Open Access Article
Open Access ArticleGenome-wide identification and characterization of an amino acid permease gene family in Nicotiana tabacum†
Yingying Zhao‡
ab,
Yalong Xu‡b,
Zhong Wangb,
Jianfeng Zhangb,
Xia Chenb,
Zhengfeng Lic,
Zefeng Lib,
Lifeng Jinb,
Pan Weib,
Lin Zhangd,
Xiaoquan Zhangd,
Ran Wang *b and
Fang Wei*a
*b and
Fang Wei*a
aSchool of Life Sciences, Zhengzhou University, No. 100 Science Road, Zhengzhou, Henan 450001, China. E-mail: zyy244792855@163.com; fangwei@zzu.edu.cn
bZhengzhou Tabacco Research Institute of CNTC, No. 2 Fengyang Street, Zhengzhou, Henan 450001, China. E-mail: xuyalong2007@163.com; zhongwang1123@126.com; zhangjianfeng83@outlook.com; chenxia0372@163.com; 379359757@qq.com; jin_lf@126.com; weipan83@126.com; wangr@ztri.com.cn
cTechnology Center, China Tobacco Yunnan Industrial Co., Ltd., Kunming, Yunnan 650231, China. E-mail: 1208913744@qq.com
dCollege of Tobacco Science, Henan Agricultural University, No. 63 Agriculture Road, Zhengzhou, Henan 450002, China. E-mail: hnndzhang@126.com; zxq013415@163.com
First published on 2nd August 2017
Abstract
The amino acid permease (AAP) protein family is an important class of amino acid transporter. However, little information about this gene family is available in tobacco. Here, a total number of 15, 7, and 6 full-length putative AAP genes have been identified in the genomes of Nicotiana tabacum and progenitors of modern tobacco, Nicotiana sylvestris and Nicotiana tomentosiformis, respectively. We performed a multiple analyses of the AAP gene family in tobacco and reported data including phylogenetic relationships, gene structures, predicted protein structures and information about conserved motifs. These tobacco AAP family members shared high levels of similarity in their nucleotide and amino acid sequences. The gene structures of the NtAAP loci were all separated by introns. Phylogenetic analyses indicated that these AAP genes were clustered into three groups. We also performed expression profiling analyses of the NtAAP genes in various tobacco tissues/organs using quantitative PCR. The differential expression patterns of the NtAAP genes suggested that the family members might have different biological functions in tobacco plants. To study the function of NtAAP2-2, its coding sequence was cloned and overexpression/RNA interference vectors were constructed and these vectors were transformed into tobacco. Compared with wild-type tobacco, the contents of Glu, Gln, Asp and Asn were found to change in the transgenic tobacco leaves, indicating that NtAAP2-2 was involved in the transportation of Glu, Gln, Asp and Asn in tobacco. Our results can serve as fundamental research for future evolutionary and functional characterization studies of the AAP genes in tobacco.
Introduction
Nitrogen (N) is an important macronutrient and has been reported to be essential for the growth, development and reproduction of higher plants. N is used for the synthesis of essential compounds such as chlorophyll, nucleic acids (DNA and RNA), and amino acids (peptides and proteins). Limitations of the supply of N will lead to severe metabolic disturbance and growth inhibition in plants. In the majority of plants, the assimilation of nitrogen is not only in the form of inorganic-nitrogen such as nitrate and ammonium, which is converted into amino acids in root or shoot tissues, but also in the form of organic-nitrogen including urea, amino acids, etc.1 The assimilation form of nitrogen is affected by many factors, and it mainly depends on the plant species, amino acid concentrations and soil pH values.2,3 The inorganic nitrogen absorbed from the soil is used in the synthesis of amino acids, which are immediately used for metabolism, transiently stored (e.g. in the vacuole) or transported in the phloem to developing vegetative or reproductive sink tissues. In most plants, the absorption and redistribution of amino acids depend on the plasma membrane transporters. The amino acids synthesized in the leaves were transported to the sink tissues mainly throuth the phloem, but the amino acids synthesized in the roots were transported to the mature leaves with the transpiration stream through the xylem. The functions of amino acid transporters are mainly involved in three fields: notably in xylem and phloem loading, source to sink transportation of amino acid, and the uptake of amino acid from external environment.4,5 Therefore, amino acid transporters play an indispensable role in the regulation of plant growth and development.It has been demonstrated that the amino acid transporters are divided into two major superfamilies: the amino acid, polyamine and choline transporters superfamily (APC) and the amino acid transporter family (ATF).7,8 The APC superfamily consists of two families: the cationic amino acid transporters (CAT) family and gamma-aminobutyric acid transporter (GAT). The ATF superfamily comprises of at least five families: amino acid permeases (AAPs), proline transporters (AtProTs), lysine and histidine transporters (AtLHTs), aromatics and neutral amino acid transporters (AtANTs) and auxin transporters (AUXs).9 Glutamine, glutamate and asparagine are the major transport forms of organic nitrogen, comparatively, aspartate is transported by most AAPs with an extremely low affinity and efficiency.10,11 In Arabidopsis, there are at least 53 genes annotated as amino acid transporters, which are present at the plasma membrane or tonoplast.6 Among all the transporters, 14 APC-type genes have been identified. 9 APC proteins (CAT1–CAT9) have a 14 transmembrane (TM) topology, while the remaining 5 only have a 12 TM topology.8 Furthermore, all the characterized ATF transporters are proton coupled, and have a 9 to 11 transmembrane (TM) domain topology.6
