 5.1.1 package56 together with the GROMOS96 53A6 force field. Newton's classical equations of motion were integrated using the Verlet Leapfrog algorithm with a 2 fs time step.57 All short-range non-bonded interactions were cut off at 1.4 nm, with dispersion correction applied to energy and pressure terms to account for the truncation of van der Waals interactions. Long-range electrostatic interactions were calculated with the smooth particle mesh Ewald method58 via cubic-spline interpolation with a Fourier grid spacing of approximately 0.12 nm. The neighbor list was updated every five simulation steps. All bond lengths were constrained using the LINCS algorithm59 with a relative geometric tolerance of 10−4. Initial velocities were assigned according to a Maxwell distribution. Each system was run in triplicate for validation of the different starting coordinates and velocities. The atomic coordinates were saved every 50 ps for subsequent analyses. The auxiliary programs provided with the GROMACS 5.1.1 package were used to analyze the simulation trajectories. The programs include gmx rms for the comparison of any two structures by computing the root-mean-square deviation of Cα atoms (Cα-RMSD) of the hIAPP pentamer, gmx mindist for the contacts between brazilin and residues within 0.6 nm, and gmx hbond for the hydrogen bond interactions between brazilin and the hIAPP pentamer. The secondary structures of the hIAPP monomer and pentamer were assigned using the dictionary secondary structure of proteins (DSSP) program developed by Kabsch and Sander.60 The β-sheet content was defined as the ratio of the number of residues in β-sheet secondary structures to the total number of residues of the hIAPP pentamer (i.e., 37 × 5 = 185). In the presence of brazilin, the data of four structural parameters of the hIAPP pentamer (i.e., Cα-RMSD, inter-chain contacts, β-sheet content and hydrogen bonds) of three MD trajectories were provided. For clarity, only averaged data were provided for the control group comprising only hIAPP. The typical binding conformations of brazilin–hIAPP were observed using visual molecular dynamics (VMD) software version 1.9.2.61 For sake of simplicity, typical snapshots of only one MD simulation were selected and shown in the manuscript. Similar results were obtained in other two MD simulations. The free energies between brazilin molecules and the hIAPP pentamer were calculated using g_mmpbsa software.62
Results and discussion
Brazilin inhibits hIAPP fibrillogenesis
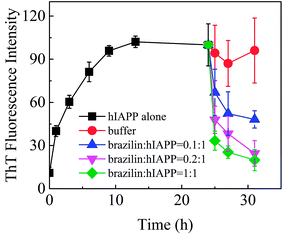
The inhibitory effect of brazilin on the fibrillogenesis of hIAPP was first examined using the ThT fluorescence assay. Fig. 2 shows the kinetics of fibrillogenesis at different molar ratios of brazilin to hIAPP measured using time-dependent ThT fluorescence. When 25 μM hIAPP was incubated alone at 25 °C, the ThT fluorescence profile showed an almost negligible lag phase, a fast growth phase within the initial 10 h, and a steady equilibrium phase after 13 h, which was consistent with earlier studies.26 By contrast, the fibrillization of hIAPP was suppressed by brazilin in a dose-dependent manner. For example, brazilin at a molar ratio to hIAPP of 0.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 markedly reduced fibril assembly, and an equimolar concentration reduced the ThT fluorescence to less than 10% of the untreated reference value, demonstrating that brazilin has a significant inhibitory effect on the fibrillogenesis of hIAPP. The inhibitory capacity of brazilin was significantly stronger than those of coffee components, such as caffeine, caffeic acid and chlorogenic acid.26 The stronger inhibitory effect of brazilin at lower concentrations was considered more favorable because it would be difficult for the compounds to reach very high concentrations in the focus of T2DM.
1 markedly reduced fibril assembly, and an equimolar concentration reduced the ThT fluorescence to less than 10% of the untreated reference value, demonstrating that brazilin has a significant inhibitory effect on the fibrillogenesis of hIAPP. The inhibitory capacity of brazilin was significantly stronger than those of coffee components, such as caffeine, caffeic acid and chlorogenic acid.26 The stronger inhibitory effect of brazilin at lower concentrations was considered more favorable because it would be difficult for the compounds to reach very high concentrations in the focus of T2DM.
 | ||
| Fig. 2 Time-dependent ThT fluorescence changes during hIAPP (25 μM) fibrillogenesis, when incubated with different concentrations of brazilin. Error bars represent the averages of three experiments. | ||
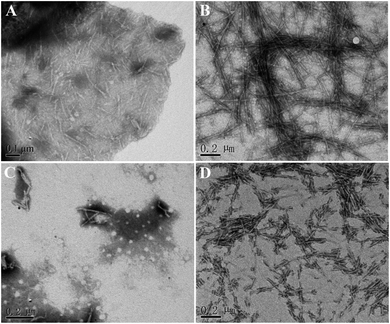
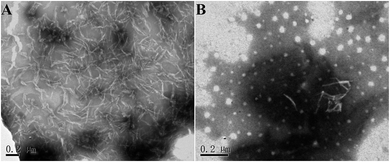
Furthermore, the morphology of hIAPP aggregates formed in the presence and absence of brazilin were observed by TEM. When hIAPP was incubated alone, abundant short fibrils and protofibrils were observed after 2 h of incubation (Fig. 3A). These aggregates elongated and got entangled with each other, forming bunches of long mature fibrils within 12 h (Fig. 3B). This was consistent with both the results of the ThT assay, and with previously published studies.13,16,26,63 By contrast, brazilin significantly reduced the length and quantity of hIAPP fibrils. For example, when brazilin was added at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, there were only a few protofibrils and some amorphous aggregates found in the TEM image taken at 2 h (Fig. 3C). Furthermore, after 12 h of incubation, rod-like aggregates as shown in Fig. 3D were observed. This indicates that the preformed fibrils were converted into rod-like aggregates that did not bind ThT molecules to generate an observable ThT fluorescence. These results confirmed that brazilin could inhibit the fibrillogenesis of hIAPP while promoting the formation of short rod-like aggregates. The TEM data therefore clearly support the results of the ThT fluorescence assay. Moreover, similar results have also been found in previous studies.13,31,64 Thus it can be concluded that brazilin prevents the amyloidogenesis of hIAPP and leads to the generation of short rod-like aggregates.
