Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Use of a next generation maleimide in combination with THIOMAB™ antibody technology delivers a highly stable, potent and near homogeneous THIOMAB™ antibody-drug conjugate (TDC)†
João P. M. Nunesa,
Vessela Vassilevab,
Eifion Robinsona,
Maurício Morais a,
Mark E. B. Smitha,
R. Barbara Pedleyb,
Stephen Caddicka,
James R. Baker
a,
Mark E. B. Smitha,
R. Barbara Pedleyb,
Stephen Caddicka,
James R. Baker *a and
Vijay Chudasama
*a and
Vijay Chudasama *ac
*ac
aDepartment of Chemistry, University College London, London, WC1H 0AJ, UK. E-mail: j.r.baker@ucl.ac.uk; v.chudasama@ucl.ac.uk; Tel: +44 (0)20 7679 2653 Tel: +44 (0)20 7679 2077
bUCL Cancer Institute, 72 Huntley Street, London, UK
cResearch Institute for Medicines (iMed.ULisboa), Faculty of Pharmacy, Universidade de Lisboa, Lisbon, Portugal
First published on 10th May 2017
Abstract
Herein we demonstrate that conjugation of a next generation maleimide (NGM) to engineered cysteines in a THIOMAB™ antibody delivers a THIOMAB™ antibody-drug conjugate (TDC) with a drug loading of ca. 2. This TDC is highly stable in blood serum conditions, selective and potent towards HER2 expressing cell lines and meets the current criteria for optimised antibody-drug conjugates (ADCs).
Antibody-drug conjugates (ADCs) have emerged as targeted therapeutics against cancer by combining the selectivity of the antibody component with the potency of cytotoxic drugs. There are two FDA-approved ADCs already on the market (Adcetris™ and Kadcyla™) and over 40 additional ADCs currently undergoing clinical trials.1–4
Classical methods of conjugation for producing ADCs include lysine modification5 and cysteine modification via reduction of solvent accessible disulfide bonds followed by reaction with maleimides.6 It has been recognised that both methods generate heterogeneous mixtures of ADCs with different drug loadings and varied sites of attachment, resulting in sub-optimal therapeutic indices and higher clearance rates.7–13 Formation of higher drug-to-antibody ratio species (DAR > 4) have narrower therapeutic indices.1,7,9,12,14 The latter method also results in the loss of structural disulfide bonds which can lead to poor stability.7,9,14
The next generation of conjugation methods has aimed to reduce heterogeneity by focusing on site-selectivity. Chemoenzymatic methods have been used for ADC synthesis such as glycan modification,15–17 glutamine amidation18 and cysteine oxidation to aldehyde tags.19 Site-selective disulfide modification with small molecules has been achieved using next-generation maleimides (NGMs),20–27 pyridazinediones (PDs)28–32 and bis-sulfones.33,34 Site-specific reactivity has also been accomplished through incorporation of non-natural aminoacids into proteins for bioorthogonal conjugation.10,35–39
The THIOMAB™ antibody technology platform uses site-directed mutagenesis to incorporate cysteines into the antibody. Conjugation to a classical maleimide bearing the drug of choice affords a THIOMAB™ antibody-drug conjugate (TDC) with full control over site reactivity and a precise drug-to-antibody ratio (DAR).12,40–42 The technology also avoids issues with some chemoenzymatic methods that can result in over oxidation of aminoacids43 and aldehyde tag conversion to unreactive gem-diols.44 TDCs have been demonstrated to improve therapeutic indices and pharmacodynamics over ADCs prepared by classical cysteine and lysine modification.11,12,42
Maleimides are rapid thiol-selective conjugation reagents.45,46 However, the succinimide conjugates thus formed can undergo retro-Michael reactions and scavenging with albumin,47 resulting in premature drug release during blood circulation.10,41,48 N-Alkyl succinimide conjugates can be stabilised against retro-Michael reactions by hydrolysis to succinamic acid derivatives,49–51 which resulted in ADCs with improved pharmacokinetics and in vivo efficacy.41,51,52 Most N-alkyl thiosuccinimide ADCs require prolonged hydrolysis (>16 h) at high pH (>8)41,51 which is not desirable from a process view and can lead to deamidation and loss of payload.51 Hydrolysis has been facilitated to under an hour by the inclusion of a basic amine on the N-alkyl substituent near the point of attachment to the thiosuccinimide52 or by using N-aryl substituents, but these also come with limitations.1,25,49
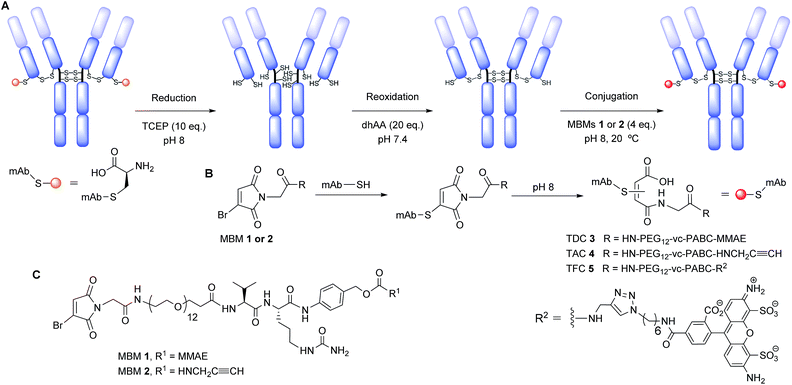
We have previously shown that ADCs prepared via disulfide bridging with NGMs can be stabilised through hydrolysis.21 We have also introduced monobromomaleimides (MBMs) as fast reagents for conjugation to single cysteines.53 We envisaged introducing a glycine spacer between the MBM and the valine–citrulline self-immolative linker carrying monomethyl auristatin E (vc-PABC-MMAE) to enhance hydrolysis of the resulting maleimide conjugate. Conjugation of this reagent to the THIOMAB™ antibody should create a highly stable and homogeneous TDC with a DAR of ca. 2 without a distinct hydrolysis step.
We chose to functionalise a thio-trastuzumab variant directed against HER2 with an engineered cysteine on each light chain (LC-V205C), obtained from Genentech (in a 3.67 mg mL−1 solution in 50 mM Tris acetates solution). This THIOMAB™ antibody displays enhanced rate of thiosuccinimide hydrolysis due to the positively charged environment surrounding the mutant site.41 However, succinimide conjugates can still undergo retro-Michael before hydrolysing. By contrast NGMs mechanistically cannot undergo retro-Michael by elimination. Thus, NGMs offer the advantage of a stable conjugation during hydrolysis. Modification of thio-trastuzumab follows a 3 step process: reduction of all solvent accessible disulfide bonds, mild reoxidation of native interchain disulfide bonds and conjugation with MBMs 1 and 2 to afford TDC 3 and thio-trastuzumab alkyne conjugate (TAC) 4, respectively (Scheme 1, see ESI for details†). Functionalisation of TAC 4 with AlexaFluor® 488 affords thio-trastuzumab fluorophore conjugate (TFC) 5. Unlike previous protocols, it was envisaged that hydrolysis of these NGM conjugates would be fast, and in situ, under our mildly basic conjugation conditions (pH 8).
MBM reagents 1 and 2 were prepared in a convergent synthesis. Monobromomaleic anhydride was condensed with glycine to afford 2-(3-bromo-2,5-dioxo-2,5-dihydro-1H-pyrrol-1-yl)acetic acid. Commercially available BocHN-vc-PABC-PNP carbonate was coupled to MMAE or propargylamine by an adapted protocol.6 Boc deprotection with TFA was followed by amide bond formation with a bifunctional polyethylene glycol spacer (PEG12). Another Boc deprotection and coupling to 2-(3-bromo-2,5-dioxo-2,5-dihydro-1H-pyrrol-1-yl)acetic acid afforded MBM reagents 1 and 2 (see ESI for further details†).
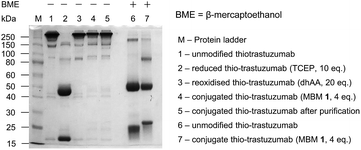
We functionalised the thio-trastuzumab by adapting a literature protocol.12 Reduction of all solvent accessible disulfide bonds was carried out with tris-2-carboxyethyl phosphine hydrochloride (TCEP), followed by reoxidation of native interchain disulfide bonds with dehydroascorbic acid (dhAA). Ellman's analysis following reduction and oxidation steps revealed the expected 10 and 2 sulfhydryls per antibody respectively, indicating there are two available cysteines per antibody for conjugation (see ESI, Table S1†). Conjugation was carried out using a modest excess of MBM 1 (4 eq., 2 eq. per cysteine) at pH 8 for 1 h at room temperature to afford TDC 3. Ellman's analysis after conjugation and purification revealed negligible free sulfhydryls (see ESI, Table S1†).
Non-reducing SDS-PAGE gel revealed complete reduction and reoxidation in the first two steps (lanes 2 and 3, Fig. 1). No fragmentation is visible following conjugation and purification (lanes 4 and 5), suggesting that the resulting TDC 3 consists of full mAb species alone. A reducing SDS-PAGE gel reveals that the light chain of TDC 3 is shifted to a higher mass indicating that conjugation has taken place on the light chain (lane 7, Fig. 1). Furthermore, it demonstrates that the conjugation is resistant to harsh reducing conditions.
Non-reducing LC-MS analysis of TDC 3 revealed a thio-trastuzumab conjugated with two maleamic acid PEG12-vc-PABC-MMAE payloads (151![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 824 Da, calculated mass 151
824 Da, calculated mass 151![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 819 Da, Fig. 2). TDC 3 was also analysed by LC-MS following reduction of the interchain disulfide bonds (TCEP, 10 eq.) and capping of free sulfhydryls with maleimide (100 eq.). This revealed a heavy chain with 3 maleimides (50
819 Da, Fig. 2). TDC 3 was also analysed by LC-MS following reduction of the interchain disulfide bonds (TCEP, 10 eq.) and capping of free sulfhydryls with maleimide (100 eq.). This revealed a heavy chain with 3 maleimides (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 888 Da) and two light chain species: one modified with one maleimide and one maleamic acid PEG12-vc-PABC-MMAE payload as proton (25
888 Da) and two light chain species: one modified with one maleimide and one maleamic acid PEG12-vc-PABC-MMAE payload as proton (25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 418 Da, expected mass 25
418 Da, expected mass 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 416 Da) and sodium (25
416 Da) and sodium (25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 443 Da, expected 25
443 Da, expected 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 438 Da) adducts, and another light chain modified with maleimide alone (23
438 Da) adducts, and another light chain modified with maleimide alone (23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 638 Da, expected mass 23
638 Da, expected mass 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 638 Da). Peak integration demonstrates that almost all of the light chains were successfully conjugated to MBM 1 and practically all of the conjugates hydrolysed to maleamic acid. From the extent of light chain conjugation we calculated a DAR of 1.8.
638 Da). Peak integration demonstrates that almost all of the light chains were successfully conjugated to MBM 1 and practically all of the conjugates hydrolysed to maleamic acid. From the extent of light chain conjugation we calculated a DAR of 1.8.
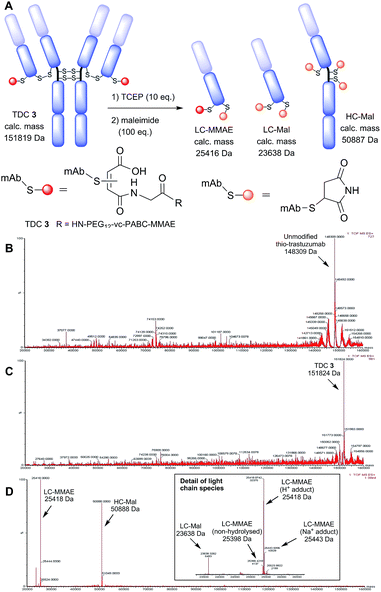 | ||
| Fig. 2 Characterisation of TDC 3 by LC-MS. (A) Fragments generated for reducing LC-MS; deconvoluted MS data for (B) unmodified thio-trastuzumab, (C) unreduced TDC 3 and (D) reduced TDC 3. | ||
This value is in agreement with results from Ellman's analysis following conjugation (see ESI, Table S1†). This confirms conjugation with MBM 1 took place on light chain alone (Fig. 2). It also demonstrates that the maleimide conjugate hydrolysed to the maleamic acid form as required for enhanced stability during conjugation conditions without any further steps required.
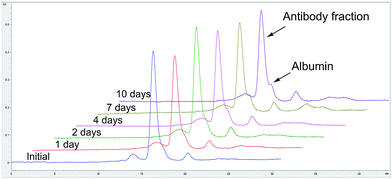
Size exclusion chromatography of TDC 3 showed that the conjugate is composed predominantly of monomeric species (>98%, see ESI, Fig. S5†). TDC 3 retains binding activity to HER2 (expressed in human cells, obtained from Sino Biological), as determined by ELISA assay, demonstrating that conjugation does not impact on receptor binding (see ESI, Fig. S6†). We also prepared TFC 5 by conjugating TAC 4 with MBM 2, followed by copper catalysed alkyne–azide cycloaddition (CuAAC) to AlexaFluor® 488. TFC 5 was incubated in blood serum (human serum, from human male plasma AB, USA origin, sterile-filtered, obtained from Sigma-Aldrich) at 37 °C over 10 days and analysed by fluorescence SEC-HPLC to assess the stability of conjugation. No significant transfer to albumin or decrease in antibody fluorescence were observed over time, demonstrating that our method of conjugation affords a highly serum-stable TDC (Fig. 3).
In possession of TDC 3 bearing MMAE, we next assessed in vitro cytotoxicity by the MTT assay. HER2-positive human breast cancer cell lines SKBR-3 and HCC-1954 were chosen and HER2-negative cell line MCF-7 was used as a negative control (all cell lines were obtained from the American Type Culture Collection (ATCC)). All cell lines were sensitive to MMAE, with IC50 values of 0.19 nM, 0.21 nM and 0.01 nM for SKBR-3, HCC-1954 and MCF-7 respectively. Gratifyingly, TDC 3 was highly potent against HER2-positive cell lines, with IC50 values of 0.14 nM and 0.31 nM for SKBR-3 and HCC-1954 respectively. TDC 3 displayed minimal activity against the HER-2 negative MCF-7 cell line. This demonstrates that TDC 3 displays potent cell-killing ability in a targeted manner for HER2 expressing cell lines (Fig. 4).
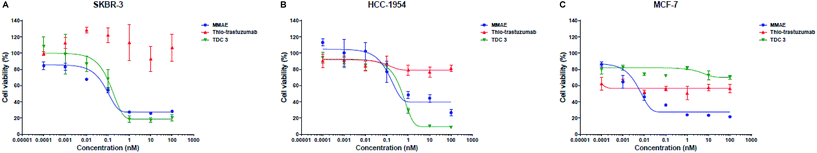 | ||
| Fig. 4 In vitro potency of TDC 3 and thio-trastuzumab on (A) SKBR-3 cells, (B) HCC-1954 cells, and (C) MCF-7. Cell viability was plotted against TDC concentration. | ||
Conclusions
In conclusion, we have applied NGM technology as MBM compounds for the functionalisation of single cysteines in a thio-trastuzumab variant. We demonstrate that this application affords a TDC with a controlled DAR as full mAb species that is highly stable, retains binding activity towards HER2 and displays potent and selective cell kill ability. Moreover, the mechanism for conjugate stabilisation via hydrolysis is shown to naturally take place during standard conjugation conditions, revealing an advantage from a process and efficiency point of view. NGM technology is entirely compatible with the current generation of superior engineered cysteine THIOMAB™ antibody variants. The TDC thus prepared with NGM technology fits all current criteria for industry-relevant ADCs.Acknowledgements
The authors gratefully acknowledge Genentech for the supply of the thio-trastuzumab variant. We also acknowledge Dr Erik Årstad and Dr Kerstin Sanders for support with fluorescence SEC-HPLC. This work was supported by the EPSRC, BBSRC, BRC, Wellcome Trust, HEFCE, SBC, MRC, UCL and UCLB for support of our programme. Dr Vessela Vassileva is funded by Cancer Research UK and the Pancreatic Cancer Research Fund.Notes and references
- J. R. Adair, P. W. Howard, J. A. Hartley, D. G. Williams and K. A. Chester, Expert Opin. Biol. Ther., 2012, 12, 1191–1206 CrossRef CAS PubMed.
- S. C. Alley, N. M. Okeley and P. D. Senter, Curr. Opin. Chem. Biol., 2010, 14, 529–537 CrossRef CAS PubMed.
- A. Beck and J. M. Reichert, mAbs, 2014, 6, 15–17 CrossRef PubMed.
- V. Chudasama, A. Maruani and S. Caddick, Nat. Chem., 2016, 8, 114–119 CrossRef CAS PubMed.
- A. A. Wakankar, M. B. Feeney, J. Rivera, Y. Chen, M. Kim, V. K. Sharma and Y. J. Wang, Bioconjugate Chem., 2010, 21, 1588–1595 CrossRef CAS PubMed.
- S. O. Doronina, et al., Nat. Biotechnol., 2003, 21, 778–784 CrossRef CAS PubMed.
- Y. T. Adem, K. A. Schwarz, E. Duenas, T. W. Patapoff, W. J. Galush and O. Esue, Bioconjugate Chem., 2014, 25, 656–664 CrossRef CAS PubMed.
- C. A. Boswell, et al., Bioconjugate Chem., 2011, 22, 1994–2004 CrossRef CAS PubMed.
- K. J. Hamblett, et al., Clin. Cancer Res., 2004, 10, 7063–7070 CrossRef CAS PubMed.
- D. Jackson, et al., PLoS ONE, 2014, 9, e83865 Search PubMed.
- J. R. Junutula, et al., Clin. Cancer Res., 2010, 16, 4769–4778 CrossRef CAS PubMed.
- J. R. Junutula, et al., Nat. Biotechnol., 2008, 26, 925–932 CrossRef CAS PubMed.
- D. S. Wilbur, M.-K. Chyan, H. Nakamae, Y. Chen, D. K. Hamlin, E. B. Santos, B. T. Kornblit and B. M. Sandmaier, Bioconjugate Chem., 2012, 23, 409–420 CrossRef CAS PubMed.
- N. S. Beckley, K. P. Lazzareschi, H.-W. Chih, V. K. Sharma and H. L. Flores, Bioconjugate Chem., 2013, 24, 1674–1683 CrossRef CAS PubMed.
- X. Li, T. Fang and G.-J. Boons, Angew. Chem., 2014, 126, 7307–7310 CrossRef.
- A. C. Stan, D. L. Radu, S. Casares, C. A. Bona and T.-D. Brumeanu, Cancer Res., 1999, 59, 115–121 CAS.
- Q. Zhou, et al., Bioconjugate Chem., 2014, 25, 510–520 CrossRef CAS PubMed.
- P. Strop, et al., Chem. Biol., 2013, 20, 161–167 CrossRef CAS PubMed.
- P. M. Drake, et al., Bioconjugate Chem., 2014, 25, 1331–1341 CrossRef CAS PubMed.
- L. Castañeda, A. Maruani, F. F. Schumacher, E. Miranda, V. Chudasama, K. A. Chester, J. R. Baker, M. E. B. Smith and S. Caddick, Chem. Commun., 2013, 49, 8187–8189 RSC.
- J. P. M. Nunes, et al., Chem. Commun., 2015, 51, 10624–10627 RSC.
- H. Pye, et al., Photochem. Photobiol. Sci., 2016, 15, 1227–1238 CAS.
- C. P. Ryan, M. E. B. Smith, F. F. Schumacher, D. Grohmann, D. Papaioannou, G. Waksman, F. Werner, J. R. Baker and S. Caddick, Chem. Commun., 2011, 47, 5452–5454 RSC.
- F. F. Schumacher, M. Nobles, C. P. Ryan, M. E. B. Smith, A. Tinker, S. Caddick and J. R. Baker, Bioconjugate Chem., 2011, 22, 132–136 CrossRef CAS PubMed.
- F. F. Schumacher, J. P. M. Nunes, A. Maruani, V. Chudasama, M. E. B. Smith, K. A. Chester, J. R. Baker and S. Caddick, Org. Biomol. Chem., 2014, 12, 7261–7269 CAS.
- F. F. Schumacher, et al., Sci. Rep., 2013, 3, 1525 CrossRef PubMed.
- (a) M. E. B. Smith, F. F. Schumacher, C. P. Ryan, L. M. Tedaldi, D. Papaioannou, G. Waksman, S. Caddick and J. R. Baker, J. Am. Chem. Soc., 2010, 132, 1960–1965 CrossRef CAS PubMed; (b) M. Morais, J. P. M. Nunes, K. Karu, N. Forte, I. Benni, M. E. B. Smith, S. Caddick, V. Chudasama and J. R. Baker, Org. Biomol. Chem., 2017, 15, 2947–2952 RSC.
- V. Chudasama, M. E. B. Smith, F. F. Schumacher, D. Papaioannou, G. Waksman, J. R. Baker and S. Caddick, Chem. Commun., 2011, 47, 8781–8783 RSC.
- M. T. W. Lee, A. Maruani, J. R. Baker, S. Caddick and V. Chudasama, Chem. Sci., 2016, 7, 799–802 RSC.
- A. Maruani, S. Alom, P. Canavelli, M. T. W. Lee, R. E. Morgan, V. Chudasama and S. Caddick, Chem. Commun., 2015, 51, 5279–5282 RSC.
- A. Maruani, H. Savoie, F. Bryden, S. Caddick, R. Boyle and V. Chudasama, Chem. Commun., 2015, 51, 15304–15307 RSC.
- (a) A. Maruani, M. E. B. Smith, E. Miranda, K. A. Chester, V. Chudasama and S. Caddick, Nat. Commun., 2015, 6, 6645 CrossRef CAS PubMed; (b) E. Robinson, J. P. M. Nunes, V. Vassileva, A. Maruani, J. C. F. Nogueira, M. E. B. Smith, R. B. Pedley, S. Caddick, J. R. Baker and V. Chudasama, RSC Adv., 2017, 7, 9073–9077 RSC.
- G. Badescu, et al., Bioconjugate Chem., 2014, 25, 1124–1136 CrossRef CAS PubMed.
- P. Bryant, et al., Mol. Pharm., 2015, 12, 1872–1879 CrossRef CAS PubMed.
- J. Y. Axup, et al., Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 16101–16106 CrossRef CAS PubMed.
- J. C. Kern, et al., J. Am. Chem. Soc., 2016, 138, 1430–1445 CrossRef CAS PubMed.
- J. C. Kern, et al., Bioconjugate Chem., 2016, 27, 2081–2088 CrossRef CAS PubMed.
- F. Tian, et al., Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 1766–1771 CrossRef CAS PubMed.
- E. S. Zimmerman, et al., Bioconjugate Chem., 2014, 25, 351–361 CrossRef CAS PubMed.
- S. C. Jeffrey, et al., Bioconjugate Chem., 2013, 24, 1256–1263 CrossRef CAS PubMed.
- B. Q. Shen, et al., Nat. Biotechnol., 2012, 30, 184–189 CrossRef CAS PubMed.
- P. Thompson, et al., J. Controlled Release, 2016, 236, 100–116 CrossRef CAS PubMed.
- W. Wang, J. Vlasak, Y. Li, P. Pristatsky, Y. Fang, T. Pittman, J. Roman, Y. Wang, T. Prueksaritanont and R. Ionescu, Mol. Immunol., 2011, 48, 860–866 CrossRef CAS PubMed.
- D. Rabuka, J. S. Rush, G. W. deHart, P. Wu and C. R. Bertozzi, Nat. Protoc., 2012, 7, 1052–1067 CrossRef CAS PubMed.
- F. Saito, H. Noda and J. W. Bode, ACS Chem. Biol., 2015, 10, 1026–1033 CrossRef CAS PubMed.
- P. Schelté, C. Boeckler, B. Frisch and F. Schuber, Bioconjugate Chem., 2000, 11, 118–123 CrossRef.
- A. D. Baldwin and K. L. Kiick, Bioconjugate Chem., 2011, 22, 1946–1953 CrossRef CAS PubMed.
- S. C. Alley, D. R. Benjamin, S. C. Jeffrey, N. M. Okeley, D. L. Meyer, R. J. Sanderson and P. D. Senter, Bioconjugate Chem., 2008, 19, 759–765 CrossRef CAS PubMed.
- R. J. Christie, et al., J. Controlled Release, 2015, 220, 660–670 CrossRef CAS PubMed.
- S. D. Fontaine, R. Reid, L. Robinson, G. W. Ashley and D. V. Santi, Bioconjugate Chem., 2015, 26, 145–152 CrossRef CAS PubMed.
- L. N. Tumey, et al., Bioconjugate Chem., 2014, 25, 1871–1880 CrossRef CAS PubMed.
- R. P. Lyon, et al., Nat. Biotechnol., 2014, 32, 1059–1062 CrossRef CAS PubMed.
- L. M. Tedaldi, M. E. B. Smith, R. I. Nathani and J. R. Baker, Chem. Commun., 2009, 6583–6585 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Conjugation and analysis protocols, synthetic procedures, 1H and 13C NMR spectra for synthesised compounds, LC-MS spectra, Ellman's analysis, SEC analysis, ELISA analysis and other supplementary results. See DOI: 10.1039/c7ra04606e |
| This journal is © The Royal Society of Chemistry 2017 |