The AAPs have been suggested to be involved in a number of physiological process in plants. Up to now, 8, 11, 19 AAP genes have been identified in Arabidopsis thaliana, Medicago sativa, Oryza sativa, respectively.12,13 AtAAP1 belongs to a family of eight members (AtAAP1-8) in Arabidopsis, and yeast complementation experiment showed that AAP1 mediated transport of neutral and acidic amino acids.14 The expression of PsAAP1 in Pisum sativum L. was found in the seed coat, cotyledon surface and the phloem throughout the plant. It was suggested the PsAAP1 might be the major amino acid transporter in the phloem, and might also function on seed development and the accumulation of storage protein in seeds,15 since transgenic PsAAP1 in pea limited seed protein levels.16 The importance of StAAP1 in long-distance transport of amino acids from source leaves to sink organs11 has also been pointed out in Solanum tuberosum. The Arabidopsis amino acid transporters display tissue- and organ-specific expression patterns during the development. AtAAP1 is localized to the root tip and epidermis cells including root hairs, and it is important in the uptake of neutral and acidic amino acids when soil solution concentration is higher than 50 mM.17 Zhang et al.18 demonstrated that AtAAP2 located to the phloem throughout the plant, and was important for amino acid transportation from xylem to phloem and the sink N/C supply. AtAAP3 is presented in the root phloem, and transports neutral and basic amino acids, but ataap3 mutants showed no obvious phenotypic defects.19 AtAAP4 is expressed in leaves, stems, and flowers, whereas AtAAP5 transcripts is found in all tissues, but with different levels. Furthermore, AtAAP6 and AtAAP8 were characterized as high affinity amino acid transporters. The expression of AtAAP8 was mainly in the veins of young flowers, peduncle, siliques and in very young seeds. The ataap8 mutants showed a reduction in seed numbers per silique, suggesting that AtAAP8 was responsible for the import of amino acid into the seed.10,20
Tobacco is one of the most important economic crops in the world. The content of amino acids will affect the quality of tobacco. Since the functions of tobacco AAP genes are unknown, it is necessary to identify and characterize the AAP gene family. In our study, we identified fifteen AAP genes in the allotetraploid Nicotiana tabacum (N.tabacum), and 7, 6 AAP genes in the diploid tobacco progenitor Nicotiana sylvestris (N.sylvestris) and Nicotiana tomentosiformis (N.tomentosiformis), respectively. A systematic analysis of the NtAAP genes in N. tabacum was presented, including the location of each member of NtAAP gene family in the chromosome, the organization of their intron/exon, their evolutionary relationships and the expression profiles of the NtAAP genes in tobacco tissues. The findings of this study will serve as a foundation for further studies of NtAAP genes at physiological and molecular levels.
Experimental section
Materials
N. tabacum (Honghua Dajinyuan) was used in the analysis of the expression profiles of the NtAAP genes. Tissue materials including roots, stems, leaves at tobacco resetting stage, and vigorous stage, flowering bud stage, flowering stage, topping stage, and middle leaf maturing stage, were collected from the experimental field in the Yunnan province of China. In addition, sepals, corollas, anthers, capillament, stigmas, styles and ovarys at flowering period were also harvested. All the plant materials were immediately frozen in the liquid N2 after collection, and then stored at −80 °C prior to RNA extraction.To clarify the function of the NtAAP2 gene under N starvation, tobacco seedlings were grown under N deficient treatment for two weeks. The N stress nutrient solution contained the following compounds: KH2PO4 0.136 g L−1, K2SO4 0.261 g L−1, CaCl2 0.222 g L−1, MgSO4 0.493 g L−1, H3BO3 6.2 mg L−1, KI 0.83 mg L−1, MnSO4 22.3 mg L−1, ZnSO4 8.6 mg L−1, Na6M07O24 0.25 mg L−1, CuSO4 0.025 mg L−1, CoCl2 0.025 mg L−1, Fe-EDTA 0.042 g L−1, 0.101 g L−1 KNO3, 0.0264 g L−1 (NH4)2SO4. Tobacco seedlings, were treated for 2 weeks, the fifth leaf was used for total RNA isolation and the analysis of amino acid content.
Tobacco AAP gene identification
We downloaded 8 AtAAPs and 19 OsAAPs protein sequences from Arabidopsis thaliana genome database website TAIR (http://arabidopsis.org) and Oryza sativa genome database website TIGR (http://rice.plantbiology.msu.Edu) as the query sequences, respectively. We then performed BLASTP search against N.tabacum, N.sylvestris and N.tomentosiformis protein sequences in China Tobacco Genome Database (V3.0) respectively, with the parameter E-value 1.0 × 10−5. Non-redundant candidate AAP were obtained from the blastp result. The Pfam database (http://pfam.sanger.ac.uk/search) and SMART (http://smart.embl-heidelberg.de/) were used to further verify the reliability of these candidate sequences and confirm each candidate NtAAP protein as a member of AAP family. The AAP protein sequences of other corresponding plant species were collected by searching the NCBI GeneBank database using “amino acid permease” as a query keyword.Gene structure analysis
The gene structures were drawn by the Gene Structure Display Server (GSDS, http://gsds.cbi.pku.edu.cn/) using mRNA and genomic sequence obtained from the genome sequences.Protein sequences of NtAAP genes characteristic analysis