1, there were only a few protofibrils and some amorphous aggregates found in the TEM image taken at 2 h (Fig. 3C). Furthermore, after 12 h of incubation, rod-like aggregates as shown in Fig. 3D were observed. This indicates that the preformed fibrils were converted into rod-like aggregates that did not bind ThT molecules to generate an observable ThT fluorescence. These results confirmed that brazilin could inhibit the fibrillogenesis of hIAPP while promoting the formation of short rod-like aggregates. The TEM data therefore clearly support the results of the ThT fluorescence assay. Moreover, similar results have also been found in previous studies.13,31,64 Thus it can be concluded that brazilin prevents the amyloidogenesis of hIAPP and leads to the generation of short rod-like aggregates.
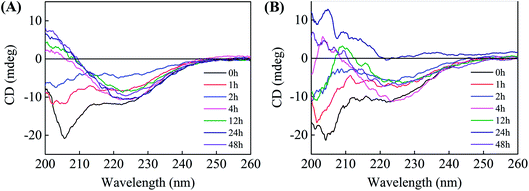
Effects of brazilin on the secondary structure of hIAPP
To gain insight into the effects of brazilin on the conformational transition of hIAPP upon aggregation, far-UV CD spectroscopy was utilized to investigate the influence of brazilin on the secondary structure of hIAPP over time (Fig. 4). As a control, we observed that the initial secondary structure of hIAPP was predominantly α-helical, with representative double minima at 207 nm and 222 nm. As the incubation time was prolonged, the negative bands around 207 and 222 nm gradually diminished, while a negative band at 225 nm and a positive band at 200 nm emerged and gradually increased (Fig. 4A), indicating that the secondary structure of hIAPP had transformed from its initial predominantly α-helical to a β-sheet-dominated form during the incubation. Typically, the β-sheet-rich amyloid fibrils of hIAPP show a minimum at ∼215 nm. However, a major negative valley at 225 nm appeared in our studies, which was similar to spectra from other studies.26,65,66 It is mainly caused by the intrinsic weakness of secondary structures determined by CD spectroscopy, and it has been proven that it arises from the structural and spectral diversity of β-sheet structures.67 It was found that this structural diversity manifests in variations of length, extent, direction, and degree of twist and distortion of the β-sheets and orientation of neighboring β-strands, i.e., the antiparallel and parallel nature. Moreover, it is known that aggregates with β-sheet structures are highly sensitive to external factors like temperature, concentration, and other solution conditions.67 Therefore, the results of CD can be considered reasonable.Judging by Fig. 4A, which was recorded in the absence of brazilin, the conformational transition of hIAPP from its initial, predominantly α-helical to the β-sheet form was mostly complete within 4 h. By contrast, brazilin at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 markedly delayed the conformational transition of hIAPP (Fig. 4B), so that by 12 h, the CD spectrum still showed a complex of β-sheet and random coil structures, as indicated by two broad negative minima around 203 and 225 nm, consistent with previous studies.68,69 This conformational stabilization effect could also be reflected in the inhibitory effect of brazilin. Until 48 h, a CD spectrogram of corresponding to β-sheet conformation was observed in the presence of brazilin, although the values of the peak and valley had slightly changed compared with pure hIAPP.
1 markedly delayed the conformational transition of hIAPP (Fig. 4B), so that by 12 h, the CD spectrum still showed a complex of β-sheet and random coil structures, as indicated by two broad negative minima around 203 and 225 nm, consistent with previous studies.68,69 This conformational stabilization effect could also be reflected in the inhibitory effect of brazilin. Until 48 h, a CD spectrogram of corresponding to β-sheet conformation was observed in the presence of brazilin, although the values of the peak and valley had slightly changed compared with pure hIAPP.
In order to further characterize the inhibitory effect of brazilin on the conformational transition of hIAPP, the secondary structure of a hIAPP monomer was calculated using the program gmx do_dssp. The time evolution of the secondary structures of the hIAPP monomer in the presence and absence of brazilin is shown in Fig. S2.† From Fig. S2A,† it is clear that the initial α-helix of residues 20–27 in water changed into a turn/β-bridge hybrid structure after 10 ns. After 20 ns, the initial α-helix spanning residues 9–19 uniformly evolved a 5-helix, although the α-helix spanning residues 5–9 persisted throughout the 100 ns simulation (Fig. S2A†). It should be noted that the residues 31–33 and 35–37 adopted a short β-sheet structure after 27 ns. Similar phenomena were observed in two other parallel MD simulations (Fig. S2B and C†), and the initial α-helix structures were transformed into 5-helix, coil and β-turn structures. Fig. S2D–F† show the conformational transition profiles of the hIAPP monomer in the presence of brazilin (brazilin/hIAPP = 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). As for the three MD trajectories, it is clear that the initial α-helix structure was well preserved in the entire 100 ns simulation, although it also decreased somewhat as the simulation time increased. Notably, throughout the three repetitive MD simulations, no β-sheet structure was observed during the whole MD simulation, which is a prerequisite for the aggregation of hIAPP.
1). As for the three MD trajectories, it is clear that the initial α-helix structure was well preserved in the entire 100 ns simulation, although it also decreased somewhat as the simulation time increased. Notably, throughout the three repetitive MD simulations, no β-sheet structure was observed during the whole MD simulation, which is a prerequisite for the aggregation of hIAPP.
The above experimental and simulation results imply that the conformational transition from initial α-helix to β-sheet is postponed by brazilin, which again was consistent with the previous CD results. A similar phenomenon was also found for other small molecule inhibitors.26,70 However, it is distinctly different from some other potent inhibitors, such as EGCG25 and resveratrol,10 which have been demonstrated to completely inhibit the conformational transition of hIAPP. It can thus be concluded that brazilin greatly retards the conformational transition of hIAPP from its initial predominantly α-helical to the aggregation-prone β-sheet-dominated form, which implies that brazilin indeed has a direct inhibitory effect on hIAPP aggregation.
Effects of brazilin on hIAPP-induced cytotoxicity
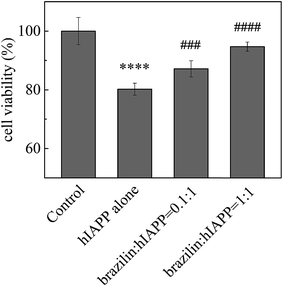
In order to determine whether brazilin is able to modulate hIAPP-induced cell death, the pancreatic β-cell line INS-1 was used to study the countering effects of brazilin on the cytotoxicity of hIAPP aggregates. The INS-1 cell line has been widely used as a model in studies of cytotoxicity related to T2DM.71 In order to first determine if brazilin alone shows significant cytotoxicity, it was added at different concentrations to the cell cultures to determine its maximal tolerance dose (Fig. S3†). In these experiments, the MTT absorbance of the cell media containing only INS-1 cells without hIAPP or brazilin was measured, and the value was defined as 100% cell viability. From Fig. S3,† it is clear that brazilin at concentrations between 0.8 and 32 μM, with the exception of 16 μM, showed almost no cytotoxicity towards the INS-1 cells, as evidenced by the ∼100% cell viability, the same as the untreated control (Fig. S3†). Interestingly, a significant increase in viability was observed with 16 μM brazilin. As for SH-SY5Y cells, no cytotoxicity was observed at lower brazilin concentrations (≤10 μM). However, when the SH-SY5Y cells were treated with 30 μM brazilin, 35% cytotoxicity was observed.31 Therefore, a significant increase in viability with 16 μM brazilin should be biologically relevant.Subsequently, different concentrations of brazilin were co-incubated with hIAPP, after which the resulting species were applied to the cells and their viability determined via MTT assay. Fig. 5 illustrates the inhibitory effect of brazilin on hIAPP-induced cytotoxicity. Upon 24 h of incubation of INS-1 cells with 25 μM hIAPP without brazilin, there was a significant reduction in cell viability, amounting to ∼78% viability relative to the untreated control (Fig. 5). Conversely, brazilin mitigated the observed cytotoxicity of hIAPP in a concentration-dependent manner. For example, cell viability improved by 88% and 96% at molar ratios of brazilin to hIAPP of 0.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, respectively. These results therefore show that brazilin can protect INS-1 cells against hIAPP-induced cytotoxicity.
1, respectively. These results therefore show that brazilin can protect INS-1 cells against hIAPP-induced cytotoxicity.
Brazilin disassembles preexisting hIAPP fibrils
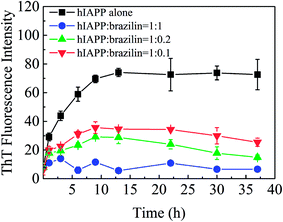
The removal or remodeling of amyloid fibrils is another central therapeutic target in the treatment of amyloid-associated diseases. Recent experimental evidence shows that the toxic oligomeric species are predominantly formed from monomers through a fibril-catalyzed secondary nucleation after a critical concentration of amyloid fibrils has been exceeded.72 Furthermore, mature fibrils of hIAPP might catalyze the aggregation of hIAPP as nuclei.73,74 Therefore, in addition to a strong inhibitory effect on fibrillogenesis, the ability to disassemble mature fibrils into nontoxic aggregates is also greatly desired in the search for promising drug candidates. Accordingly, the ability to eliminate mature fibrils is also an essential feature of a potential drug for T2DM. Hence, the effects of brazilin on the mature hIAPP fibrils were investigated.To do so, monomeric hIAPP was first incubated for 24 h to form mature fibrils, after which different concentrations of brazilin were added to the resulting solutions. The effects of brazilin on the mature hIAPP fibrils were monitored using the ThT fluorescence assay. From Fig. 6, it is clear that the fluorescence intensity of the control group contacted only with buffer remained unchanged, implying that no disaggregation happened with the addition of buffer, as expected. On the other hand, upon incubation of brazilin with the mature fibrils at different molar ratios (0.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 0.2
1, 0.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), the corresponding ThT profiles showed that brazilin induced the disassembly of the mature fibrils in a dose-dependent manner. For example, when hIAPP fibrils were exposed to an equimolar concentration of brazilin, the ThT fluorescence intensity decreased by more than 90% within 3 h. This suggests that the pre-existing fibrils were converted into species that do not bind ThT molecules, thus they did not generate ThT fluorescence.
1), the corresponding ThT profiles showed that brazilin induced the disassembly of the mature fibrils in a dose-dependent manner. For example, when hIAPP fibrils were exposed to an equimolar concentration of brazilin, the ThT fluorescence intensity decreased by more than 90% within 3 h. This suggests that the pre-existing fibrils were converted into species that do not bind ThT molecules, thus they did not generate ThT fluorescence.
When pure buffer was added to the mature hIAPP fibrils, there was almost no effect on the well-developed fibrillar morphology after 24 h, so that long and bundled fibrils were also observed (Fig. 7A). By contrast, if the fibrils were treated with an equimolar concentration of brazilin for 24 h, the mature fibrils completely disassembled into granular and amorphous aggregates, in addition to a small number of protofibrils (Fig. 7B). These images indicate that brazilin significantly modifies the morphology of hIAPP fibrils due to its remodeling effects, which was in good agreement with the ThT fluorescence data (Fig. 6).
Furthermore, the cytotoxicity of the aggregates disassembled from hIAPP fibrils was examined using the MTT assay. Brazilin-treated and untreated hIAPP fibrils were added to INS-1 cells and their viability was measured after 24 h of incubation. When different concentrations of brazilin were added to the hIAPP solutions, they all protected INS-1 cells from hIAPP-induced apoptosis to some extent, and the protective effect was dose-dependent (Fig. 8). Compared to the hIAPP-induced cell viability of 78%, brazilin-treated cell viability rates were improved by 12% at a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of brazilin to hIAPP. This indicates that brazilin disassembled the hIAPP fibrils into less toxic aggregates.
1 ratio of brazilin to hIAPP. This indicates that brazilin disassembled the hIAPP fibrils into less toxic aggregates.
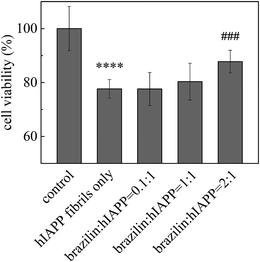 | ||
| Fig. 8 Detoxifying effect of brazilin on mature fibrils of hIAPP. ****p < 0.0001 compared to the control group; ###p < 0.001 compared to the hIAPP-treated group. | ||
Brazilin directly interacts with the fibrillar hIAPP pentamer
The above experimental results have confirmed that brazilin inhibits hIAPP fibrillogenesis, disrupts the mature fibrils and alleviates the corresponding cytotoxicity. However, molecular interactions between brazilin and different hIAPP species are not yet well understood. In order to explore the inhibitory mechanism of brazilin on hIAPP fibrillogenesis, an all-atom molecular dynamics simulation was performed to study the interactions between the hIAPP species and brazilin molecules. Herein, the fibrillar pentamer was selected to represent the hIAPP aggregates (Fig. S1A†). The stability of the hIAPP pentamer was first monitored, and the structural parameters including Cα-RMSD, the total inter-chain contacts and β-sheet content of the hIAPP pentamer in water were computed, as shown in Fig. S4.† For clarity, only the averaged values are provided for the control group comprising only the hIAPP pentamer. From Fig. S4A† it can be seen that in the absence of brazilin, the Cα-RMSD values fluctuate in a very small range after 80 ns after an initial rapid increase, indicating the structural stability of the hIAPP pentamer and convergence of these MD trajectories. Moreover, the values of Cα-RMSD of the hIAPP pentamer in the presence of brazilin were larger than those of the control group. By contrast, the inter-chain contacts and β-sheet content of hIAPP pentamer with brazilin was lower than in the control group (Fig. S4B and C†). This implies that the addition of brazilin induced large conformational fluctuations in the hIAPP pentamer and disrupted the preexisting hIAPP fibrils, which was consistent with the experimental results.The free energies of binding between the hIAPP pentamer and brazilin molecules were first calculated using g_mmpbsa software, and the results are shown in Table 1. It is clear that both hydrophobic and electrostatic energies greatly contribute to the interactions between brazilin and the hIAPP pentamer, although the absolute value of the hydrophobic energies (−429.1 kJ mol−1) is more than three times larger than that of the electrostatic energies (−128.5 kJ mol−1). Brazilin is a hydrophobic compound with two aromatic rings and four hydroxyl groups (Fig. 1), while hIAPP contains many hydrophobic residues, such as Thr, Ala, Leu, Phe, Val, Ile and Tyr (Fig. S1C†). Moreover, brazilin has zero net charge and only contains four hydroxyl groups, suggesting that it interacts with hIAPP by hydrogen bonds via its hydroxyl groups. The number of hydrogen bonds between brazilin and the hIAPP pentamer was calculated (Fig. S5†), revealing that there were more than 10 hydrogen bonds. It can thus be concluded that the direct contacts are dominated by hydrophobic interactions and hydrogen bonding, which is consistent with the results reported for the brazilin-Aβ42 pentamer.75 Therefore, hydrophobic interactions contribute more than 75% to the free energy of binding in the brazilin–hIAPP complex, while electrostatic interactions play a secondary role (<25%). The results suggest that the direct interactions between hIAPP and brazilin lead to its inhibitory effects on hIAPP fibrillogenesis and remodeling effects on mature fibrils of hIAPP.
Residues important for the binding of brazilin to the fibrillar hIAPP pentamer
To further identify whether brazilin preferentially interacts with certain residues of the hIAPP pentamer, the average number of contacts between brazilin and each residue was calculated, as shown in Fig. 9. Herein, the same criterion of 10 contacts between brazilin and each residue used in previous work was also employed to identify the important interacting residues of the hIAPP pentamer.75 From Fig. 9, it is clear that brazilin exhibited strong preferential interactions with the following 11 residues: Asn3, Thr4, Thr9, Arg11, Asn14, Phe15, His18, Ser19, Ser20, Asn21 and Phe23. Particularly, residues from the N-terminus and turn region interacted more strongly with brazilin than those from the C-terminus. Importantly, some of these residues, such as Arg11, Phe15 and Ser19, were reported to play an important role in the self-assembly of hIAPP.76,77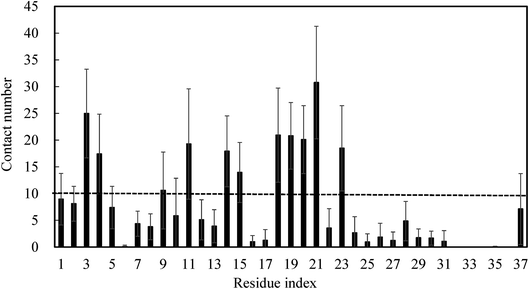 | ||
| Fig. 9 The number of contact between brazilin and each residue was calculated. Data were averaged over all three simulations, with error bars representing the standard deviations. | ||
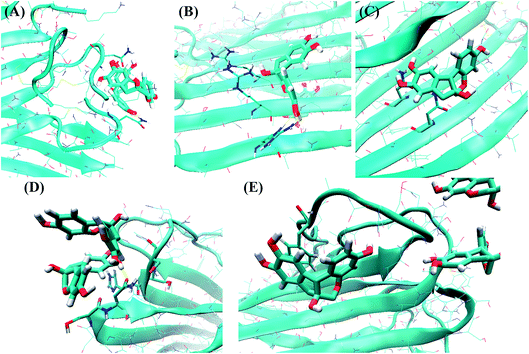
Within the N-terminal region, the six residues Asn3, Thr4, Thr9, Arg11, Asn14 and Phe15 were identified as being important. Fig. 10A shows a snapshot of two brazilin molecules interacting with the Asn3 of two monomers via hydrogen bonds. For example, the hydrogen atom of the hydroxyl group of brazilin can interact with the oxygen atom of the carboxyl group of Asn3 to form a hydrogen bond. At the same time, the oxygen atom of the hydroxyl group of another brazilin molecule can interact with the hydrogen atom of the amide group of Asn3, forming another hydrogen bond. Since brazilin molecules interact with the residue Asn3 and disrupt the intermolecular interactions between Asn3 and other residues of hIAPP pentamer, the atomic contacts between Asn3 and other residues of hIAPP pentamer are used to represent their interactions, and are displayed as a function of simulation time in Fig. S6.† It is clear that the contact number between residue Asn3 and other residues of the isolated hIAPP pentamer of the control group was larger than those of brazilin-treated groups. That is, brazilin can competitively interact with Asn3 via hydrogen bonds and disrupt the interactions of Asn3 with other residues of the hIAPP pentamer. Similarly, interactions between brazilin and the charged residue Arg11 of hIAPP are also dominated by hydrogen bonds (Fig. 10B).
From Fig. S7A,† it is clear that the residue Asn14 is located in the hydrophobic groove of the hIAPP pentamer and forms one hydrogen bond with Ser28 of the hIAPP pentamer. That is, the residues Asn14 and Ser28 are located in the steric zipper section of the hIAPP pentamer (Fig. S7A†). The distance between Asn14 and Ser28 of monomer C was studied in the absence and presence of brazilin (Fig. S8†). For clarity, only resides Asn14 and Ser28 of monomer C were studied. In the absence of brazilin, the distance between Asn14 and Ser28 remained stable at around 0.3 nm, except for some transient increases. That is, hydrogen bonds between Asn14 and Ser28 were stable in water. In the presence of brazilin, however, the distance between Asn14 and Ser28 became greater than 0.6 nm after approximately 1 ns, and fluctuated at around 0.8 nm in the last 50 ns (Fig. S8†). Therefore, the interactions between Asn14 and Ser28 were disrupted by brazilin molecules, and finally the preformed aggregates were gradually disrupted.
Fig. 10C shows the direct interactions between brazilin and Asn14 of hIAPP via one hydrogen bond. However, in the presence of brazilin, one brazilin molecule broke the initial interactions of residues Asn14 and Ser28, and formed a strong hydrogen bond with Asn14 (Fig. 10C and S7B†). Consequently, the parallel conformation of the pentamer was disrupted and some of the residues located in the hydrophobic groove were exposed (Fig. S7B†), after which more brazilin molecules can interact with the exposed residues from the hydrophobic groove and make the β-sheet move from its initial position. In the end, the intermolecular interactions of hIAPP were being interfered with, and the marginal chain was gradually disarranged, so that the β-sheet structures of two lateral monomers were partly disrupted (Fig. S7†), which in turn induced the formation of unstructured aggregates. This observation agrees well with the ThT and TEM results.
As for the turn region, brazilin molecules preferentially interacted with the four hydrophilic residues His18, Ser19, Ser20 and Asn21 (Fig. 9). The direct interactions between these important residues and brazilin are shown in Fig. 10D. It is clear that one of two brazilin molecules directly interacts with these residues via three hydrogen bonds. In addition, π–π stacking is another important interaction between brazilin and the hIAPP pentamer. In this system, brazilin molecules specifically interact with aromatic residues (i.e., Phe15 and Phe23) via π–π stacking interactions. Fig. 10E shows a snapshot of one brazilin molecule interacting with the important aromatic residue Phe23. There are π–π stacking interactions between the indene group of brazilin and the benzene ring of Phe23. A similar interaction type was also found between EGCG and hIAPP oligomers.25 Previous studies have illustrated that aromatic residues (e.g., Phe, Trp and Tyr) play a key role in the formation and inhibition of amyloid fibrils.78,79 Therefore, π–π stacking and hydrogen bonding between brazilin and the hIAPP pentamer disturbed the inter- and intra-molecular interactions of hIAPP, which inhibited fibrillogenesis or disassembled mature fibrils.
Conclusions
In this study, we found that brazilin, a natural compound extracted from Caesalpinia sappan, can inhibit hIAPP fibrillogenesis, disaggregate mature fibril and decreases their corresponding cytotoxicity. ThT fluorescence analysis results confirmed that brazilin prevents hIAPP fibrillogenesis in a dose-dependent manner, and an equimolar concentration of brazilin to hIAPP reduced the ThT fluorescence to ∼10% of the non-treated control. Based on TEM pictures, it is clear that the fibrillar form of hIAPP remodeled to form amorphous aggregates. The conformational transition from its initial predominantly α-helical structure to the β-sheet form was greatly delayed by brazilin based on the CD results. Furthermore, the cell viability data show that co-incubation of hIAPP with brazilin greatly decreases hIAPP-induced cytotoxicity. In addition, brazilin not only disrupted the mature fibrils, inducing the formation of amorphous aggregates, but also protected the cultured cells from hIAPP-induced toxicity. Finally, MD simulations have revealed that the molecular interactions between brazilin and hIAPP were mainly hydrophobic and electrostatic interactions, including hydrogen bonds. It was found that 11 important residues Asn3, Thr4, Thr9, Arg11, Asn14, Phe15, His18, Ser19, Ser20, Asn21 and Phe23 contribute most of the interactions between brazilin and hIAPP. The results therefore demonstrate that brazilin is an inhibitor of hIAPP fibrillogenesis with multiple effects in amyloid inhibition, delay of conformational transition, cell protection and fibril disruption. These findings will help our comprehensive understanding of the aggregation mechanisms of hIAPP and the development of more promising treatments for T2DM.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank for Prof. Robet Tycko from National Institutes of Health, USA, for kindly providing the all-atom model of the hIAPP pentamer. This work was funded by the National Natural Science Foundation of China (No. 21576199).References
- A. Aguzzi and T. O'Connor, Nat. Rev. Drug Discovery, 2010, 9, 237–248 CrossRef CAS PubMed.
- A. Lorenzo, B. Razzaboni, G. C. Weir and B. A. Yankner, Nature, 1994, 368, 756–760 CrossRef CAS PubMed.
- P. Jiang, L. Wei, K. Pervushin and Y. G. Mu, J. Phys. Chem. B, 2010, 114, 10176–10183 CrossRef CAS PubMed.
- L. Caillon, A. R. Hoffmann, A. Botz and L. Khemtemourian, J. Diabetes Res., 2016, 2016, 5639875 Search PubMed.
- A. C. Susa, C. Wu, S. L. Bernstein, N. F. Dupuis, H. Wang, D. P. Raleigh, J. E. Shea and M. T. Bowers, J. Am. Chem. Soc., 2014, 136, 12912–12919 CrossRef CAS PubMed.
- P. Westermark, A. Andersson and G. T. Westermark, Physiol. Rev., 2011, 91, 795–826 CrossRef CAS PubMed.
- L. C. Lopez, O. Varea, S. Navarro, J. A. Carrodeguas, N. Sanchez de Groot, S. Ventura and J. Sancho, Int. J. Mol. Sci., 2016, 17, 964 CrossRef PubMed.
- M. F. Engel, C. C. vandenAkker, M. Schleeger, K. P. Velikov, G. H. Koenderink and M. Bonn, J. Am. Chem. Soc., 2012, 134, 14781–14788 CrossRef CAS PubMed.
- Y. Mo, J. Lei, Y. Sun, Q. Zhang and G. Wei, Sci. Rep., 2016, 6, 33076 CrossRef CAS PubMed.
- Q. Wang, L. Ning, Y. Niu, H. Liu and X. Yao, J. Phys. Chem. B, 2015, 119, 15–24 CrossRef CAS PubMed.
- F. Lolicato, A. Raudino, D. Milardi and C. La Rosa, Eur. J. Med. Chem., 2015, 92C, 876–881 CrossRef PubMed.
- Z. X. Xu, Q. Zhang, G. L. Ma, C. H. Chen, Y. M. He, L. H. Xu, Y. Zhang, G. R. Zhou, Z. H. Li, H. J. Yang and P. Zhou, J. Diabetes Res., 2016, 2016, 1867059 Search PubMed.
- Z. X. Xu, G. L. Ma, Q. Zhang, C. H. Chen, Y. M. He, L. H. Xu, G. R. Zhou, Z. H. Li, H. J. Yang and P. Zhou, ChemPhysChem, 2017, 18, 1611–1619 CrossRef CAS PubMed.
- J. Kellock, G. Hopping, B. Caughey and V. Daggett, J. Mol. Biol., 2016, 428, 2317–2328 CrossRef CAS PubMed.
- K. Sivanesam, I. Shu, K. N. L. Huggins, M. Tatarek-Nossol, A. Kapurniotu and N. H. Andersen, FEBS Lett., 2016, 590, 2575–2583 CrossRef CAS PubMed.
- H. Wang, Z. Ridgway, P. Cao, B. Ruzsicska and D. P. Raleigh, Biochemistry, 2015, 54, 6704–6711 CrossRef CAS PubMed.
- P. C. Rosas, G. M. Nagaraja, P. Kaur, A. Panossian, G. Wickman, L. R. Garcia, F. A. Al-Khamis and A. A. Asea, PLoS One, 2016, 11, e0149409 Search PubMed.
- P. Nedumpully-Govindan, E. N. Gurzov, P. Chen, E. H. Pilkington, W. J. Stanley, S. A. Litwak, T. P. Davis, P. C. Ke and F. Ding, Phys. Chem. Chem. Phys., 2016, 18, 94–100 RSC.
- E. N. Gurzov, B. Wang, E. H. Pilkington, P. Chen, A. Kakinen, W. J. Stanley, S. A. Litwak, E. G. Hanssen, T. P. Davis, F. Ding and P. C. Ke, Small, 2016, 12, 1615–1626 CrossRef CAS PubMed.
- C. Airoldi, E. Sironi, C. Dias, F. Marcelo, A. Martins, A. P. Rauter, F. Nicotra and J. Jimenez-Barbero, Chem.–Asian J., 2013, 8, 596–602 CrossRef CAS PubMed.
- P. Velander, L. Wu, W. K. Ray, R. F. Helm and B. Xu, Biochemistry, 2016, 55, 4255–4258 CrossRef CAS PubMed.
- Q. Sun, J. Zhao, Y. Zhang, H. J. Yang and P. Zhou, RSC Adv., 2016, 6, 105690–105698 RSC.
- C. H. Chen, T. Yao, Q. Zhang, Y. M. He, L. H. Xu, M. Zheng, G. R. Zhou, Y. Zhang, H. J. Yang and P. Zhou, RSC Adv., 2016, 6, 15240–15246 RSC.
- F. L. Meng, A. Abedini, A. Plesner, C. B. Verchere and D. P. Raleigh, Biochemistry, 2010, 49, 8127–8133 CrossRef CAS PubMed.
- Q. Wang, J. Guo, P. Jiao, H. Liu and X. Yao, PLoS One, 2014, 9, e94796 Search PubMed.
- B. A. Cheng, X. R. Liu, H. Gong, L. Q. Huang, H. Chen, X. Zhang, C. Z. Li, M. Y. Yang, B. J. Ma, L. H. Jiao, L. Zheng and K. Huan, J. Agric. Food Chem., 2011, 59, 13147–13155 CrossRef CAS PubMed.
- M. Y. Lim, J. H. Jeon, E. Y. Jeong, C. H. Lee and H. S. Lee, Food Chem., 2007, 100, 1254–1258 CrossRef CAS.
- D. Y. Lee, M. K. Lee, G. S. Kim, H. J. Noh and M. H. Lee, Molecules, 2013, 18, 2449–2457 CrossRef CAS PubMed.
- S. Y. Choi and C. K. Moon, Planta Med., 1997, 63, 400–404 CrossRef CAS PubMed.
- H. Lee, S. W. Kang, H. S. Byun, J. Jeon, K. A. Park, K. Kang, W. Seo, M. Won, J. H. Seok, M. D. Han, H. M. Shen and G. M. Hur, PLoS One, 2015, 10, e0136122 Search PubMed.
- W. J. Du, J. J. Guo, M. T. Gao, S. Q. Hu, X. Y. Dong, Y. F. Han, F. F. Liu, S. Jiang and Y. Sun, Sci. Rep., 2015, 5, 7992 CrossRef CAS PubMed.
- B. O'Nuallain, A. D. Williams, P. Westermark and R. Wetzel, J. Biol. Chem., 2004, 279, 17490–17499 CrossRef PubMed.
- E. Andreetto, L. M. Yan, M. Tatarek-Nossol, A. Velkova, R. Frank and A. Kapurniotu, Angew. Chem., Int. Ed., 2010, 49, 3081–3085 CrossRef CAS PubMed.
- A. Jones, P. Kulozik, A. Ostertag and S. Herzig, J. Alzheimer's Dis., 2009, 16, 787–808 CrossRef PubMed.
- J. H. Luo, S. K. T. S. Warmlander, A. Graslund and J. P. Abrahams, ACS Chem. Neurosci., 2016, 7, 269–274 CrossRef CAS PubMed.
- C. Sims-Robinson, B. Kim, A. Rosko and E. L. Feldman, Nat. Rev. Neurol., 2010, 6, 551–559 CrossRef CAS PubMed.
- M. R. Nicolls, Curr. Alzheimer Res., 2004, 1, 47–54 CrossRef CAS PubMed.
- K. Jackson, G. A. Barisone, E. Diaz, L. W. Jin, C. DeCarli and F. Despa, Ann. Neurol., 2013, 74, 517–526 CrossRef CAS PubMed.
- J. Miklossy, H. Qing, A. Radenovic, A. Kis, B. Vileno, F. Laszlo, L. Miller, R. N. Martins, G. Waeber, V. Mooser, F. Bosman, K. Khalili, N. Darbinian and P. L. McGeer, Neurobiol. Aging, 2010, 31, 1503–1515 CrossRef CAS PubMed.
- L. M. Yan, A. Velkova and A. Kapurniotu, Curr. Pharm. Des., 2014, 20, 1182–1191 CrossRef CAS PubMed.
- W. M. Berhanu, F. Yasar and U. H. Hansmann, ACS Chem. Neurosci., 2013, 4, 1488–1500 CrossRef CAS PubMed.
- R. Hu, M. Zhang, H. Chen, B. Jiang and J. Zheng, ACS Chem. Neurosci., 2015, 6, 1759–1768 CrossRef CAS PubMed.
- M. Zhang, R. Hu, B. Ren, H. Chen, B. Jiang, J. Ma and J. Zheng, ACS Chem. Neurosci., 2017, 8, 524–537 CrossRef CAS PubMed.
- M. Zhang, R. Hu, H. Chen, Y. Chang, J. Ma, G. Liang, J. Mi, Y. Wang and J. Zheng, Phys. Chem. Chem. Phys., 2015, 17, 23245–23256 RSC.
- B. Cheng, H. Gong, X. C. Li, Y. Sun, H. Chen, X. Zhang, Q. Wu, L. Zheng and K. Huang, Proteins: Struct., Funct., Bioinf., 2013, 81, 613–621 CrossRef CAS PubMed.
- D. C. Rodriguez Camargo, K. Tripsianes, K. Buday, A. Franko, C. Gobl, C. Hartlmuller, R. Sarkar, M. Aichler, G. Mettenleiter, M. Schulz, A. Boddrich, C. Erck, H. Martens, A. K. Walch, T. Madl, E. E. Wanker, M. Conrad, M. H. de Angelis and B. Reif, Sci. Rep., 2017, 7, 44041 CrossRef CAS PubMed.
- S. Luca, W. M. Yau, R. Leapman and R. Tycko, Biochemistry, 2007, 46, 13505–13522 CrossRef CAS PubMed.
- K. B. Koziara, M. Stroet, A. K. Malde and A. E. Mark, J. Comput.-Aided Mol. Des., 2014, 28, 221–233 CrossRef CAS PubMed.
- C. Oostenbrink, A. Villa, A. E. Mark and W. F. Van Gunsteren, J. Comput. Chem., 2004, 25, 1656–1676 CrossRef CAS PubMed.
- F. F. Liu, X. Y. Dong and Y. Sun, J. Mol. Graphics Modell., 2008, 27, 421–429 CrossRef CAS PubMed.
- Q. Q. Wang, S. Y. Zhou, W. Wei, X. J. Yao, H. X. Liu and Z. D. Hu, Phys. Chem. Chem. Phys., 2015, 17, 29103–29112 RSC.
- F. F. Liu, X. Y. Dong, L. He, A. P. Middelberg and Y. Sun, J. Phys. Chem. B, 2011, 115, 11879–11887 CrossRef CAS PubMed.
- J. A. Lemkul and D. R. Bevan, Biochemistry, 2012, 51, 5990–6009 CrossRef CAS PubMed.
- E. P. Raman, T. Takeda and D. K. Klimov, Biophys. J., 2009, 97, 2070–2079 CrossRef CAS PubMed.
- B. Hess, C. Kutzner, D. van der Spoel and E. Lindahl, J. Chem. Theory Comput., 2008, 4, 435–447 CrossRef CAS PubMed.
- M. J. Abraham, T. Murtola, R. Schulz, S. Pall, J. C. Smith, B. Hess and E. Lindahl, SoftwareX, 2015, 1, 19–25 CrossRef.
- L. Verlet, Phys. Rev., 1967, 159, 98–103 CrossRef CAS.
- U. Essman, L. Perera, M. L. Berkowitz, T. Darden, H. Lee and L. G. Pedersen, J. Chem. Phys., 1995, 103, 8577–8593 CrossRef.
- B. Hess, J. Chem. Theory Comput., 2008, 4, 116–122 CrossRef CAS PubMed.
- W. Kabsch and C. Sander, Biopolymers, 1983, 22, 2577–2637 CrossRef CAS PubMed.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics Modell., 1996, 14, 33–38 CrossRef CAS.
- R. Kumari, R. Kumar and A. Lynn, J. Chem. Inf. Model., 2014, 54, 1951–1962 CrossRef CAS PubMed.
- S. Bedrood, Y. Li, J. M. Isas, B. G. Hegde, U. Baxa, I. S. Haworth and R. Langen, J. Biol. Chem., 2012, 287, 5235–5241 CrossRef CAS PubMed.
- N. Xiong, X. Y. Dong, J. Zheng, F. F. Liu and Y. Sun, ACS Appl. Mater. Interfaces, 2015, 7, 5650–5662 CAS.
- B. A. Cheng, H. Gong, X. C. Li, Y. Sun, X. Zhang, H. Chen, X. R. Liu, L. Zheng and K. Huang, Biochem. Biophys. Res. Commun., 2012, 419, 495–499 CrossRef CAS PubMed.
- F. L. Palhano, J. Lee, N. P. Grimster and J. W. Kelly, J. Am. Chem. Soc., 2013, 135, 7503–7510 CrossRef CAS PubMed.
- A. Micsonai, F. Wien, L. Kernya, Y. H. Lee, Y. Goto, M. Refregiers and J. Kardos, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E3095–E3103 CrossRef CAS PubMed.
- J. M. Khan, A. Qadeer, S. K. Chaturvedi, E. Ahmad, S. A. A. Rehman, S. Gourinath and R. H. Khan, PLoS One, 2012, 7, e29694 CAS.
- V. L. Anderson and W. W. Webb, Biophys. J., 2012, 102, 897–906 CrossRef CAS PubMed.
- F. Attanasio, M. Convertino, A. Magno, A. Caflisch, A. Corazza, H. Haridas, G. Esposito, S. Cataldo, B. Pignataro, D. Milardi and E. Rizzarelli, ChemBioChem, 2013, 14, 583–592 CrossRef CAS PubMed.
- J. Sjolander, E. Byman, K. Kulak, S. C. Nilsson, E. M. Zhang, U. Krus, G. T. Westermark, P. Storm, B. C. King, E. Renstrom and A. M. Blom, J. Biol. Chem., 2016, 291, 21644–21655 CrossRef PubMed.
- S. I. Cohen, S. Linse, L. M. Luheshi, E. Hellstrand, D. A. White, L. Rajah, D. E. Otzen, M. Vendruscolo, C. M. Dobson and T. P. Knowles, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 9758–9763 CrossRef CAS PubMed.
- M. A. Rubio, D. E. Schlamadinger, E. M. White and A. D. Miranker, Biochemistry, 2015, 54, 987–993 CrossRef CAS PubMed.
- D. E. Schlamadinger and A. D. Miranker, Biophys. J., 2014, 107, 2559–2566 CrossRef CAS PubMed.
- Y. L. Tu, S. Ma, F. F. Liu, Y. Sun and X. Y. Dong, J. Phys. Chem. B, 2016, 120, 11360–11368 CrossRef CAS PubMed.
- M. Z. Zhang, R. D. Hu, H. Chen, Y. Chang, X. Gong, F. F. Liu and J. Zheng, Phys. Chem. Chem. Phys., 2015, 17, 10373–10382 RSC.
- M. Zhang, R. Hu, H. Chen, X. Gong, F. Zhou, L. Zhang and J. Zheng, J. Chem. Inf. Model., 2015, 55, 1628–1639 CrossRef CAS PubMed.
- P. Nedumpully-Govindan, A. Kakinen, E. H. Pilkington, T. P. Davis, P. C. Ke and F. Ding, Sci. Rep., 2016, 6, 19463 CrossRef CAS PubMed.
- L. Wang, A. I. Ilitchev, M. J. Giammona, F. Li, S. K. Buratto and M. T. Bowers, J. Phys. Chem. B, 2016, 120, 11905–11911 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Additional Fig. S1–S9. See DOI: 10.1039/c7ra05742c |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |