Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Biotransformation of dehydroepiandrosterone (DHEA) by environmental strains of filamentous fungi†
Ewa Kozłowska *a,
Monika Urbaniakb,
Anna Kancelistac,
Monika Dymarskaa,
Edyta Kostrzewa-Susłowa,
Łukasz Stępień
*a,
Monika Urbaniakb,
Anna Kancelistac,
Monika Dymarskaa,
Edyta Kostrzewa-Susłowa,
Łukasz Stępień b and
Tomasz Janeczko
b and
Tomasz Janeczko *a
*a
aDepartment of Chemistry, Wrocław University of Environmental and Life Sciences, Norwida 25, 50-375 Wrocław, Poland. E-mail: e.a.kozlowska@gmail.com; janeczko13@interia.pl
bDepartment of Pathogen Genetics and Plant Resistance, Institute of Plant Genetics, Polish Academy of Sciences, Strzeszyńska 34, 60-479 Poznań, Poland
cDepartment of Biotechnology and Food Microbiology, Wrocław University of Environmental and Life Sciences, Chełmońskiego 37, 51-630 Wrocław, Poland
First published on 21st June 2017
Abstract
Microbial transformations of steroids are an important method of obtaining new steroid derivatives of potential pharmaceutical activity, which follows the principles of green chemistry. We studied the ability of selected filamentous fungus species to transform dehydroepiandrosterone (DHEA) and interesting DHEA derivatives were obtained. Twenty-five strains of filamentous fungi were isolated from soil and air in Wroclaw and they were used as biocatalysts. Strains of the genus Penicillium transformed the substrate into the products of Baeyer–Villiger lactonization in the D ring. Biotransformation of DHEA by Fusarium acuminatum KCh S1 strain led to a stereoselective hydroxylation at 7α (the product was obtained with 97% conversion). Androst-5-ene-3β,7α,17α-triol, compound with high anticancer activity towards glioblastoma and lymphoma cell lines was obtained with 65% yield in the culture of Mucor hiemalis KCh W2 strain. In the culture of Aspergillus versicolor KCh TJ1 strain, DHEA was transformed into androst-1,4-diene-3,17-dione (ADD) with high isolated yield.
Introduction
Steroid drugs, besides antibiotics, are the largest group of medicines that are used to treat and prevent various diseases. These compounds have anti-inflammatory, anti-allergic, immunomodulatory, anti-androgenic, anti-estrogenic, antitumor, antiviral, antibacterial, and hypotensive properties, as well as being widely known sex hormones.1–6Dehydroepiandrosterone (DHEA) is a mammalian hormone, which is transformed in vivo into androgens and estrogens.7 The majority of DHEA found in blood is produced in the zona reticularis of the adrenal cortex. Very small amounts may be produced also in testicles, ovaries and brain cells.8 The majority of DHEA in mammalian blood is in the form of biologically inactive hydrophilic sulphate, which after hydrolysis by steroid sulphatase can be further transformed.9
DHEA's action is a result of combined influence on various receptors and enzymatic pathways. It is proven that DHEA and its metabolites can modulate the level of nitric oxide, reactive oxygen species and interleukin-2, as well as may interact with Sigma-1 receptor, Gα12 and Gα13 receptors, peroxisome proliferator-activated receptor (PPAR)-α, receptors for pregnane X (PXR), androstanol, estrogen β (ERβ), γ-aminobutyric acid type A (GABAA) and N-methyl-D-aspartate (NMDA).8,10–12 This widespread mechanism of action can lead to multiple applications in the treatment of e.g.: elderly, menopause, osteoporosis, ovarian dysfunction, Crohn's disease, systemic lupus erythrematosus, asthma and Alzheimer's disease.8,13,14 However, more side effects are recognised because of low DHEA specificity of action. Conversion of DHEA to androgens in female body leads to elevated testosterone level, which cause acne, increased facial, axillary and pubic hair growth.7,8,15 A solution could be the application of derivatives with greater specificity, higher activity than their precursors and no-sex hormone activity.6
Due to widespread application of steroid compounds in medicine, there is a need for an efficient method for production of active pharmaceutical ingredients (APIs) and their key intermediates, and also for new derivatives with stronger biological activity.16 Biotransformations are good alternative to chemical synthesis in obtaining steroid compounds, allowing regio-and stereoselective introduction of substituents in non-activated positions and the possibility to replace a several-step synthesis with a single microbial transformation.7 The most known example of using biotransformations in steroid synthesis was the improvement of a 39-step synthesis of cortisone from bile acids, which resulted in lowering production cost from 200$ to 1$.1 A very popular microbial transformation that found a large-scale application is the side-chain degradation in phytosterols derived from soybeans by bacteria of the genera Mycobacterium, Moraxella, and Rhodococcus, leading to progesterone or ADD (androst-1,4-diene-3,17-dione).17 Steroid functionalization by hydroxylation leads to new derivatives with higher polarity and usually higher biological activity.18,19 The most popular positions of hydroxylation of steroid skeleton are as follows: 6α, 6β, 7α, 7β, 9α, 11α, 11β, 12α, 12β, 14α, 15α and 15β. Other carbons can be also hydroxylated. Fungi can also provide dehydrogenated and lactonised derivatives.1,17,20–22 Biotransformations of DHEA may become a low-cost and quick method to receive biologically active DHEA derivatives.23,24
Experimental
Materials
The substrate – dehydroepiandrosterone (DHEA) was purchased from Sigma-Aldrich. The microorganisms used were: Penicillium chrysogenum KCh S4, Fusarium acuminatum KCh S1, Mucor hiemalis KCh W2, Penicillium commune KCh W7 and Aspergillus versicolor KCh TJ1. All strains were obtained from the Department of Chemistry of the Wrocław University of Environmental and Life Sciences and were isolated using the method described below.Isolation of strains
Samples of soil from two landfill sites in Wrocław: Nadodrze district (S) and Słodowa Island, Old Town (W) were placed in sterile tubes. Under aseptic conditions 1 g of the soil was suspended in 10 cm3 of sterile water and shaken. Then 1 cm3 of the supernatant was taken out and serial decimal dilution was made. The inoculum from each of the dilutions was transferred into Sabouraud solid growth medium (3% glucose, 1% aminobac, 0.8% agar). From the (S) soil sample five pure cultures were isolated, whereas from the (W) soil sample seven strains were isolated. The strains that were the most effective in the screening tests were identified using molecular biology techniques and used in the reactions on a preparative scale. The results presented in this paper concern only the identified strains. A sample of air from the autoclave room in the Department of Chemistry (TJ) was used to inoculate agar plates using Koch sedimentation method. From the grown colonies 4 pure strains were obtained, from which the most effective one was the strain molecularly identified as Aspergillus versicolor KCh TJ1. The strain was maintained on Sabouraud 4% agar slopes and freshly subcultured before use in the transformation experiments.DNA extraction and molecular identification of fungal strains
Genomic DNA was extracted using a modified CTAB (cetyl hexadecyltrimethylammonium bromide) protocol, by the method described earlier.25 The DNA extracts were stored at −20 °C until used. Species identification was done on the basis of the sequence of the Internal Transcribed Spacers of the ribosomal DNA region (ITS1-ITS2) (Table 1). PCR was done in 20 μL volumes using C-1000 thermal cyclers (Bio-Rad, Hercules, CA, USA). Each reaction tube contained 0.5 μL of DreamTaq Green DNA polymerase (Thermo Scientific, Espoo, Finland), 2.5 μL of 10 × DreamTaq Green Buffer, 12.5 pmol of forward/reverse primers, 2.5 mM of each dNTP, and about 20 ng of fungal DNA. PCR conditions were as follows: 5 min at 94 °C; 35 cycles of 45 s at 94 °C, 45 s at 56 °C, and 1 min at 72 °C; and 5 min at 72 °C.| Marker | 5′ > 3′ sequence | Amplicon size (bp) | Reference |
|---|---|---|---|
| ITS4 | TCCTCCGCTTATTGATATGC | 600 | 26 |
| ITS5 | GGAAGTAAAAGTCGTAACAAGG |
Amplicons were electrophoresed in 1.5% agarose gel (Invitrogen) with GelGreen Nucleic Acid Stain (Biotium, Inc.).
For sequence analysis, PCR-amplified DNA fragments were purified with exonuclease I (Thermo Scientific) and shrimp alkaline phosphatase (Thermo Scientific) using the following programme: 30 min at 37 °C and 15 min at 80 °C. DNA fragments were labeled using forward primer and the BigDye Terminator 3.1 kit (Applied Biosystems, Foster City, CA, USA), according to producer's recommendations and precipitated with 96% ethanol. Sequence reading was performed using Applied Biosystems equipment. Sequences were analysed using BLASTn algorithm.
Screening procedure
Erlenmeyer flasks (300 mL), each containing 100 mL of the cultivation medium consisting of 3 g glucose and 1 g aminobac dissolved in water, were inoculated with a suspension of microorganisms and then incubated for 3 days at 25 °C on a rotary shaker. After full growth of the culture 10 mg of a substrate dissolved in 1 mL of THF was added. After 1, 3 and 7 days of incubation under the above conditions, portions of 10 mL of the transformation mixture were extracted with CHCl3 (3 × 10 mL). The extracts were dried over MgSO4, concentrated in vacuo and analyzed by GC and TLC. All experiments were repeated three times.Preparative biotransformation
The same transformations were performed on the preparative scale in 2000 mL flasks, each containing 500 mL of the cultivation medium. The cultures were incubated under the same conditions and then 100 mg of DHEA dissolved in 2 mL of THF was added to the grown cultures. After complete transformation of the substrate the mixtures were extracted with CHCl3 (3 × 300 mL), dried (MgSO4) and concentrated in vacuo. The transformation products were separated by preparative TLC and analyzed (TLC, GC).Analytical methods
The course of biotransformation was controlled by means of TLC. Composition of product mixtures was established by GC. Products were purified using preparative TLC (silica gel GF, 20 × 20 cm, 500 μm, Analtech) and hexane/acetone mixture (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) as eluent. After elution products were detected under UV light (365 nm), then scraped from the plate and eluted with acetone to give fraction. Analytical TLC was performed on silica gel G plates (Merck). Compounds were detected by spraying the plates with H2SO4/CH3OH mixture (1
1, v/v) as eluent. After elution products were detected under UV light (365 nm), then scraped from the plate and eluted with acetone to give fraction. Analytical TLC was performed on silica gel G plates (Merck). Compounds were detected by spraying the plates with H2SO4/CH3OH mixture (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) and visualized under UV (254 nm). GC analysis was performed using a Hewlett-Packard 5890A (Series II) GC instrument fitted with a flame ionization detector (FID). The DB-5MS (crosslinked phenyl methyl siloxane) capillary column (30 m × 0.32 mm × 0.25 μm) was used to determine the composition of product mixtures. The following temperature programme was used: 220 °C (1 min)/4 °C min−1/260 °C (1 min)/30 °C min−1/300 °C (5 min). For GC-MS analysis, GCMS-QP2010 (Shimadzu) was used with a ZB-5 (crosslinked phenyl methyl siloxane) capillary column (30 m × 0.25 mm × 0.25 μm). The following temperature programme was used: 180 °C (3 min)/10 °C min−1/300 °C (5 min). The NMR spectra were recorded on DRX 500 MHz Bruker spectrometer and measured in CDCl3.
1, v/v) and visualized under UV (254 nm). GC analysis was performed using a Hewlett-Packard 5890A (Series II) GC instrument fitted with a flame ionization detector (FID). The DB-5MS (crosslinked phenyl methyl siloxane) capillary column (30 m × 0.32 mm × 0.25 μm) was used to determine the composition of product mixtures. The following temperature programme was used: 220 °C (1 min)/4 °C min−1/260 °C (1 min)/30 °C min−1/300 °C (5 min). For GC-MS analysis, GCMS-QP2010 (Shimadzu) was used with a ZB-5 (crosslinked phenyl methyl siloxane) capillary column (30 m × 0.25 mm × 0.25 μm). The following temperature programme was used: 180 °C (3 min)/10 °C min−1/300 °C (5 min). The NMR spectra were recorded on DRX 500 MHz Bruker spectrometer and measured in CDCl3.
Results and discussion
Spectral data of isolated metabolites
Product structures were determined by means of elemental analysis, 1H NMR, 13C NMR and correlation spectroscopy. 13C NMR spectra of all the products obtained are summarized in Table 2.| Atom number | Products | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| AD | ADD | 6βOH-AD | 6βOH-ADD | Testololactone | 7αOH-DHEA | 3β,7α,17α-Triol | 3βOH-lactone | 7Oxo-DHEA | |
| 1 | 35.05 | 155.43 | 37.34 | 156.86 | 35.46 | 37.07 | 37.22 | 37.04 | 36.60 |
| 2 | 33.81 | 127.82 | 34.28 | 127.03 | 33.84 | 31.39 | 31.10 | 31.59 | 31.25 |
| 3 | 199.21 | 186.33 | 200.40 | 186.65 | 199.40 | 71.25 | 71.41 | 71.61 | 70.44 |
| 4 | 124.04 | 124.24 | 126.58 | 126.17 | 124.07 | 42.06 | 42.19 | 42.01 | 41.99 |
| 5 | 170.26 | 168.44 | 168.03 | 165.52 | 169.63 | 146.64 | 146.28 | 140.79 | 166.27 |
| 6 | 32.46 | 32.65 | 72.83 | 73.70 | 32.35 | 123.67 | 123.99 | 120.70 | 126.06 |
| 7 | 30.66 | 32.41 | 37.34 | 38.86 | 30.37 | 64.37 | 65.63 | 31.20 | 201.20 |
| 8 | 35.58 | 35.22 | 29.52 | 29.93 | 37.92 | 37.32 | 37.98 | 34.58 | 44.47 |
| 9 | 53.70 | 52.40 | 53.76 | 51.96 | 52.45 | 42.72 | 42.49 | 49.11 | 50.22 |
| 10 | 38.56 | 43.54 | 38.17 | 43.62 | 38.46 | 37.63 | 37.60 | 36.92 | 38.54 |
| 11 | 20.23 | 22.20 | 20.37 | 22.04 | 21.82 | 20.19 | 20.43 | 22.09 | 20.72 |
| 12 | 31.19 | 31.30 | 31.37 | 31.34 | 38.93 | 31.18 | 31.51 | 39.04 | 30.85 |
| 13 | 47.41 | 47.79 | 47.74 | 47.87 | 82.87 | 47.23 | 45.08 | 83.30 | 48.00 |
| 14 | 50.71 | 50.53 | 51.00 | 50.63 | 45.62 | 45.05 | 42.13 | 46.85 | 45.88 |
| 15 | 21.67 | 22.02 | 21.81 | 21.96 | 19.82 | 22.02 | 24.89 | 20.03 | 24.31 |
| 16 | 35.67 | 35.74 | 35.88 | 35.80 | 28.53 | 35.91 | 32.75 | 28.95 | 35.77 |
| 17 | 220.35 | 220.03 | 220.69 | 220.15 | 171.44 | 221.30 | 80.00 | 171.64 | 220.52 |
| 18 | 13.62 | 13.92 | 13.88 | 14.01 | 17.40 | 13.39 | 16.76 | 20.20 | 13.89 |
| 19 | 17.29 | 18.83 | 19.65 | 20.59 | 20.11 | 18.38 | 18.43 | 19.42 | 17.57 |
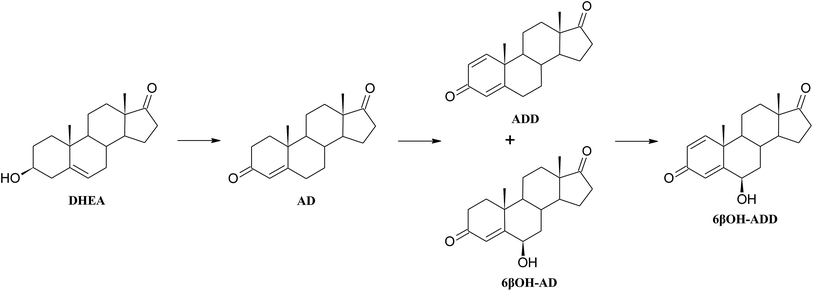
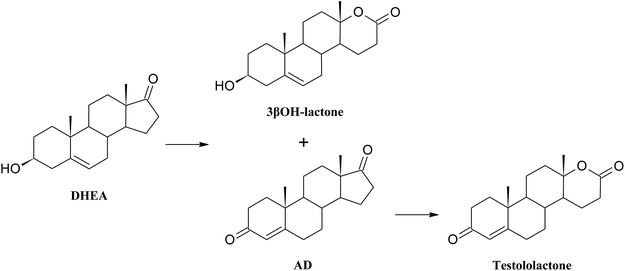
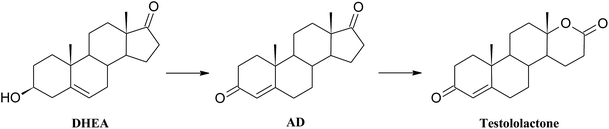
As a result of a 3 day transformation of DHEA (100 mg) with the strain Aspergillus versicolor KCh TJ1 androst-4-ene-3,17-dione (AD) (65.0 mg, 65%) was isolated. After seven days of diene-3,17-dione (6βOH-ADD) (3.9 mg, 4%) and 6β-hydroxyandrost-4-ene-3,17-dione (6βOH-AD) (2.3 mg, 2%) were isolated. After a 3 day transformation with the strain Penicillium commune KCh W7 3β-hydroxy-17a-oxa-D-homo-androst-5-ene-17-one (3βOH-lactone) (8.1 mg, 8%) and 17a-oxa-D-homo-androst-4-ene-17-one (testololactone) (40.8 mg, 41%) were isolated from the post-cultivation medium. Whereas, a 3 day transformation with the strain Penicillium chrysogenum KCh S4 led to isolation of androst-4-ene-3,17-dione (AD) (15.9 mg, 16%) and 17a-oxa-D-homo-androst-4-ene-17-one (testololactone) (34.2 mg, 34%).
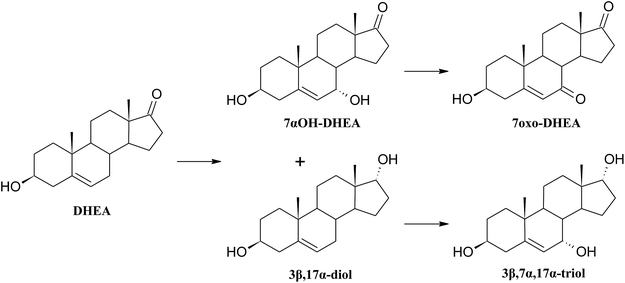
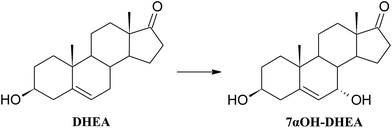
A 3 day transformation of DHEA in the culture of Mucor hiemalis KCh W2 allowed for the isolation of androst-5-ene-3β,7α,17α-triol (3β,7α,17α-triol) (37.3 mg, 38%), 3β,7α-dihydroxyandrost-5-ene-17-one (7αOH-DHEA) (25.3 mg, 25%) and 3β-hydroxyandrost-5-ene-7,17-dione (7Oxo-DHEA) (2.2 mg, 2%). Whereas, 7 days of transformation with the strain Fusarium acuminatum KCh S1 gave 3β,7α-dihydroxyandrost-5-ene-17-one (7αOH-DHEA) (71.8 mg, 72%).
Discussion of results
In order to find new biocatalysts for effective transformations of DHEA, samples of soil and air were collected from the area of Wrocław (Poland). Using the method of serial dilutions, 25 pure strains were isolated. All the strains were tested with respect to their ability to transform DHEA. The most effective strains were identified using molecular techniques. The strains selected for biotransformation on a large scale transformed the substrate with high conversion efficiency (Table 4). All the products were characterized based on their spectral data: 1H and 13C NMR (Table 2), HMBC and HMQC correlation spectra and analysis by GC and TLC.| Name of fungal strain | Identified fungal species | Sequence identity | Sequence |
|---|---|---|---|
| KCh S1 | Fusarium acuminatum | 99% identity to the Fusarium acuminatum, acc. numbers: KT965739.1, KR051403.1, KR051401.1 | TCTACTGATCCGAGGTCACATTCAGAAGTTGGGGTTTTACGGCATGGCCGCGCCGCGTTCCAGTTGCGAGGTGTTAGCTACTACGCAATGGAGGCTGCAGCGAGACCGCCAATGTATTTCGGGGGCGGCACCGCCCAGAAGGGCAGAGCCGATCCCCAACACCAAACCCGGGGGCTTGAGGGTTGAAATGACGCTCGAACAGGGCATGCCCGCCGGAATACCAGCGGGCGCAATGTGCGTTCAAAGATTCGATGATTCACTGAATTCTGCAATTCACATTACTTATCGCATTTTGCTGCGTTCTTCATCGATGCCAGAACCAAGAGATCCGTTGTTGAAAGTTTTGATTTATTTGTTTGTTTTACTCAGAAGTTACAATAAGAAACATTAGAGTTTGGGTCCTCTGGCGGGCCGTCCCGTTTTACGGGGCGCGGGCTGATCCGCCGAGGCAACATTAAGGTATGTTCACAGGGGTTTGGGAGTTGTAAACTCGGTAATGATCCCTCCGCTGGTTCACCAAC |
| KCh W2 | Mucor hiemalis | 100% identity to the Mucor hiemalis acc. numbers: KT583167.1, KP942918.1, KM280062.1 | ATATGCTTAAGTTCAGCGGGTAATCCCGCCTGATTTCAGATCAAATTTAAGAATGATTTATTTGGGAGGCCCCAGAGATAGTCTTTTTACTAGAGCATTCCTTTATATTAAAAAAATGTTCAGGCAGAAAGAACAATAGTTCAGGCCTAATAATTTTAAAGAATTCAGCAATAAAGCCGAAACTCTCATTTCCATTCACAACAAAATTATGAATGTGGGGTGTTTTTGATACTGAAACAGGCGTGCTCAATGGAATACCATTGAGCGCAAGTTGCGTTCAAAGACTCGATGATTCACTGAATATGCAATTCACACTAGTTATCGCACTTTGCTACGTTCTTCATCGATGCGAGAACCAAGAGATCCGTTGTTAAAAGTTGTTTTATAAGTTTTTTACGCTTATGTTACAATTATAATACTGAATTCTTTTGGTAAATAATTAATAGGATACCAGGCCTAAGCCTGACTTTCAGCTCGGTTAACATTCTAGTAGCCTATCCCTATGGCTAAAAGACATTCCTCAGACGCTGAAAAATTAAAACAGTTCACAGTAAAAAAGAATGAACCATTCGCTAGAAAACTAGCAAAGGCCATCTAAATTATTTAT |
| KCh S4 | Penicillium chrysogenum | 99% identity to the Penicillium chrysogenum acc. numbers: KY218708.1, KY218694.1, KY218692.1 | TGCGGAGGACATTACCGAGTGAGGGCCCTCTGGGTCCAACCTCCCACCCGTGTTTATTTTACCTTGTTGCTTCGGCGGGCCCGCCTTAACTGGCCGCCGGGGGGCTTACGCCCCCGCTGCCTTTCGGGCCCGTCCCCCGGGATCGGAGGACGGGGCCCAACACACAAGCCGTGCTTGAGGGCAGAAATGACGCTCGGACAGGCATGCCCCCCGGAATACCAGGgGGCGCAATGTGCGTTCAAAGACTCGATGATT |
| KCh W7 | Penicillium commune | 100% identity to the Penicillium commune, acc. numbers: KP120886.2, JN368450.1, HQ652873.1 | CTGATCGAGGTCACCTGGATAAAAATTTGGGTTGATCGGCAAGCGCCGGCCGGGCCTACAGAGCGGGTGACAAAGCCCCATACGCTCGAGGACCGGACGCGGTGCCGCCGCTGCCTTTCGGGCCCGTCCCCCGGGATCGGAGGACGGGGCCCAACACACAAGCCGTGCTTGAGGGCAGCAATGACGCTCGGACAGGCATGCCCCCCGGAATACCAGGGGGCGCAATGTGCGTTCAAAGACTCGATGATTCACTGAATTTGCAATTCACATTACGTATCGCATTTCGCTGCGTTCTTCATCGATGCCGGAACCAAGAGATCCGTTGTTGAAAGTTTTAAATAATTTATATTTTCACTCAGACTACAATCTTCAGACAGAGTTCGAGGGTGTCTTCGGCGGGCGCGGGCCCGGGGGCGTAAGCCCCCCGGCGGCCAGTTAAGGCGGGCCCGCCGAAGCAACAAGGTAAAATAAACACGGGTGGGAGGTTGGACCCAGAGG |
| KCh TJ1 | Aspergillus versicolor | 99% identity to the Aspergillus versicolor acc. numbers: KF691800.1, LC105698.1, LC105694.1 | CGGAGGACATTACTGAGTGCGGGCTGCCTCCGGGCGCCCAACCTCCCACCCGTGAATACCTAACACTGTTGCTTCGGCGGGGAACCCCCTCGGGGGCGAGCCGCCGGGGACTACTGAACTTCATGCCTGAGAGTGATGCAGTCTGAGTCTGAATATAAAATCAGTCAAAACTTTCAACAATGGATCTCTTGGTTCCGGCATCGATGAAGAACGCAGCGAACTGCGATAAGTAATGTGAATTGCAGAATTCAGTGAATCATCGAGTCTTTGAACGCACATTGCGCCCCCTGGCATTCCGGGGGGCATGCCTGTCCGAGCGTCATTGCTGCCCATCAAGCCCGGCTTGTGTGTTGGGTCGTCGTCCCCCCCGGGGGACGGGCCCGAAAGGCAGCGGCGGCACCGTGTCCGGTCCTCGAGCGTATGGGGCTTTGTCACCCGCTCGACTAGGGCCGGCCGGGCGCCAGCCGACGTCTCCAACCATTTTTCTTCAGGTTGA |
| Strain | Compounds found in the reaction mixture | Retention time by GC [min] | Retention factor by TLC | Biotransformation timea [days] | ||
|---|---|---|---|---|---|---|
| 1 | 3 | 7 | ||||
| a Compounds composition in the reaction mixture [%]. | ||||||
| Aspergillus versicolor KCh TJ1 | DHEA | 4.9 | 0.62 | 89 | — | — |
| AD | 6.0 | 0.65 | 5 | 88 | 6 | |
| ADD | 6.4 | 0.60 | — | 2 | 56 | |
| 6βOH-AD | 7.8 | 0.53 | — | 4 | 24 | |
| 6βOH-ADD | 8.3 | 0.47 | — | — | 2 | |
| Penicillium commune KCh W7 | DHEA | 4.9 | 0.62 | 65 | 0 | 0 |
| AD | 6.0 | 0.65 | 22 | 0 | 0 | |
| 3βOH-lactone | 8.6 | 0.48 | 3 | 11 | 9 | |
| Testololactone | 10.2 | 0.50 | 1 | 85 | 77 | |
| Penicillium chrysogenum KCh S4 | DHEA | 4.9 | 0.62 | 26 | 0 | — |
| AD | 6.0 | 0.65 | 61 | 22 | — | |
| Testololactone | 10.2 | 0.50 | 7 | 62 | — | |
| Mucor hiemalis KCh W2 | DHEA | 4.9 | 0.65 | 24 | 4 | 0 |
| 3β,17α-Diol | 6.0 | 0.60 | 29 | 6 | 0 | |
| 3β,7α,17α-Triol | 6.1 | 0.17 | 10 | 65 | 62 | |
| 7αOH-DHEA | 6.9 | 0.35 | 12 | 34 | 43 | |
| 7Oxo-DHEA | 7.4 | 0.54 | 0 | 2 | 3 | |
| Fusarium acuminatum KCh S1 | DHEA | 4.9 | 0.62 | 76 | 12 | 0 |
| 7αOH-DHEA | 6.9 | 0.35 | 21 | 85 | 97 | |
Selected five fungal strains that were used in this study. All strains were genetically identified by determination of the ITS1-ITS2 rDNA sequence (Table 3). The DNA fragments were amplified using PCR with ITS4-ITS5 primers (Table 1), and sequenced. The complete sequences of these products were compared with reference ITS sequences deposited in the GenBank Database (Table 3).
Introduction of a double bond between C1 and C2 of a steroid may increase biological activity of the newly formed compound. Chemical methods of obtaining 1-ene-steroids are usually multi-step processes and may lead to spontaneous aromatisation of the A ring. In contrast, biological methods are efficient and selective.27 The majority of literature reports on microbial dehydrogenation in C1–C2 positions relate to bacteria of the genera Mycobacterium, Rhodococcus, Nocardia, and Arthrobacter.28,29 Unfortunately, introduction of a double bond to the A ring by bacteria is the beginning of the steroid degradation pathway.30 There are many reports of using bacterial 3-ketosteroid Δ1-dehydrogenase enzymes fom Arthrobacter simplex or Gordonia neofelifaecis NRRL B-59395 in transformation of various 3-on-4-en steroids, although conversion of 3β-ol-4-en was unsuccessful.27,31–34 It has been found that steroid compounds (cholesterol, progesterone, pregnenolone, AD, DHEA and testosterone) were transformed to ADD by the Fusarium fungi, however, with lower yields since ADD was an intermediate product.16,35 Also Trichoderma hamatum strain has ability to transform different 3-one-4-en steroid substrates to 1,4-diene derivatives.36 Obtained with high yield ADD may be a key intermediate in synthesis of many sex hormones, aromatase inhibitors and also immunomodulatory drugs.37–39
High ability to perform Baeyer–Villiger oxidation in a D ring of steroids was observed for filamentous fungi of the genera Aspergillus and Penicillium and, in smaller degree, for Beauveria, Chaetomium, Fusarium, Mucor, and Rhizopus.40 Conversions of cyclic ketones to corresponding lactones are catalysed by Baeyer–Villiger monooxygenases (BVMO) (E.C. 1.14.13.x).41 These class of enzymes introduce a single atom of molecular oxygen into the substrate while the other one is reduced to water. They use a FAD or FNM as a prosthetic group and require NADPH or NADH as a cofactor.41
An analysis of the available literature reports revealed three possible transformations pathways:
1. DHEA may be transformed directly to 3βOH-lactone: Penicillium lilacinum AM111, Aspergillus tamarii KITA;42,43
2. At first isomerization and oxidation of 3-hydroxy-5-ene moiety to 3-oxo-4-ene occure, and then BV oxidation of the D ring takes place – Penicillium griseopurpureum, Penicillium lanosocoeruleum;44,45
3. At first BV oxidation to 3βOH-lactone occurs and then oxidation of the hydroxyl group at C-3 takes place, leading to formation of testololactone: Penicillium simplicissimum WY134-2, Aspergillus parasitic.46,47
According to the literature, it is possible that the two reactions occur simultaneously, as in the case of Penicillium glabrum and Penicillium camemberti AM83.44,48 In the culture of Penicillium commune KCh W7 strain lactonization was observed both in the substrate (5-ene-3-ol moiety), and in the formed in biotransformation AD (5-ene-3-one moiety). Moreover, 3βOH-lactone was not transformed to testololactone. Literature review indicates that isolated Baeyer–Villiger monooxygenases (BVMO) can transform DHEA to 3β-hydroxy-17a-oxa-D-homo-androst-5-ene-17-one in higher yield than AD to 17a-oxa-D-homo-androst-4-ene-17-one.49 Additionally in the culture of Beauveria bassiana, Penicillium camemberti AM83 and P. lilacinum AM111 AD was transformed slower than DHEA into corresponding lactones.42,48,50
3β,7α,17α-Trihydroxyandrost-5-ene shows high anticancer activity toward lymphoma.55 It has been proved that androstentriols with such location of the hydroxyl groups have the most potent anticancer activity. A chemical method of obtaining 3β,7α,17α-trihydroxyandrost-5-ene from 3β,17α-dihydroxyandrost-5-ene by selective oxidation of allylic carbon atom and reduction of the newly formed carbonyl group is known.55 However, to the best of our knowledge, there is no literature report on microbial method of obtaining 3β,7α,17α-trihydroxyandrost-5-ene.
An example of identical transformation of DHEA and some other 5-ene steroids to 7α-hydroxyderivatives by the strain Fusarium culmorum has been reported.56 Similar reactions of selective hydroxylation were also performed by Fusarium moniliforme verticillioides, F. oxysporum, Gelasinospora retispora and Mucor silvaticus,57–60 as well as by non-selective ones: Absidia coerulea AM93![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 61 and Mortierella isabellina AM212.62 7-Hydroxyderivatives of DHEA are interesting due to their neuroprotective63 and immunostimulatory properties,64 and also anti-inflammatory activity without the glucocorticoid action.5,19 7-Hydroxyderivatives of DHEA may also be regarded as epiandrosterone derivative with proven neuroprotective activity both in vitro and in vivo.65
61 and Mortierella isabellina AM212.62 7-Hydroxyderivatives of DHEA are interesting due to their neuroprotective63 and immunostimulatory properties,64 and also anti-inflammatory activity without the glucocorticoid action.5,19 7-Hydroxyderivatives of DHEA may also be regarded as epiandrosterone derivative with proven neuroprotective activity both in vitro and in vivo.65
Conclusions
Among 25 fungal strains isolated from the area of Wrocław, we selected 5 strains which were the most efficient in transformation of dehydroepiandrosterone and we used them for the biotransformation on larger scale, in order to identify the metabolites. Additionally, the strains were subjected to genetic identification.All the products were purified by chromatographic methods. Based on GC, TLC analysis and 1H NMR, 13C NMR spectra (including correlation spectra) we established the structures of nine products.
Three compounds were found to be new metabolites of DHEA. To the best of our knowledge, there are no reports in literature on transformations of DHEA leading to: adrost-1,4-diene-3,17-dione, 6β-hydroxyandrost-1,4-diene-3,17-dione and androst-5-ene-3β,7α,17α-triol.
The majority of tested biocatalysts showed excellent selectivity in the transformation of DHEA. Due to this feature, they possibly may be used for industrial production of bioactive DHEA metabolites provided basic genetic engineering techniques successfully increase the content of specific enzymes in these biocatalysts.
Acknowledgements
Publication supported by the Wroclaw Centre of Biotechnology under the Leading National Research Centre (KNOW) programme for years 2014–2018. Special thanks to Jarosław Popłoński for language correction.References
- M. V. Donova and O. V. Egorova, Appl. Microbiol. Biotechnol., 2012, 94, 1423–1447 CrossRef CAS PubMed.
- M. M. Chen, F. Q. Wang, L. C. Lin, K. Yao and D. Z. Wei, Appl. Microbiol. Biotechnol., 2012, 96, 133–142 CrossRef CAS PubMed.
- R. M. Loria, Steroids, 2002, 67, 953–966 CrossRef CAS PubMed.
- O. Hennebert, M. A. Pelissier, S. Le Mee, E. Wülfert and R. Morfin, J. Steroid Biochem. Mol. Biol., 2008, 110, 255–262 CrossRef CAS PubMed.
- M. A. Pélissier, C. Muller, M. Hill and R. Morfin, Steroids, 2006, 71, 240–248 CrossRef PubMed.
- B. Brazzini and N. Pimpinelli, Am. J. Clin. Dermatol., 2002, 3, 47–58 CrossRef PubMed.
- M. Panjari and S. R. Davis, Hum. Reprod. Update, 2007, 13, 239–248 CrossRef CAS PubMed.
- K. Rutkowski, P. Sowa and J. R.-T. A. Kuryliszyn-Moskal, Drugs, 2014, 74, 1195–1207 CrossRef CAS PubMed.
- F. Hammer, S. Subtil, P. Lux, C. Maser-Gluth, P. M. Stewart, B. Allolio and W. Arlt, J. Clin. Endocrinol. Metab., 2005, 90, 3600–3605 CrossRef CAS PubMed.
- D. Liu and J. S. Dillon, J. Biol. Chem., 2002, 277, 21379–21388 CrossRef CAS PubMed.
- X. Ding, D. Wang, L. Li and H. Ma, Int. J. Biochem. Cell Biol., 2016, 70, 126–139 CrossRef CAS PubMed.
- T. Suzuki, N. Suzuki, R. A. Daynes and E. G. Engleman, Clin. Immunol. Immunopathol., 1991, 61, 202–211 CrossRef CAS PubMed.
- M. Maggio, F. De Vita, A. Fisichella, E. Colizzi, S. Provenzano, F. Lauretani, M. Luci, G. Ceresini, E. Dall, P. Caffarra, G. Valenti and G. Paolo, J. Steroid Biochem. Mol. Biol., 2015, 145, 281–292 CrossRef CAS PubMed.
- D. T. Villareal, J. O. Holloszy and W. M. Kohrt, Clin. Endocrinol., 2000, 53, 561–568 CrossRef CAS PubMed.
- M. Panjari, R. J. Bell, F. Jane, J. Adams, C. Morrow and S. R. Davis, Maturitas, 2009, 63, 240–245 CrossRef CAS PubMed.
- H. Zhang, J. Ren, Y. Wang, C. Sheng, Q. Wu, A. Diao and D. Zhu, Tetrahedron, 2013, 69, 184–189 CrossRef CAS.
- P. Fernandes, A. Cruz, B. Angelova, H. M. Pinheiro and J. M. S. Cabral, Enzyme Microb. Technol., 2003, 32, 688–705 CrossRef CAS.
- R. Hampl, O. Lapčík, M. Hill, J. Klak, A. Kasal, A. Nováček, I. Šterzl, J. Šterzl and L. Stárka, Physiol. Res., 2000, 49 Search PubMed.
- M.-A. Pelissier, C. Trap, M.-I. Malewiak and R. Morfin, Steroids, 2004, 69, 137–144 CrossRef CAS PubMed.
- K. Kristan and T. L. Rižner, J. Steroid Biochem. Mol. Biol., 2012, 129, 79–91 CrossRef CAS PubMed.
- H. Pellissier and M. Santelli, Org. Prep. Proced. Int., 2001, 33, 1–58 CrossRef CAS.
- T. Janeczko, N. Milecka and E. Kostrzewa-Susłow, Przem. Chem., 2012, 91, 767–771 CAS.
- C. N. Ahlem, T. M. Page, D. L. Auci, M. R. Kennedy, K. Mangano, F. Nicoletti, Y. Ge, Y. Huang, S. K. White, S. Villegas, D. Conrad, A. Wang, C. L. Reading and J. M. Frincke, Steroids, 2011, 76, 145–155 CrossRef CAS PubMed.
- D. L. Auci, C. L. Reading and J. M. Frincke, Autoimmun. Rev., 2009, 8, 369–372 CrossRef CAS PubMed.
- Ł. Stępień, M. Jestoi and J. Chełkowski, World Mycotoxin J., 2013, 6, 399–409 CrossRef.
- T. J. White, T. Bruns, S. Lee and J. Taylor, in PCR Protocols: a Guide to Methods and Applications, ed. M. A. Innis, D. H. Gelfand, J. J. Sninsky and T. J. White, Academic Press, San Diego, USA, 1990, pp. 315–322 Search PubMed.
- Y. Li, F. Lu, T. Sun and L. Du, Lett. Appl. Microbiol., 2007, 44, 563–568 CrossRef CAS PubMed.
- L. Fernández De Las Heras, R. Van Der Geize, O. Drzyzga, J. Perera and J. María Navarro Llorens, J. Steroid Biochem. Mol. Biol., 2012, 132, 271–281 CrossRef PubMed.
- M. Kisiela, A. Skarka, B. Ebert and E. Maser, J. Steroid Biochem. Mol. Biol., 2012, 129, 31–46 CrossRef CAS PubMed.
- J. K. Capyk, I. Casabon, R. Gruninger, N. C. Strynadka and L. D. Eltis, J. Biol. Chem., 2011, 286, 40717–40724 CrossRef CAS PubMed.
- K. P. Choi, I. Molnár, M. Yamashita and Y. Murooka, J. Biochem., 1995, 117, 1043–1049 CrossRef CAS PubMed.
- Y. Li, F. Lu, T. Sun and L. Du, Lett. Appl. Microbiol., 2007, 44, 563–568 CrossRef CAS PubMed.
- W. Wei, S. Y. Fan, F. Q. Wang and D. Z. Wei, World J. Microbiol. Biotechnol., 2014, 30, 1947–1954 CrossRef CAS PubMed.
- Q. Zhang, Y. Ren, J. He, S. Cheng, J. Yuan, F. Ge, W. Li, Y. Zhang and G. Xie, Ann. Microbiol., 2015, 65, 1961–1971 CrossRef CAS.
- L. A. R. Sallam, A. M. El-Refai and H. A. El-Minofi, Process Biochem., 2005, 40, 203–206 CrossRef CAS.
- A. Bartmańska and J. Dmochowska-Gładysz, Enzyme Microb. Technol., 2007, 40, 1615–1621 CrossRef.
- L. R. Lehman and J. D. Stewart, Curr. Org. Chem., 2001, 5, 439–470 CrossRef CAS.
- I. Ahmad and Shagufta, Eur. J. Med. Chem., 2015, 102, 375–386 CrossRef CAS PubMed.
- M. R. Yadav, M. A. Barmade, R. S. Tamboli and P. R. Murumkar, Eur. J. Med. Chem., 2015, 105, 1–38 CrossRef CAS PubMed.
- L. Butinar, M. Mohorčič, V. Deyris, K. Duquesne, G. Iacazio, M. Claeys-Bruno, J. Friedrich and V. Alphand, Phytochemistry, 2015, 117, 144–153 CrossRef CAS PubMed.
- V. Alphand and R. Wohlgemuth, Curr. Org. Chem., 2010, 14, 1928–1965 CrossRef CAS.
- T. Kołek, A. Szpineter and A. Świzdor, Steroids, 2008, 73, 1441–1445 CrossRef PubMed.
- A. C. Hunter, E. Coyle, F. Morse, C. Dedi, H. T. Dodd and S.-J. Koussoroplis, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2009, 1791, 110–117 CrossRef CAS PubMed.
- L. H. Huang, J. Li, G. Xu, X. H. Zhang, Y. G. Wang, Y. L. Yin and H. M. Liu, Steroids, 2010, 75, 1039–1046 CrossRef CAS PubMed.
- A. Świzdor, Molecules, 2013, 18, 13812–13822 CrossRef PubMed.
- B. Yang, Y. Wang, X. Chen, J. Feng, Q. Wu and D. Zhu, Tetrahedron, 2014, 70, 41–46 CrossRef CAS.
- M. L. Mascotti, M. A. Palazzolo, F. R. Bisogno and M. Kurina-Sanz, Steroids, 2016, 109, 44–49 CrossRef CAS PubMed.
- T. Kołek, A. Szpineter and A. Świzdor, Steroids, 2009, 74, 859–862 CrossRef PubMed.
- E. Beneventi, G. Ottolina, G. Carrea, W. Panzeri, G. Fronza and P. C. K. Lau, J. Mol. Catal. B: Enzym., 2009, 58, 164–168 CrossRef CAS.
- A. Świzdor, T. Kołek, A. Panek and A. Białońska, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2011, 1811, 253–262 CrossRef PubMed.
- H.-M. Liu, H. Li, L. Shan and J. Wu, Steroids, 2006, 71, 931–934 CrossRef CAS PubMed.
- G. Moeller and J. Adamski, Mol. Cell. Endocrinol., 2006, 248, 47–55 CrossRef CAS PubMed.
- P. Lukacik, K. L. Kavanagh and U. Oppermann, Mol. Cell. Endocrinol., 2006, 248, 61–71 CrossRef CAS PubMed.
- M. Shao, X. Zhang, Z. Rao, M. Xu, T. Yang, H. Li, Z. Xu and S. Yang, Green Chem., 2016, 18, 1774–1784 RSC.
- M. R. Graf, W. Jia, M. L. Lewbart and R. M. Loria, Chem. Biol. Drug Des., 2009, 74, 625–629 CAS.
- T. Kołek, J. Steroid Biochem. Mol. Biol., 1999, 71, 83–90 CrossRef.
- A. C. Cotillon and R. Morfin, J. Steroid Biochem. Mol. Biol., 1999, 68, 229–237 CrossRef CAS PubMed.
- P. Durairaj, S. Malla, S. P. Nadarajan, P.-G. Lee, E. Jung, H. H. Park, B.-G. Kim and H. Yun, Microb. Cell Fact., 2015, 14, 45 CrossRef PubMed.
- M. Koshimura, T. Utsukihara, A. Hara, S. Mizobuchi, C. A. Horiuchi and M. Kuniyoshi, J. Mol. Catal. B: Enzym., 2010, 67, 72–77 CrossRef CAS.
- Y. Wang, D. Sun, Z. Chen, H. Ruan and W. Ge, Biocatal. Biotransform., 2013, 31, 168–174 CrossRef CAS.
- N. Milecka-Tronina, T. Kołek, A. Świzdor and A. Panek, Bioorg. Med. Chem., 2014, 22, 883–891 CrossRef CAS PubMed.
- T. Kołek, N. Milecka, A. Świzdor, A. Panek and A. Białońska, Org. Biomol. Chem., 2011, 9, 5414–5422 Search PubMed.
- M. Schumacher, S. Weill-Engerer, P. Liere, F. Robert, R. J. M. Franklin, L. M. Garcia-Segura, J. J. Lambert, W. Mayo, R. C. Melcangi, A. Parducz, U. Suter, C. Carelli, E. E. Baulieu and Y. Akwa, Prog. Neurobiol., 2003, 71, 3–29 CrossRef CAS PubMed.
- R. Morfin, P. Lafaye, A. C. Cotillon and F. Nato, Ann. N. Y. Acad. Sci., 2000, 917, 971–982 CrossRef CAS PubMed.
- A. K. Pringle, W. Schmidt, J. K. Deans, E. Wulfert, K. G. Reymann and L. E. Sundstrom, Eur. J. Neurosci., 2003, 18, 117–124 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: 1H, 13C NMR, HSQC, COSY and MS data of all products. See DOI: 10.1039/c7ra04608a |
| This journal is © The Royal Society of Chemistry 2017 |