The conserved motifs in the protein sequences were identified with the Multiple Em (Expectation Maximization) for Motif Elicitation (MEME) program Version 4.11.2 (http://meme-suite.org/tools/meme). The MEME program was employed using the following parameters: number of repetitions, zero or one occurrence per sequence; maximum number of motifs, 10; optimum motif width set to >40 and <120. The protein structure of each NtAAP protein was confirmed by SMART (http://smart.embl-heidelberg.de/). In addition, protein sequences of NtAAP genes were also analyzed by using the online ExPASy programs (http://www.expasy.org-/tools/protparam.html). The number of amino acids, amino acid composition, molecular weight, theoretical isoelectric point (pI), and instability index were obtained by this tool. Furthermore, the topology of NtAAP in the N.tabacum was predicted by combining the results from multiple sequence alignment, secondary structure prediction, and transmembrane helix prediction. Multiple sequence alignment was performed using DNAMAN with default parameters. Accordingly, hydrophobicity analysis was performed using HMMTOP (http://www.enzim.hu/hmmtop/) and TMRPres2D.Phylogenetic analysis
To examine the evolutionary relationships among the AAP genes of tobacco and other plant species, multiple sequences alignment was performed using Clustal W with the AAP protein sequences of N. tabacum, Arabidopsis thaliana, Zea mays, Brassica rapa and Oryza sativa. Gaps and ambiguously aligned regions were removed. Next, an unroot phylogenetic tree was constructed based on the alignment using ML method implemented in MEGAC5.1. Bootstrap testing was performed with 1000 re-samplings, and then colored with the iTOL tool (http://itol.embl.de/).Gene expression analysis
Total RNA was extracted using a Superpure Plantploy RNA kit (Gene Answer), and then treated by RNA-free DNase I. The concentration and quality of RNA samples were evaluated with Nanodrop 2000 (Thermo), and the integrity of RNA was checked by 1% agarose gel electrophoresis. The first-stand of cDNA was synthesized from 1 μg total RNA using Reverse Transcript M-MLV (Gene Answer) with Random Primer. The concentration of cDNA was measured by Nandrop 2000, and then the volume was diluted to the concentration of 50 ng μL−1.For quantitative PCR analysis, the gene specific primers were designed based on the tobacco coding sequence, and 26 s rRNA was used as a reference gene. The primer sequences were listed in Table S1.† qPCR was performed using a 2× SYBR Green qPCR Mix (Gene Answer) on a Bio-Rad CFX96 TM instrument (Bio-Rad). The qPCR program was set as follows: 95 °C for 3 min, 40 cycles of 95 °C for 30 s, 60 °C for 30 s and 72 °C for 40 s. Each reaction was carried out in 25 μL reaction volume containing 12.5 μL SYBR Green Mix, 150 ng cDNA sample, 1 μL upstream/downstream primer. Three replicates were set for each sample. The average value of the three Cps was obtained and then calculated with the method of 2−ΔΔCp.21
Cloning of NtAAP2-2, vector construction, plant transformation and confirmation
Primers for NtAAP2-2 cloning were designed by DNAMAN (V6.0) software, and the coding sequence of NtAAP2-2 were cloned by PCR amplification with high-fidelity DNA polymerase (TransGen Biotech). The 50 μL PCR amplification reaction mixture contained 10× PCR buffer II, 10 mM dNTP mix, 1 μM of each primer, 1 μL Hifiase, and 150 ng of template cDNA. PCR amplification was performed on a Bio-Rad DNA thermal cycler.To clarify the physiological role of NtAAP2-2 in N. tabacum, we designed gene-specific primers (NtAAP2-2-CDS list in Table S1†) to amplify the full length CDS of NtAAP2-2 gene. The sequence was then cloned into the overexpression (OE) vector (Super pCAMBIA1300) with SpeI and KpnI restriction sites. Besides, we constructed the NtAAP2-2-RNAi (RNA interference) vector. The primers with attB adapters were designed from the partial CDS of NtAAP2-2, listed in the ESI Table S1.† After amplification, the fragment was then integrated into the expression vector pHellsgate2 by BP Clonase enzyme mix (invitrogen).
NtAAP2-2-RNAi and NtAAP2-2-OE plasmids were all transformed into Agrobacterium strain GV3101, which were used to generate N. tabacum transgenic lines. The transgenic method was described in the previous reports.16,22 Wild-type (K326) plants were used as controls in the below experiments. T0 transgenic plants were grown in the greenhouse until seed harvesting. The NtAAP2-2-OE T1 transgenic plants were obtained through the selection on MS medium containing 250 mg L−1 cephalosporin and 5 mg L−1 hygromycin. The NtAAP2-2-RNAi T1 transgenic plants were selected by 150 mg L−1 kanamycin and 250 mg L−1 cephalosporin. To confirm the integration of transgenic plants, PCR was used to screen the genomic DNA. The NtAAP2-2-RNAi transgenic plants were selected by using primers of marker gene Kanamycin (nptII). The NtAAP2-2-OE transgenic plants were selected by hygromycin primers, NtAAP2-2 flanking primers and flag-R, listed in Table S1.† The amplification program as list: 94 °C, 3 min; 94 °C, 30 s, 55 °C, 30 s, 72 °C, 2 min, 28 cycles; 72 °C, 10 min; 4 °C.
Free amino acid analysis
The amino acid contents in tobacco leaves from transgenic individuals and wild type plants were evaluated by amino acid analyzer method. Free amino acid was extracted from 0.1 g freeze-dried leaves with 2 mL lithium citrate. The mixture was extracted with ultrasonic for 30 min followed by centrifugation (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 10 min), the supernatant was collected and filtered through a 0.22 μm filter, and then transferred to amino acid analyzer. The standard amino acid mixture was supplemented with 2.5 μmol mL−1 asparagine, asparagine, alanine, arginine, glycine, glutamic, glutamine, histidine, isoleucine, leucine, lysine, benzene alanine, proline, serine, threonine, tyrosine and tryptophan.
000 rpm, 10 min), the supernatant was collected and filtered through a 0.22 μm filter, and then transferred to amino acid analyzer. The standard amino acid mixture was supplemented with 2.5 μmol mL−1 asparagine, asparagine, alanine, arginine, glycine, glutamic, glutamine, histidine, isoleucine, leucine, lysine, benzene alanine, proline, serine, threonine, tyrosine and tryptophan.
Results and discussion
Tobacco putative AAP gene identification
A total of 18 AAP potential genes were identified from the N.tabacum from the tobacco genome databases, and then were used for multiple sequence alignment by DNAMAN software. Three NtAAP potential genes (the information of these genes were showed in Table S2†) were removed for their obvious lower similarity (less than 40%) compared to other gene sequences, and finally 15 putative NtAAP genes were retained (Table 1). Among the 15 putative NtAAP sequences, NtAAP3, NtAAP6 and NtAAP2 have 7, 6 and 2 allelic haplotypes, respectively. The allelic haplotypes of each AAPs were indicated additional “−1” to “−7” behind the gene name. Sequence identity analysis showed that the amino acid sequences of NtAAPs shared 69.96% identity, and the nucleotide identity was up to 71.04% (Table S3†). NtAAP proteins were analyzed by ExPASy programs tool. As shown in Table 1, almost all the putative NtAAP CDS were about 1500 bp in length, which gave rise to about 500 amino acid sequences of putative NtAAP proteins. But the gene and CDS length of NtAAP6-3 was 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 586 bp and 1332 bp, respectively. Instability index of all the predicted proteins were inferior to 40. Isoelectric point was estimated among 8.36 to 9.52, and the tobacco putative NtAAP proteins were all alkaline. Furthermore, we also identified 7 and 6 putative AAP genes in diploid ancestor N.sylvestris and N.tomentosiformis, respectively.
586 bp and 1332 bp, respectively. Instability index of all the predicted proteins were inferior to 40. Isoelectric point was estimated among 8.36 to 9.52, and the tobacco putative NtAAP proteins were all alkaline. Furthermore, we also identified 7 and 6 putative AAP genes in diploid ancestor N.sylvestris and N.tomentosiformis, respectively.
| Gene ID | Gene name | MW (kDa) | PI | Amino acid size (bp) | CDS size (bp) | gDNA size (bp) | Instability index | Conserved domains | |
|---|---|---|---|---|---|---|---|---|---|
| N.tabacum | Ntab0475230 | NtAAP3-1 | 52.73 | 8.69 | 480 | 1443 | 4376 | 31.47 | 1 Aa_trans |
| Ntab0186340 | NtAAP3-2 | 52.73 | 8.69 | 480 | 1443 | 4440 | 31.47 | 1 Aa_trans | |
| Ntab0484720 | NtAAP3-3 | 52.49 | 8.69 | 478 | 1437 | 4583 | 32.12 | 1 Aa_trans | |
| Ntab0049050 | NtAAP3-4 | 52.48 | 8.69 | 478 | 1437 | 2303 | 32.12 | 1 Aa_trans | |
| Ntab0126970 | NtAAP3-5 | 52.75 | 8.52 | 480 | 1443 | 2384 | 30.68 | 1 Aa_trans | |
| Ntab0889300 | NtAAP3-6 | 52.76 | 8.36 | 480 | 1443 | 2384 | 30.68 | 1 Aa_trans | |
| Ntab0329990 | NtAAP3-7 | 52.55 | 8.36 | 479 | 1440 | 2383 | 31.81 | 1 Aa_trans | |
| Ntab0126360 | NtAAP6-1 | 55.04 | 9.09 | 501 | 1506 | 4922 | 38.55 | 2 Aa_trans | |
| Ntab0759460 | NtAAP6-2 | 51.97 | 8.73 | 475 | 1428 | 5056 | 36.3 | 1 Aa_trans | |
| Ntab0648520 | NtAAP6-3 | 48.62 | 9.07 | 443 | 1332 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 586 586 |
37.65 | 2 Aa_trans | |
| Ntab0439400 | NtAAP6-4 | 53.87 | 8.99 | 487 | 1464 | 4407 | 37.08 | 1 Aa_trans | |
| Ntab0331750 | NtAAP6-5 | 51.23 | 8.85 | 464 | 1395 | 3864 | 32.61 | 1 Aa_trans, 1Trp_Tyr_perm | |
| Ntab0927350 | NtAAP6-6 | 52.02 | 9.10 | 472 | 1419 | 5189 | 32.91 | 1 Aa_trans | |
| Ntab0821150 | NtAAP2-1 | 56.99 | 9.52 | 513 | 1542 | 3914 | 31.04 | 1 Aa_trans | |
| Ntab0800540 | NtAAP2-2 | 56.79 | 9.45 | 512 | 1539 | 3748 | 31.49 | 1 Aa_trans | |
| N.sylvestris | Nsyl0356170 | NsyAAP3-1 | 52.48 | 8.56 | 478 | 1537 | 2304 | 32.12 | 1 Aa_trans |
| Nsyl0306370 | NsyAAP3-2 | 52.55 | 8.36 | 479 | 1440 | 2371 | 31.81 | 1 Aa_trans | |
| Nsyl0116290 | NsyAAP6-1 | 55.04 | 9.09 | 501 | 1506 | 4932 | 38.55 | 2 Aa_trans | |
| Nsyl0231930 | NsyAAP6-2 | 53.75 | 8.63 | 489 | 1470 | 4525 | 37.84 | 1 Aa_trans | |
| Nsyl0227040 | NsyAAP6-3 | 51.36 | 9.22 | 466 | 1401 | 5187 | 31.03 | 1 Aa_trans, 1Trp_Tyr_perm | |
| Nsyl0178970 | NsyAAP6-4 | 51.36 | 9.22 | 466 | 1401 | 5187 | 31.03 | 1 Aa_trans, 1Trp_Tyr_perm | |
| Nsyl0101440 | NsyAAP2 | 56.99 | 9.52 | 513 | 1542 | 3914 | 31.04 | 1 Aa_trans | |
| N.tomento-siformis | Ntom0167920 | NtomAAP3-1 | 52.51 | 8.69 | 478 | 1437 | 1915 | 31.45 | 1 Aa_trans |
| Ntom0127780 | NtomAAP3-2 | 52.73 | 8.52 | 480 | 1443 | 2382 | 30.85 | 1 Aa_trans | |
| Ntom0030730 | NtomAAP6-1 | 51.97 | 8.84 | 475 | 1428 | 5074 | 36.30 | 1 Aa_trans | |
| Ntom0331850 | NtomAAP6-2 | 28.88 | 9.07 | 269 | 810 | 2001 | 43.93 | 1 Aa_trans | |
| Ntom0300680 | NtomAAP6-3 | 51.43 | 9.00 | 466 | 1401 | 4613 | 31.69 | 1 Aa_trans, 1Trp_Tyr_perm | |
| Ntom0297890 | NtomAAP2 | 56.82 | 9.25 | 512 | 1539 | 3747 | 32.29 | 1 Aa_trans |
Exon–intron structure and chromosome location of NtAAP genes
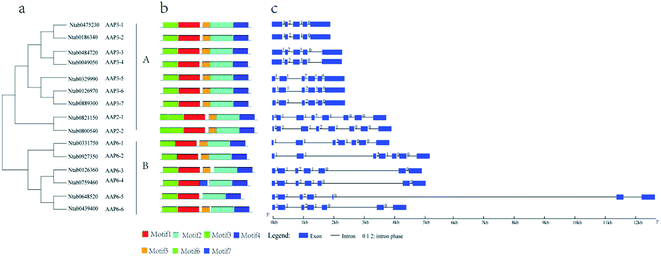
Based on phylogenetic analysis, the AAP genes in the tobacco could be divided into two major clades (Fig. 1a). The exon–intron structures of NtAAP genes were analyzed by Gene Structure Display Server (GSDS), which were showed in the Fig. 1c. All of the NtAAP genes were disrupted by introns. In the clade A, the CDS of NtAAP3-1 to NtAAP3-4 were comprised by 5 exons, and the first exon was larger than other NtAAP3 exons, which had 6 exons. NtAAP6 had 7 exons that belonged to the clade B, in addition, NtAAP2 had 8 exons. Comparative analysis among the NtAAP family suggested that the first and the last exon of NtAAP3 were more likely to be splitted by introns. In addition, we ranked the intron positions relative to the conserved exons, and finally identified 7 putative positions for introns, among which 5 introns (except introns 1 and 7) were found in most of the NtAAP sequences (Fig. S1†). The lack of one or more introns, which mainly occurred in the 1th, 2th, 7th intron positions, lead to the formation of larger exons, including the 333 bp and/or 653 bp exons in some of the NtAAP genes (Fig. S1†).Structure and motif analysis of the NtAAP proteins
We also analyzed the amino acid sequences of the NtAAPs using the MEME motif search tool, and identified a total of seven distinct motifs. The length and sequence information of these 7 motifs were listed in Table S4.† The position of each motif in the NtAAP protein was listed in Fig. 1b. Motifs 1–4 were transmembrane domain with transmembrane transporter activity, and were present in all NtAAPs. Motif 5 was existed in all the NtAAP proteins except for NtAAP6-6. Motif 6 was only presented in the NtAAP2, and was located in its N-terminus. Moreover, the motif 5 and 6 were functional unknown domains, since there was no hit when searching the sequence in the InterProScan. The presence of the same type of conserved motifs indicated that the NtAAP family members might have similar functions. In addition, all the NtAAP protein sequences were searched against the SMART database to predict the putative protein structures, and the results revealed that each of the proteins contained at least one Aa_trans domains (Table 1).Transmembrane structure of the NtAAPs
The hydrophobic transmembrane domain of the NtAAPs was predicted by HMMTOP. It was shown that the family members contained 9–12 transmembrane helices (Fig. S2†). According to the position of the N/C-terminal and the number of predicted transmembrane helices, the transmembrane structure could be divided into 3 types: N/C terminals were all predicted in cytoplasmic contained 10 TMs, including NtAAP3-1 to NtAAP3-4, NtAAP6-1, NtAAP6-5 and NtAAP6-6; N/C terminals were all predicted in extracellular contained 12 TMs, including NtAAP2, and NtAAP3-5 to NtAAP3-7; N/C-terminals of the other NtAAP proteins were predicted in the extracellular and cytoplasmic, respectively, containing 9 TMs (Fig. S2†). Multiple sequences alignment displayed that the limits of the TM 1 segment (H, I, I, T, A, V, I, G, S, G, V, L, S, L, A, W, A) were presented in all the NtAAPs (Fig. S3†), therefore, this region was highly conserved.Phylogenetic analysis of the tobacco AAP genes
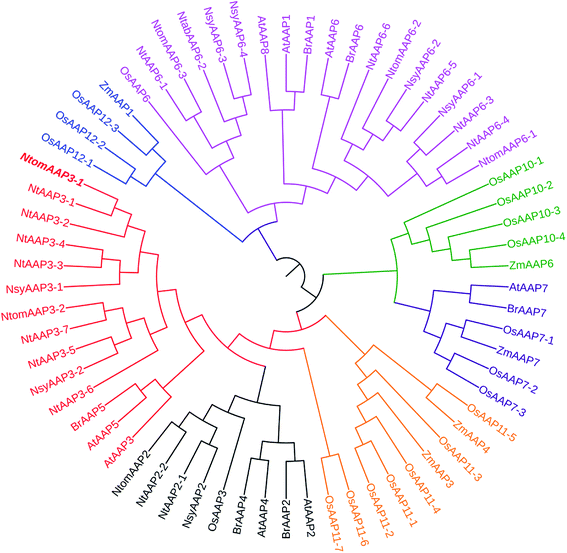
Classification of genes and phylogenetic relationship are important for the functional analysis of a gene family. To examine the evolutionary relationship among the AAP genes of tobacco and other plant species, multiple sequences alignment was performed using Clustal W, and an unroot tree was constructed based on the alignment using ML method implemented in MEGA 5.1. Based on the phylogenetic analysis, the AAP proteins from N. tabacum, Arabidopsis thaliana, Zea mays, Brassica rapa and Oryza sativa were classified into 4 main clusters (1, 2, 3, 4), which was consistent with the previous report.13 The cluster 1 was sub-divided into three subclusters (A, B, C), and the cluster 4 was sub-divided into two subclusters (A and B) (Fig. 2). The NtAAPs were divided into clusters 1 and clusters 4, which contained 9 and 6 NtAAP proteins, respectively. All of the NtAAP3 and NtAAP2 proteins belonged to the cluster 1A and cluster 1C, respectively. It is well known that gene structural diversity drives the evolution of multigene families. In Cluster 1A, NtAAP3-1 to NtAAP3-4 in the same branch contained different exons numbers with other NtAAP3 genes (Fig. 1c), indicating that these NtAAP3 genes might have different evolutionary processes. In addition, cluster 1A also included Arabidopsis AtAAP3 and AtAAP5. Cluster 1C only contained one rice AAP protein (and multiple eudicot sequences), which was distributed to the same branch with NtAAP2 proteins in tobacco. Notes that NtAAP2 gene has been appeared before the evolution of dicotyledonous and monocotyledonous. All of these genes in cluster 1C appeared to be involved in the loading of amino acid into phloem.13 NtAAP6 proteins in tobacco were divided into cluster 4B, which also contained the Arabidopsis AtAAP1, AtAAP6, AtAAP8, and OsAAP6. NtAAP6-1/NtAAP6-2 and OsAAP6, other NtAAP6 proteins and AtAAP1, AtAAP6, AtAAP8 were branched into a group, respectively. These genes might function in the transportation of amino acid from xylem to phloem.23 Interestingly, cluster 1B and cluster 2 only contained monocots protein sequences, it is possible that this large group of AAPs might be evolved after monocot–dicot divergence, and might have specialized roles in monocots or dicots.Expression pattern analysis of the NtAAP genes in tobacco tissues
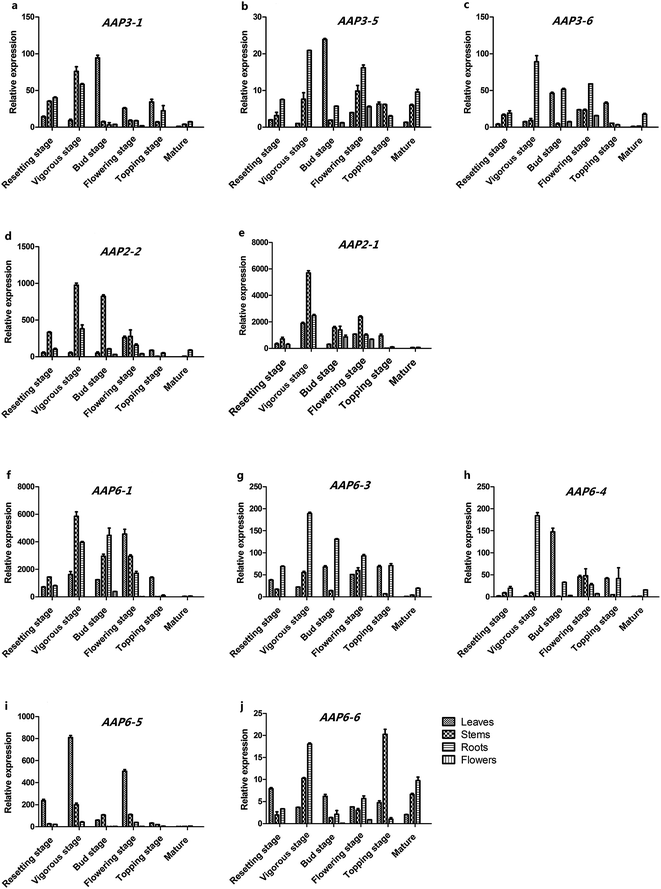
To detect the relative expression levels of each NtAAP gene in the tobacco leaves, stems, roots and flowers from different developmental stages, a comprehensive quantitative PCR analysis was further performed. As shown in Fig. 3, NtAAP6-1, NtAAP6-3 and NtAAP6-6 were highly expressed in stems and root, whereas NtAAP6-5 was mainly expressed in leaves. NtAAP2 was highly expressed in stems at resetting stage and vigorous stage. In addition, NtAAP2 gene was also ubiquitously expressed in the tissues of other growth stages. NtAAP3-1, NtAAP3-6 and NtAAP3-7 were highly expressed in roots and stems at resetting, vigorous and flowering stage (Fig. 3). However, their expression levels at flowering bud and topping stage were higher in leaves than those in other tissues. Furthermore, NtAAP3 gene was also expressed in the floral organ.Phylogenetic analysis and expression patterns can be used to predict the molecular functions of genes involved in different physiological processes. AtAAP3, AtAAP5 and NtAAP3 are assigned to the cluster 1A, and these two AAP genes in Arabidopsis appear to be associated with loading of amino acid into phloem. The function of AtAAP3 in loading amino acid into phloem seems to be restricted to the roots.24 AtAAP5 is associated with root acquisition of amino acid.25 In our analysis, the transcripts of NtAAP3-6 and NtAAP3-7 genes are not only detected in the stems and roots at resetting and vigorous stage, but also in flowers at bud and flower stage. Therefore, NtAAP3 might play a role not only in phloem loading, but also in importation of amino acids into sink tissues. In addition, AtAAP2, AtAAP4 and NtAAP2 are classified into cluster 1C. AtAAP2 functions in phloem loading of amino acids along the transport pathway, and is essential for N supply to seed sinks.18 NtAAP2 is highly expressed in the stems at resetting stage and vigorous stage, and was also detected in the floral organ. Therefore, NtAAP2 may be involved in the delivery of amino acids to seed sinks. NtAAP6 genes, AtAAP1, 6 and 8 are assigned into cluster 4B. AtAAP6 is mainly expressed in sink tissues, such as roots and sink leaves. GUS expression studies revealed that AtAAP6 is also expressed in the xylem parenchyma cell. It's predicted to be important for xylem to phloem transfer of amino acids in Arabidopsis.10,26 Although AAP1, AAP5, LHT1 have been demonstrated to affect root uptake of amino acids, only AAP1 has been shown to be localized to the root epidermis and root hairs, and function in the import of glutamate and neutral amino acids into root hairs and epidermis at relatively high amino acid concentrations.17,25 In addition, AtAAP1-mediated uptake of amino acids by the embryo is important for the storage protein synthesis and seed yield.27 AtAAP8 plays a crucial role for the uptake of amino acids into the endosperm, and supplies the developing embryo with amino acids during early embryogenesis.20 The transcripts of NtAAP6-1, NtAAP6-3, and NtAAP6-6 are detected in the root at relatively high level, therefore, these NtAAP6 genes might have similar functions with the AAP genes in Arabidopsis.
Functional analysis of NtAAP2 in amino acid transportation
We successfully cloned the coding sequence and the gene sequence of NtAAP2-2 from tobacco (Honghua Dajinyuan) (Fig. S4A†). Bioinformatics analysis indicated that the CDS of NtAAP2-2 was 1539 bp, encoding 512 amino acids with molecular weight 56.79 kDa and the pI 9.45 (Table 1). Based on the CDS of NtAAP2-2, we successfully constructed NtAAP2-2 overexpression and RNA interference vectors (Fig. S4A†). To better understand the biological functions of NtAAP2, we constructed the overexpression (OE) and RNAi mutant lines for the NtAAP2-2. When we obtained the T1 generation transgenic lines and controls, DNA was extracted and used as templates for PCR identification. NtAAP2-2-Flag and nptt were used as primers for RT-PCR. The NtAAP2-2-RNAi lines had a specific 659 bp band, and the NtAAP2-2-OE lines had a specific 320 bp band, while these two bands were not present in the controls (Fig. S4B and D†). Six confirmed NtAAP2-2-OE and RNAi transgenic T1 lines were chosen for further analysis, respectively. Moreover, the total RNA of transgenic T1 lines and controls were isolated. The first strand cDNA were synthesized and then used to analyze the expression of NtAAP2-2. The expression level of NtAAP2-2 was dramatically increased in the NtAAP2-2-OE transgenic plants, but significantly reduced in the NtAAP2-2-RNAi lines (Fig. S4C and E†).Tobacco is an important economic crop in China. It is also widely used as a model plant in gene transformation researches owing to its mature genetic transformation system. Amino acids are essential organic nitrogen compounds in plants. The content of amino acids in tobacco leaves not only affects the growth of tobacco, but also affects the quality and flavor of tobacco. Transgenic NtAAP2-2-RNAi and OE lines exhibited obvious phenotypic modulations compared to those of WT plants. The leaf area increased in the NtAAP2-2-OE lines, but decreased in the NtAAP2-2-RNAi lines Fig. 4A. To further explore the function of NtAAP2-2 under N stress, NtAAP2-2-RNAi, OE lines and controls were treated with low N nutrient solution. After one week of N-deficiency treatment, NtAAP2-2-RNAi transgenic plants showed slightly yellow leaves. While the OE lines and controls grown well. After 3 weeks of treatment, as shown in the Fig. 4B, NtAAP2-2-OE transgenic lines still had no phenotypic changes, while the leaves of WT began to turn yellow, the plant growth and leaf growth of WT lines was seriously retarded, but the NtAAP2-2-OE transgenic lines still showed healthy growth (Fig. 4B), while NtAAP2-2-RNAi transgenic plants showed significant dwarfism.
 | ||
| Fig. 4 The phenotype of NtAAP2-2 transgenic lines. (A) The phenotype of NtAAP2-2 transgenic lines under normal condition. (B) The phenotype of NtAAP2-2 transgenic lines under N stress. | ||
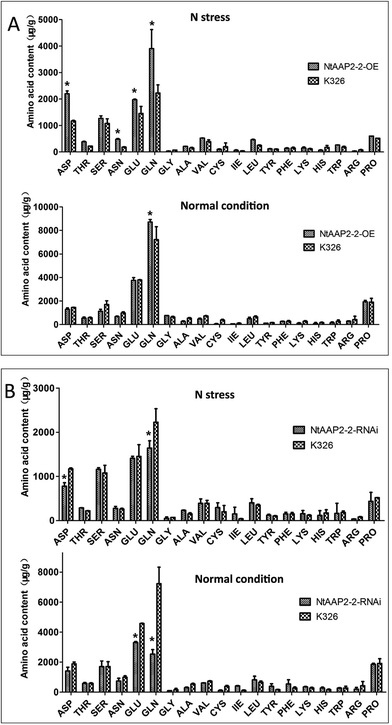
As amino acid permease are principal mediators of N uptake and distribution in plants, we evaluated amino acid content from individual transgenic lines and wild type plants under normal growth conditions and N stressed treatment, respectively. Under normal growth condition, the content of Gln increased by 20% in NtAAP2-2-OE transgenic lines than that of control plants (Fig. 5A), the levels of other amino acid were slightly changed. However, under N stress growth condition, the content of Asp in NtAAP2-2-OE transgenic lines increased by 53.3%, Asn increased by 60%, Glu increased by 26%, Gln content increased 53%. In contrast, the Gln content in NtAAP2-2-RNAi transgenic lines decreased by 64.8% under normal growth conditions, while the Glu content decreased by 27.7%. The content of Asp decreased by 33.3%, and Gln content decreased by 26.3% in N stressed NtAAP2-2-RNAi transgenic lines (Fig. 5B). The results indicated that NtAAP2 might participate in the transport of Gln and Glu. In plants, glutamic and glutamine are not only raw materials for the synthesis of other amino acids, but also involved in the synthesis of other N-containing compounds, such as chlorophyll, pyridine nucleic acid.28 The decrease of Gln and Glu in NtAAP2-2-RNAi transgenic lines may lead to the decrease of chlorophyll content, which further leads to plant leaves loss of green. The contents of Gln in NtAAP2-2-OE transgenic lines were increased, which could be further used to synthesize other amino acids and N-containing compounds. Under N stress, overexpression of NtAAP2-2 could provide more N sources, which enhanced plant resistance to N stress.
Conclusions
In the present study, 15 NtAAP proteins containing conserved domain Aa_trans were identified and characterized. Phylogenetic analyses revealed that the NtAAP gene family members were divided into three clusters. The exon/intron structure, transmembrane sequence and motif compositions of the NtAAP genes and proteins are highly conserved in each class, indicating that the function was conserved. An additional comprehensive analysis of the expression profiles revealed that most NtAAP genes were expressed in different tissues under normal growth conditions. We successfully cloned the NtAAP2-2 gene from tobacco, and the overexpression vectors and the RNAi vector of NtAAP2-2 were constructed. In transgenic tobacco, the contents of Asp, Asn, Glu and Gln were changed, indicating that NtAAP2-2 was involved in the transport of Asp, Asn, Glu and Gln in tobacco plants. Thus, results of the present study provide the fundamental information for future functional investigations of this gene family.Acknowledgements
This work was supported by the Science Project of the Zhengzhou Tobacco Research Institute (902013CZ0620). We also thank Dr John Hugh Snyder and Fangfei Liu for their kind help in discussion, writing and revising the manuscript.References
- T. Näsholm, K. Kielland and U. Ganeteg, New Phytol., 2009, 182, 31 CrossRef.
- S. Jämtgård, T. Näsholm and K. Huss-Danell, Plant Soil, 2008, 302, 221–231 CrossRef.
- T. Ge, S. W. Song, P. Roberts, D. L. Jones, D. F. Huang and K. Iwasaki, Environ. Exp. Bot., 2009, 66, 357–361 CrossRef CAS.
- H. Svennerstam, U. Ganeteg and T. Näsholm, New Phytol., 2008, 180, 620 CrossRef CAS PubMed.
- M. Tegeder, Curr. Opin. Plant Biol., 2012, 15, 315–321 CrossRef CAS PubMed.
- D. Wipf, U. Ludewig, M. Tegeder, D. Rentsch, W. Koch and W. B. Frommer, Trends Biochem. Sci., 2002, 27, 139 CrossRef CAS PubMed.
- A. Ortizlopez, H. Chang and D. R. Bush, Biochim. Biophys. Acta, Biomembr., 2000, 1465, 275 CrossRef CAS.
- Y. H. Su, W. B. Frommer and U. Ludewig, Plant Physiol., 2004, 136, 3104 CrossRef CAS PubMed.
- Y. Lu, Z. Song, L. Kai, X. Lian and H. Cai, Acta Physiol. Plant., 2012, 34, 1943–1962 CrossRef CAS.
- S. Okumoto, R. Schmidt, M. Tegeder, W. N. Fischer, D. Rentsch, W. B. Frommer and W. Koch, J. Biol. Chem., 2002, 277, 45338–45346 CrossRef CAS PubMed.
- W. Koch, M. Kwart, M. Laubner, D. Heineke, H. Stransky, W. B. Frommer and M. Tegeder, Plant J., 2003, 33, 211–220 CrossRef CAS PubMed.
- D. Rentsch, Trends Plant Sci., 1998, 3, 188–195 CrossRef.
- M. Tegeder and J. M. Ward, Front. Plant Sci., 2012, 3, 1–11 Search PubMed.
- W. B. Frommer, S. Hummel and J. W. Riesmeier, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 5944–5948 CrossRef CAS.
- M. Tegeder, Q. Tan, A. K. Grennan and J. W. Patrick, Funct. Plant Biol., 2007, 34, 1019–1028 CrossRef CAS.
- L. Zhang, M. G. Garneau, R. Majumdar, J. Grant and M. Tegeder, Plant J. Cell Mol. Biol., 2015, 81(1), 134–146 CrossRef CAS PubMed.
- Y. H. Lee, J. Foster, J. Chen, L. M. Voll, A. P. Weber and M. Tegeder, Plant J., 2007, 50, 305–319 CrossRef CAS PubMed.
- L. Z. Zhang, Q. M. Tan, R. Lee, A. Trethewy, Y. H. Lee and M. Tegeder, Plant Cell, 2010, 22, 3603–3620 CrossRef CAS PubMed.
- W. N. Fischer, D. D. F. Loo, W. Koch, U. Ludewig, K. J. Boorer, M. Tegeder, D. Rentsch, E. M. Wright and W. B. Frommer, Plant J. Cell Mol. Biol., 2002, 29, 717–731 CrossRef CAS.
- R. Schmidt, H. Stransky and W. Koch, Planta, 2007, 226, 805 CrossRef CAS PubMed.
- J. S. Yang, R. Wang, J. J. Meng, Y. P. Bi, P. L. Xu, F. Guo, S. B. Wan, Q. W. He and X. G. Li, J. Plant Physiol., 2010, 167, 534–539 CrossRef CAS PubMed.
- R. B. Horsch, Science, 1985, 227, 1229–1231 CAS.
- E. Hunt, S. Gattolin, H. J. Newbury, J. S. Bale, H. M. Tseng, D. A. Barrett and J. Pritchard, J. Exp. Bot., 2010, 61, 55 CrossRef CAS PubMed.
- S. Okumoto, W. Koch, M. Tegeder, W. N. Fischer, A. Biehl, D. Leister, Y. D. Stierhof and W. B. Frommer, J. Exp. Bot., 2004, 55, 2155–2168 CrossRef CAS PubMed.
- H. Svennerstam, S. Jämtgård, I. Ahmad, K. Huss-Danell, T. Näsholm and U. Ganeteg, New Phytol., 2011, 191, 459–467 CrossRef CAS PubMed.
- E. Hunt, S. Gattolin, H. J. Newbury, J. S. Bale, H. M. Tseng, D. A. Barrett and J. Pritchard, J. Exp. Bot., 2010, 61, 55 CrossRef CAS PubMed.
- A. Sanders, R. Collier, A. Trethewy, G. Gould, R. Sieker and M. Tegeder, Plant J., 2009, 59, 540–552 CrossRef CAS PubMed.
- H. M. Lam, K. T. Coschigano, I. C. Oliveira, R. Melooliveira and G. M. Coruzzi, Annu. Rev. Plant Physiol. Plant Mol. Biol., 1996, 47, 569–593 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra05610a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |