Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Synthesis and characterization of a bifunctional nanoprobe for CGG trinucleotide repeat detection†
Xiaoqian Zhua,
Jiao Lia,
Hehong Lva,
Hanping He *ab,
Heng Liu
*ab,
Heng Liu ab,
Xiuhua Zhang
ab,
Xiuhua Zhang ab and
Shengfu Wangab
ab and
Shengfu Wangab
aMinistry of Education Key Laboratory for the Synthesis and Application of Organic Functional Molecules, College of Chemistry and Chemical Engineering, Hubei University, Youyi Road 368, Wuchang, Wuhan, Hubei 430062, PR China. E-mail: hehanping@hubu.edu.cn; Fax: +86-027-88663043; Tel: +86-027-50865319
bHubei Collaborative Innovation Center for Advanced Organic Chemical Materials, Hubei University, Youyi Road 368, Wuchang, Wuhan, Hubei 430062, PR China
First published on 20th July 2017
Abstract
A novel bifunctional nanoprobe was designed and used in an electrochemical sensor to rapidly detect CGG trinucleotide repeats. The bifunctional nanoprobe was synthesized using carboxyl-functionalized Fe3O4 magnetic nanoparticles as carriers of nucleic acids recognition molecule and electron mediator-ferrocene derivatives. Both the small molecules with the terminal amino group were immobilized on the surface of the magnetic nanoparticles via an amide bond, which resulted in double functions of recognition and electrochemical activity. An electrochemical sensor based on the new bifunctional nanoprobe was established for CGG trinucleotide repeat detection. After the target DNA was captured by the sensor, the recognition molecule on the surface of the bifunctional nanoprobe could interact with the G base of the CGG trinucleotide repeat selectively. This allowed the bifunctional nanoprobes to be enriched on the modified electrode surface, which resulted in a remarkable electrochemical signal. CGG trinucleotide repeat can be easily discriminated from other trinucleotide repeats by the sensor. Furthermore, these results demonstrated that the new sensor exhibited an excellent selectivity to the Fragile X syndrome disease biomarker CGG repeat DNA. The proposed method is simple and rapid, the target DNA is label-free and has excellent repeatability for the detection of the CGG trinucleotide repeat because of the double function of the designed nanoprobe. This sensing strategy will provide great promise for the early diagnosis of neurodegenerative diseases associated with trinucleotide repeats.
1. Introduction
The sequence detection of trinucleotide repeats has attracted significant interest because the expansion of trinucleotide repeats was correlated with various neurodegenerative diseases,1–5 such as Fragile X syndrome (CGG),6,7 myotonic dystrophy (CTG),8 Friedreich ataxia (GAA),9,10 several types of spinocerebellar ataxia (CAG)11 and Huntington's disease (CAG).12,13 In these studies, the dynamic expansion of the CGG trinucleotide repeat in the 5′-untranslated region of the first exon of the FMR1 gene engendered the Fragile X syndrome, the single most common cause of inherited mental impairment.6,7 Moreover, the study has revealed that the transcriptional silencing of FMR1 occurs together with the specific hypermethylation of a promoter CpG island and of the fully expanded CGG trinucleotide repeat. Thus, the Fragile X syndrome was closely correlated with the CGG trinucleotide repeat.2,4 Hence, selective and sensitive detection of CGG trinucleotide repeat has become very important.Electrochemical techniques have proven to be one of the most advantageous ways of determining DNA sequences because of their rapid response, simple operation, low cost and affordable instrumentation.14–16 However, the study of trinucleotide repeats was very limited because of the difficulty of repeats determination. Such determination is extremely important in early clinical diagnostic of these neurodegenerative diseases, and the development of a simple and rapid electrochemical method for selective detection of trinucleotide repeats is still a challenging and long-standing goal. Our recent studies have made a little contribution to the field. Simple electrochemical biosensors to detect the CGG trinucleotide repeat with excellent selectivity and sensitivity were reported using a new bifunctional electrochemical probe containing a recognition part of NCD and an electro-active ferrocenyl group,14 and the electrochemical impedance sensors were investigated by the recognition molecule-modified electrode.15 These studies have opened paths for the development of trinucleotide repeats-based electrochemical sensors, which are important for the early diagnosis and treatment of neurodegenerative diseases.17,18
Currently, various nanomaterials (gold, carbon, magnetic nanoparticle, semiconducting, etc.) have received much attention in the field of electrochemical sensors, for example, as capture probes, reporting molecules, target labels, electrode fabrication, and electrode coatings.19 These nanomaterials offer improved biocompatibility, additional binding sites and higher signal intensities compared to traditional electrochemical sensor materials. Among these nanomaterials, magnetic nanoparticles (MNPs) have received tremendous attention in diverse fields, such as biomedical sciences, biochemical sensing, magnetic separation, imaging and targeted drug delivery. The advantages were attributed to the unique nanoscale physicochemical properties of MNPs, particularly their magnetic properties, low toxicity, biocompatibility and easy purification.20–28 In order to introduce chemical functionalities for the conjugation of affinity ligands, increasing specificity, better biocompatibility, and reduction of risk of cytotoxicity etc., the surface of MNPs was normally modified with a variety of materials.21,27 The carboxyl modification of Fe3O4 MNPs (CMNPs) has attracted increasing attention because of the application in ligand exchange. The CMNPs were prepared through the co-precipitation method with good chemical stability and dispersion in the water phase, which is usually a further modification.29 For example, Fe3O4 MNPs functionalized with carboxyl as the carrier for carbonic anhydrase immobilization were reported to capture CO2 with high reusability.28
Except for utilizing nanomaterials to amplify the signals, another effective function is usually introducing electron mediators in electrochemical detection systems for increasing the sensitivity.19,30 Ferrocene and its derivatives were frequently adopted as electron mediators in electrochemical sensing because of their favourable electrochemical properties.31 To avoid the unstable adsorption on the electrode's surface and the leakage of the mediators, proper strategies can be used by covalently linking ferrocene derivatives with polymers, high molecular weight compounds or nanoparticles before immobilization.32,33 For example, ferrocene derivatives can be easily self-assembled on the surface of gold nanoparticles34,35 and Fe3O4@AuNPs.33,36 Thus, to design and synthesize a ferrocene derivative with the specified terminal group may be a promising proposal in biosensing techniques.
In a previous study, synthetic naphthyridine derivatives exhibited excellent recognition for specific sequences of DNA such as G bulge, G–G mismatch or trinucleotide repeats DNA because of the complementary hydrogen bonding between the naphthyridine group and the guanine base.14,15,37–41 Reasonably, the selectivity and high affinity of naphthyridine derivatives and nucleotide binding might be achieved by a combination of stacking and complementary hydrogen-bonding interaction of the naphthyridine group and the G base. Recently, other methods for the detection of CGG trinucleotide repeat were also developed.42,43 Although successful exploration results were obtained, it would be very important to design a new electrochemical probe that has excellent ability to selectively detect the CGG trinucleotide repeat and can be directly used for a facile electrochemical detection.
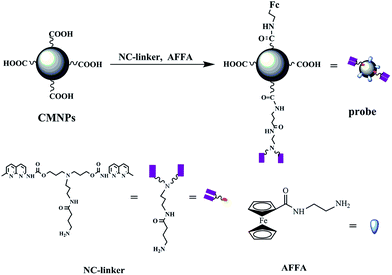
In this study, we designed a bifunctional nanoprobe using CMNPs as carriers covalently bound with nucleic acids recognition molecule (NC-linker)15 and ferrocene derivatives (AFFA), opening up wide applications in biosensors. The prepared CMNPs possessed a high surface area, good chemical stability and good water-solubility, which provided a compatible platform for maintaining the activity of the immobilized NC-linker and AFFA. More importantly the bifunctional nanoprobe was endowed with a double function, recognition domain and electrochemical activity, which can be applied in the electrochemical sensor for rapid detection of the CGG trinucleotide repeat based on the significant and selective changes in electrochemical signals. All the results obtained in this study would provide a sound basis for further exploration.
2. Experimental
2.1 Reagents and apparatus
1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC), FeCl3·6H2O, FeSO4·7H2O and NH3·H2O were obtained from Sinopharm Chemical Reagent Co., Ltd. N,N-Diisopropylethylamine (DIPEA), 6-mercapto-1-hexanol (MCH), ferrocenecarboxylic acid, N-hydroxysuccinimide (NHS), 2-(N-morpholino)ethanesulfonic acid (MES), N-(tert-butoxycarbonyl)-ethylenediamine, o-benzotriazol-1-yl-tetramethyluronium hexafluorophosphate (HBTU), oleic acid, and KMnO4 were obtained from the Aladdin Reagent Database Inc. (Shanghai, China). Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) was purchased from Sigma-Aldrich. All other reagents were analytical grade and used without further purification. PBS buffer solution (10 mM, pH = 7.4), MES buffer solution (0.1 M, pH 6.0 containing 0.5 M NaCl), and Tris–HCl (20 mM, pH = 7.4) containing 140 mM NaCl and 5 mM MgCl2 were used in the experiments. All aqueous solutions were prepared with ultrapure water (18.25 MΩ cm). All the DNA strands were synthesized and HPLC-purified by the Shanghai Sangon Biotechnology Co., Ltd. (Shanghai, China) as shown in Table S1.†Ultraviolet visible (UV-vis) absorption spectra were measured with a UV-2700 spectrophotometer (Shimadzu Co., Japan). The morphology and size of the MNPs were checked via transmission electron microscopy (TEM) (JEM-1200EX, JEOL, Japan). Fourier transform infrared (FTIR) spectra were obtained on a Fourier transform spectrometer (Spectrum one, PerkinElmer, USA). The XRD pattern was obtained using an X-ray diffractometer (Rigaku D/Max-IIIC, Japan). 1H (600 MHz) NMR spectra were recorded on a Varian INOVA 600 spectrometer and tetramethylsilane was used as the internal standard. Liquid chromatography mass spectrometry (LC-MS) was measured using an Agilent 1260-6224 LC-MS TOF instrument. All the electrochemical measurements were carried out employing a CHI660E electrochemical workstation (CH Instruments, Inc. Shanghai) using a conventional three-electrode system with a gold electrode (GE), a saturated calomel electrode (SCE), and a platinum wire as the working, reference, and the counter electrodes, respectively.
2.2 Synthesis of bifunctional nanoprobe
The bifunctional nanoprobe was synthesized using CMNPs as carriers of the nucleic acids recognition molecule (NC-linker) and the electrochemical active molecules-ferrocene derivatives (AFFA). The synthesis process of the bifunctional nanoprobe is as shown in Scheme 1.2.3 Sensor fabrication
The GE with a diameter of 2 mm was polished using 0.05 μm alumina slurry, followed by sonicating successively in both ethanol and ultrapure water to remove the residual alumina powder. Then, the electrode was electrochemically cleaned in 0.5 M H2SO4 to remove any remaining impurities and rinsed thoroughly with ultrapure water. The fresh pretreated GE was immediately incubated in 50 μL of 2 μM capture DNA containing 10 μL of 1 mM TCEP for 12 h at room temperature, followed by rinsing with ultrapure water. TCEP is used to prevent the terminal cysteine from forming disulfide bonds. Then, the electrode was immersed into 60 μL of 0.1 mM MCH for 1 h at room temperature to block the nonspecific binding sites, followed by rinsing with ultrapure water.2.4 DNA detection
In a typical DNA analysis, the capture DNA modified electrodes were incubated in 60 μL of different concentrations of trinucleotide repeat DNA for 5 min at 65 °C in a water bath, followed by naturally cooling to room temperature to perform the hybridization reaction. Then, the electrode was washed with ultrapure water three times. Next, it was further incubated with 60 μL of the bifunctional nanoprobe for 2 h at 37 °C. Finally, the electrode was washed thoroughly with PBS buffer solution to remove non-specifically bound MNPs. The analytical procedure involved SWV scanning from −0.1 to 0.6 V in PBS buffer solution. The electrochemical responses were recorded at about 0.46 V for DNA detection.3. Results and discussion
3.1 Design of bifunctional nanoprobe
In a previous study, the synthetic naphthyridine derivatives exhibited excellent recognition of the G–G mismatch DNA and CGG trinucleotide repeat selectively.14,15,37–41 It is very important to design a bifunctional probe that can selectively recognize the CGG trinucleotide repeat and posses an electrochemical response for a facile electrochemical detection. Although successful exploration results were obtained using a bifunctional probe,14 the sensitivity of the CGG trinucleotide repeat detection also could be further improved. Considering that MNPs have unique nanoscale physicochemical properties, a new bifunctional nanoprobe was designed, as shown in Scheme 1 which used CMNPs as carriers covalently bound with nucleic acids recognition molecule and ferrocene derivatives, that could amplify the signals in electrochemical detection systems for increasing the sensitivity. The bifunctional nanoprobe was endowed with a double function, recognition domain and electrochemical activity, which can be applied in an electrochemical sensor for rapid detection of the CGG trinucleotide repeat.3.2 Characterization of carboxyl-functionalized Fe3O4 MNPs
The CMNPs were prepared by synthesis of the oleic acid-coated Fe3O4 MNPs, and subsequently the oleic acid on the surface of the MNPs was oxidized by KMnO4. Then, the obtained hydrophilic monolayer CMNPs were characterized by TEM, XRD and FTIR.The morphology and size of the CMNPs were observed and analyzed via TEM (Fig. 1A). The CMNPs present a spherical morphology, are relatively uniform in size and exhibit a relatively narrow size distribution with an average diameter of 10 nm. The MNPs maintained a good stability and dispersity and could act as excellent immobilized carriers for other molecules.28 The CMNPs were further confirmed by typical XRD patterns (Fig. 1B). The Fe3O4 MNPs (curve a) exhibited diffraction peaks centred at 2θ values of 30.2°, 35.5°, 43.2°, 53.5°, 57.1°, and 62.7°, which were identical to the standard spectrum of Fe3O4 corresponding to the crystal planes of (220), (311), (400), (422), (511), and (440), respectively.28,29 The diffraction peaks of CMNPs (curve b) appeared at 30.2°, 35.7°, 43.4°, 53.8°, 57.3° and 63.0°, which were also identical to the standard spectrum of Fe3O4 corresponding to the crystal planes of (220), (311), (400), (422), (511), and (440), respectively. These results revealed that the crystal structure of Fe3O4 was substantially unchanged during the oxidation process of oleic acid-coated Fe3O4 MNPs by KMnO4.
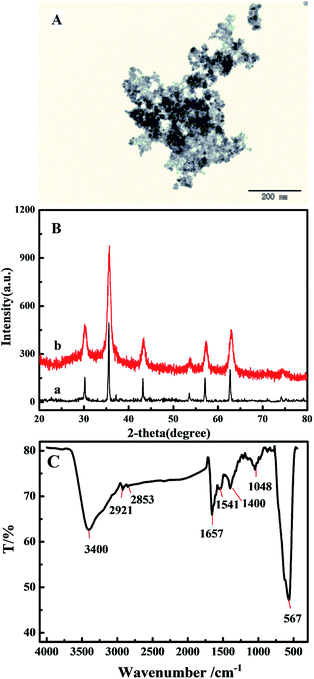 | ||
| Fig. 1 (A) TEM image of the CMNPs. (B) XRD pattern of Fe3O4 MNPs (a) and CMNPs (b). (C) FTIR spectrum of the CMNPs. | ||
FTIR spectroscopy was used to observe the surface composition of the CMNPs (Fig. 1C). The characteristic absorption peak for Fe3O4 MNPs was observed at 567 cm−1. The stretching vibration and deformation of the C–O appeared at 1048 and 1400 cm−1, respectively. The peak located at 1657 cm−1 corresponded to the bending vibration of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O. The peaks at 2921 cm−1 and 2853 cm−1 were also observed, attributed to the symmetric and asymmetric stretching vibration of CH2. In addition, a high intensity O–H stretching vibration appeared at 3400 cm−1. These results indicated that the carboxylic group was present on the surface of the Fe3O4 MNPs, which indicated the relatively successful preparation of CMNPs.28,29
O. The peaks at 2921 cm−1 and 2853 cm−1 were also observed, attributed to the symmetric and asymmetric stretching vibration of CH2. In addition, a high intensity O–H stretching vibration appeared at 3400 cm−1. These results indicated that the carboxylic group was present on the surface of the Fe3O4 MNPs, which indicated the relatively successful preparation of CMNPs.28,29
3.3 Successful preparation of bifunctional nanoprobe
After activation of carboxyl on the surface of CMNPs in the presence of EDC/NHS, NC-linker and AFFA were immobilized on the surface of the CMNPs via amide bond to get the bifunctional nanoprobe, which has both the function of recognition and contribution of electrochemical signal as the redox probe. The resulting bifunctional nanoprobe can quickly respond to an external magnetic field and quickly be redispersed homogeneously by shaking with the hand because of the magnetite content. The excellent magnetic property of the bifunctional nanoprobe can be used for efficient and simple magnetic separation.Important issues that must be considered are whether the NC-linker and AFFA were successfully immobilized on the surface of CMNPs. Thus, the UV-vis absorption spectra were used to study every step of modification of the bifunctional nanoprobe (Fig. 2A). For comparison, the NC-linker modified CMNPs and AFFA-modified CMNPs were also measured. No significant absorbance peak was observed for the CMNPs (curve a). Three extra absorption peaks were clearly observed at 225, 319 and 332 nm for the NC-linker modified CMNPs (curve b), attributed to the characteristic absorptions of the free NC-linker solution (Figure S1A,† curve b). An additional absorption peak was also clearly observed at 260 nm for the AFFA-modified CMNPs (curve c) corresponding to the characteristic absorption peaks of the free AFFA solution (Figure S1B,† curve b). All the characteristic absorption peaks of the free NC-linker and AFFA solution were observed at 225, 260, 319, and 332 nm for the bifunctional nanoprobe (curve d), indicating the successful coupling of amine of the NC-linker and AFFA with the carboxylic acids on the surface of CMNPs.
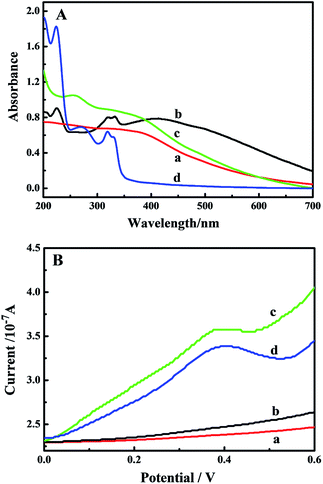 | ||
| Fig. 2 (A) UV-vis and (B) SWV spectra of the CMNPs (a), NC-linker modified CMNPs (b), AFFA modified CMNPs (c) and the bifunctional nanoprobe (d). | ||
Having an excellent electrochemical signal for the bifunctional nanoprobe is also our greatest concern. The corresponding SWV spectra were measured. As shown in Fig. 2B, the electro-active compound, AFFA solution, exhibited a typical oxidation peak at about 0.46 V. However, no significant peaks were observed for the CMNPs (curve a) and NC-linker modified CMNPs (curve b), indicating that they had no electrochemical signal. Importantly, we found a clear redox peak at about 0.40 V for the AFFA-modified CMNPs (curve c) and the bifunctional nanoprobe (curve d), which can be attributed to the redox of the ferrocene group in AFFA.14,33 The results confirmed that AFFA had been successfully modified on the surface of CMNPs. Thus, the bifunctional nanoprobe can be further used in the detection of the CGG trinucleotide repeat to give a successful electrochemical response as the redox probe.
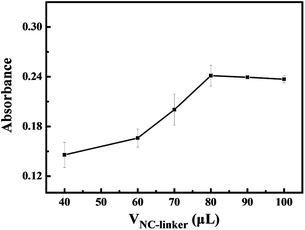
In order to obtain a large electrochemical signal, an experiment with different volumes of the NC-linker (1 mM) from 40 to 100 μL, while keeping the AFFA amount intact, was carried out. The UV absorption curves of the resulting bifunctional nanoprobe were tested. It was found that the absorbed intensity of the NC-linker on the surface of the biofunctional nanoprobes increased gradually and reached a plateau with an increase in the NC-linker volume, as shown in Fig. 3. The most significant increase in the absorbance was achieved when 80 μL of NC-linker was used. Thus, 80 μL NC-linker (1 mM) was selected as the optimal condition.
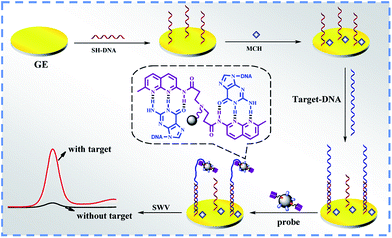
3.4 Principle of the sensor
The question of how to use the bifunctional nanoprobe for the detection of DNA-correlated diseases is another important issue. According to our previous study, the NC-linker on the surface of the bifunctional nanoprobe was able to bind to the G–G mismatch DNA and the CGG trinucleotide repeat selectively.14,15,37–41 Herein, the bifunctional nanoprobe was expected to be used for the detection of the CGG trinucleotide repeat that is correlated to neurodegenerative disease Fragile X syndrome.The principle of the bifunctional nanoprobe for electrochemical DNA biosensing is outlined in Scheme 2. SH-DNA is immobilized on a gold electrode surface via Au–S bonds. Then, MCH is further assembled on the electrode to block blank binding sites. In the presence of the target DNA, the electrode can capture the target DNA because the SH-DNA are complementary to the part of the target DNA. After incubation with the bifunctional nanoprobe, the bifunctional nanoprobe is incorporated into the CGG trinucleotide repeat on the surface of the electrode via binding of the NC-linker with the CGG trinucleotide repeat. The corresponding electrochemical responses attributed to AFFA of the bifunctional nanoprobe are used to detect the CGG trinucleotide repeat. Moreover, the poor binding of the NC-linker on the surface of the bifunctional nanoprobe with other trinucleotide repeats would result in no or a less apparent electrochemical response compared to the experimental groups.
3.5 Characterization of the biosensor
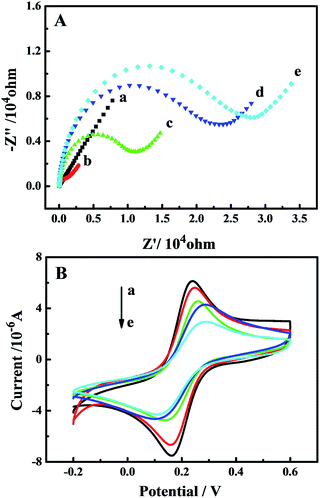
EIS is a sensitive and effective method to characterize the conductivity of the electrode surface and the increase in the semicircle diameter is equal to the increase in the electron-transfer resistance (Ret). EIS and CV were used to characterize the assembly processes of the gold electrode using [Fe(CN)6]3−/4− as the electro-active probes. As shown in Fig. 4A, the bare gold electrode (curve a) exhibited a nearly straight line, reflecting the excellent conductivity. When SH-DNA was self-assembled onto the electrode surface, the Ret increased (curve b), which suggested that SH-DNA formed an additional barrier to inhibit the electron transfer on the electrode surface because of the electrostatic repulsion between the negative charges of the SH-DNA phosphate backbone and the [Fe(CN)6]3−/4− ions. After MCH was used to block nonspecific sites, the Ret further increased (curve c) because of the insulating property of MCH. The Ret was largely increased (curve d) after the target DNA was captured on the surface of the electrode, implying that the increased negative charges of the nucleotide greatly inhibited the electron transfer. The increase in Ret indicated successful immobility of SH-DNA and hybridization of the trinucleotide repeat DNA with SH-DNA. When the sensor was incubated with bifunctional nanoprobes, the Ret significantly increased (curve e), which was attributed to the introduction of the bifunctional nanoprobe that obstructed the interfacial electron transfer. The results indicated that the biosensor was effectively and successfully fabricated.The corresponding CV curves are shown in Fig. 4B. With the stepwise construction of the sensor, the peak current decreased successively and the peak separation increased continuously, ascribed to the sequential increase in hindrance. These CV results were consistent with those obtained from the EIS measurements, confirming that the biosensing interface had been fabricated successfully.
3.6 Performance of the sensor
To test the feasibility of the bifunctional nanoprobe-based biosensor, SWV curves of the biosensor were obtained in PBS buffer solution before and after incubation of the biosensor with the bifunctional nanoprobes. As shown in Fig. 5, clearly the CGG modified biosensor did not show any electrochemical signal before incubation with the bifunctional nanoprobes (curve a). Importantly, an evident current peak was observed at about 0.47 V (curve b) because of the excellent binding ability of the bifunctional nanoprobe with the CGG trinucleotide repeat after the CGG-modified biosensor was incubated with the bifunctional nanoprobe. These facts further demonstrated the successful preparation and application of the bifunctional nanoprobes. The bifunctional nanoprobes could be used for the sensitive detection of the CGG trinucleotide repeat using the electrochemical system.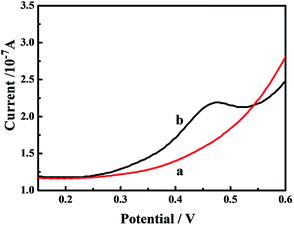 | ||
| Fig. 5 SWV response of the CGG-modified electrode before (a) and after (b) incubation with bifunctional nanoprobe. | ||
3.7 Selectivity
Another important question we must consider is the sequence selectivity in electrochemical detections. The specificity is a key issue in the early diagnosis of the Fragile X syndrome disease. Based on our previous study, the NC-linker compound and its derivative had excellent selectivity for the CGG trinucleotide repeat.14,15 The specificity of the proposed biosensor was investigated by incubating with different repeat DNA. For comparison, other trinucleotide repeats (1 μM) including CCG, GAA, TGG, CTG, CAG and ATT with multiple concentrations of the CGG trinucleotide repeat was used (Fig. 6). Different trinucleotide repeats-modified sensors were fabricated according to the same procedures with the CGG trinucleotide repeat. In order to minimize the nonspecific interaction of the bifunctional nanoprobe and DNA, the biosensors were rinsed in 2 mL PBS buffer solution before SWV analysis. After incubation with the bifunctional nanoprobe, CGG trinucleotide repeat (0.2 μM) produced an evident current response. Pleasantly, other trinucleotide repeats caused almost negligible electrochemical changes because of the poor binding of the bifunctional nanoprobe with the trinucleotide repeats. According to the above results, the bifunctional nanoprobe has the ability to bind with the CGG trinucleotide repeat selectively, clarifying that the present bifunctional nanoprobe can be used in principle to detect the CGG trinucleotide repeat. The responses of other trinucleotide repeats were almost ignored. These results suggested that the biosensor had a high sequence specificity and could be used for the detection of the CGG trinucleotide repeat.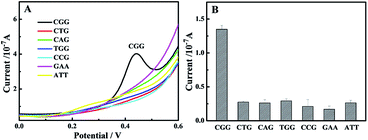 | ||
| Fig. 6 Selectivity of the proposed electrochemical biosensor to CGG at 0.2 μM and other trinucleotide repeats at the same concentration of 1 μM. | ||
Thus, the advantages of the proposed method such as fast, excellent selectivity, and low cost were clear. In addition, the method was based on the novel bifunctional nanoprobe that can selectively bind with the CGG trinucleotide repeat and was very different from other DNA sensors utilizing oligonucleotides as a probe and working either in the hybridization or denaturation modes. The studies showed that the bifunctional nanoprobe had an excellent selectivity, exhibiting promising clinical applications. These abovementioned aspects make the method useful and important for the development of CGG trinucleotide repeat detection. Next, we will try to find real samples and improve the method to explore the value and the feasibility in a clinical setting.
4. Conclusions
In summary, a bifunctional nanoprobe was successfully synthesized using CMNPs as carriers for NC-linker and AFFA immobilization via the amide bond, which was endowed with a double function, recognition domain and electrochemical activity. The characterizations of CMNPs and the bifunctional nanoprobe were studied systematically. Based on the bifunctional nanoprobe, a simple and rapid electrochemical sensor was developed for the detection of the CGG trinucleotide repeat. The results demonstrated that the new electrochemical sensor had an excellent ability to selectively detect the CGG trinucleotide repeat label-free. This new method explored the successful application of the functional MNPs in the electrochemical detection of the CGG trinucleotide repeat and would be a very promising option for the early diagnosis and treatment of neurodegenerative diseases.Acknowledgements
This study was supported by the National Natural Science Foundation of China (Grant 21575035) and Foreign Science and Technology Cooperation Fund of Hubei province, China (2015BHE025).Notes and references
- G. R. Sutherland and R. I. Richards, Proc. Natl. Acad. Sci., 1995, 92, 3636–3641 CrossRef CAS.
- R. S. Hansen, S. M. Gartler, C. R. Scott, S. H. Chen and C. D. Laird, Hum. Mol. Genet., 1992, 1, 571–578 CrossRef CAS PubMed.
- R. D. Wells, J. Biol. Chem., 1996, 271, 2875–2878 CrossRef CAS PubMed.
- S. M. Mirkin, Nature, 2007, 447, 932–940 CrossRef CAS PubMed.
- M. V. Evans-Galea, A. J. Hannan, N. Carrodus, M. B. Delatycki and R. Saffery, Trends Mol. Med., 2013, 19, 655–663 CrossRef CAS PubMed.
- R. I. Richards and G. R. Sutherland, Trends Biochem. Sci., 1997, 22, 432–436 CrossRef CAS PubMed.
- P. Weisman-Shomer, E. Cohen, I. Hershco, S. Khateb, O. Wolfovitz-Barchad, L. H. Hurley and M. Fry, Nucleic Acids Res., 2003, 31, 3963–3970 CrossRef CAS PubMed.
- C. J. Cummings and H. Y. Zoghbi, Hum. Mol. Genet., 2000, 9, 909–916 CrossRef CAS PubMed.
- V. Campuzano, L. Montermini, M. D. Moltò and L. Pianese, Science, 1996, 271, 1423 CAS.
- L. Montermini, E. Andermann, M. Labuda, A. Richter, M. Pandolfo, F. Cavalcanti, L. Pianese, L. Iodice, G. Farina, A. Monticelli, M. Turano, A. Filla, G. D. Michele and S. Cocozza, Hum. Mol. Genet., 1997, 6, 1261–1266 CrossRef CAS PubMed.
- A. Rovio, V. Tiranti, A. L. Bednarz, A. Suomalainen, J. N. Spelbrink, N. Lecrenier, A. Melberg, M. Zeviani, J. Poulton, F. Foury and H. T. Jacobs, Eur. J. Hum. Genet., 1999, 7, 140–146 CAS.
- I. Jonson, R. Ougland, A. Klungland and E. Larsen, Stem Cell Res., 2013, 11, 1264–1271 CrossRef CAS PubMed.
- D. C. Rubinsztein, Trends Genet., 2002, 18, 202–209 CrossRef CAS PubMed.
- H. P. He, J. P. Xia, X. Q. Peng, G. Chang, X. H. Zhang, Y. F. Wang, K. Nakatani, Z. W. Lou and S. F. Wang, Biosens. Bioelectron., 2013, 49, 282–289 CrossRef CAS PubMed.
- H. P. He, X. Q. Peng, M. Huang, G. Chang, X. H. Zhang and S. F. Wang, Analyst, 2014, 139, 5482–5487 RSC.
- E. Paleček and M. Bartošík, Chem. Rev., 2012, 112, 3427–3481 CrossRef PubMed.
- M. Labib, E. H. Sargent and S. O. Kelley, Chem. Rev., 2016, 116, 9001–9090 CrossRef CAS PubMed.
- A. Granzhan, N. Kotera and M. P. Teulade-Fichou, Chem. Soc. Rev., 2014, 43, 3630–3665 RSC.
- X. Q. Zhu, J. Li, H. P He, M. Huang, X. H. Zhang and S. F. Wang, Biosens. Bioelectron., 2015, 74, 113–133 CrossRef CAS PubMed.
- T. Hyeon, Chem. Commun., 2003, 927–934 RSC.
- A. Nel, T. Xia, L. Mädler and N. Li, Science, 2006, 311, 622–627 CrossRef CAS PubMed.
- U. Jeong, X. Teng, Y. Wang, H. Yang and Y. Xia, Adv. Mater., 2007, 19, 33–60 CrossRef CAS.
- Q. M. Kainz and O. Reiser, Acc. Chem. Res., 2014, 47, 667–677 CrossRef CAS PubMed.
- T. K. Jain, J. Richey, M. Strand, D. L. Leslie-Pelecky, C. A. Flask and V. Labhasetwara, Biomaterials, 2008, 29, 4012–4021 CrossRef CAS PubMed.
- M. A. M. Gijs, F. Lacharme and U. Lehmann, Chem. Rev., 2010, 110, 1518–1563 CrossRef CAS PubMed.
- N. Lee, D. Yoo, D. Ling, M. H. Cho, T. Hyeon and J. Cheon, Chem. Rev., 2015, 115, 10637–10689 CrossRef CAS PubMed.
- H. Lee, T. H. Shin, J. Cheon and R. Weissleder, Chem. Rev., 2015, 115, 10690–10724 CrossRef CAS PubMed.
- B. H. Lv, Z. R. Yang, F. J. Pan, Z. M. Zhou and G. H. Jing, Int. J. Biol. Macromol., 2015, 79, 719–725 CrossRef CAS PubMed.
- P. F. Su, G. Chen and J. Zhao, Chem. J. Chin. Univ., 2011, 32, 1472–1477 CAS.
- J. Y. Wang, L. C. Chen and K. C. Ho, ACS Appl. Mater. Interfaces, 2013, 5, 7852–7861 CAS.
- X. Q. Peng, H. P. He, J. P. Xia, Z. W. Lou, G. Chang, X. H. Zhang and S. F. Wang, Tetrahedron Lett., 2014, 55, 3541–3544 CrossRef.
- A. Shaporenko, K. Rössler, H. Lang and M. Zharnikov, J. Phys. Chem. B, 2006, 110, 24621–24628 CrossRef CAS PubMed.
- M. L. Liu, Q. Chen, C. L. Lai, Y. Y. Zhang, J. H. Deng, H. T. Li and S. Z. Yao, Biosens. Bioelectron., 2013, 48, 75–81 CrossRef CAS PubMed.
- M. L. Liu, J. H. Deng, Q. Chen, Y. Huang, L. P. Wang, Y. Zhao, Y. Y. Zhang, H. T. Li and S. Z. Yao, Biosens. Bioelectron., 2013, 41, 275–281 CrossRef CAS PubMed.
- H. W. Shu, W. Wen, H. Y. Xiong, X. H. Zhang and S. F. Wang, Electrochem. Commun., 2013, 37, 15–19 CrossRef CAS.
- J. D. Qiu, M. Xiong, R. P. Liang, H. P. Peng and F. Liu, Biosens. Bioelectron., 2009, 24, 2649–2653 CrossRef CAS PubMed.
- T. Peng, C. Dohno and K. Nakatani, Angew. Chem., Int. Ed., 2006, 45, 5623–5626 CrossRef CAS PubMed.
- H. P. He, M. Hagihara and K. Nakatani, Chem.–Eur. J., 2009, 15, 10641–10648 CrossRef CAS PubMed.
- M. Hagihara, H. P. He, M. Kimura and K. Nakatani, Bioorg. Med. Chem. Lett., 2012, 22, 2000–2003 CrossRef CAS PubMed.
- H. P. He, J. P. Xia, G. Chang, X. Q. Peng, Z. W. Lou, K. Nakatani, X. Zhou and S. F. Wang, Biosens. Bioelectron., 2013, 42, 36–40 CrossRef CAS PubMed.
- J. Li, H. P. He, X. Q. Peng, M. Huang, X. H. Zhang and S. F. Wang, Anal. Sci., 2015, 7, 663–667 CrossRef PubMed.
- H. Liu, Y. Wang, L. Zhang, Y. Shao and B. Zheng, Mater. Lett., 2015, 139, 265–267 CrossRef CAS.
- T. Shibata and K. Nakatani, ChemBioChem, 2016, 17, 1685–1688 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra05268e |
| This journal is © The Royal Society of Chemistry 2017 |