Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Dependence between cytotoxicity and dynamic subcellular localization of up-conversion nanoparticles with different surface charges†
Xiaodan Liab,
Ying Tanga,
Lijun Xu*a,
Xianggui Kong b,
Li Zhanga,
Yulei Chang
b,
Li Zhanga,
Yulei Chang b,
Huiying Zhao*a,
Hong Zhang
b,
Huiying Zhao*a,
Hong Zhang bc and
Xiaomin Liu*b
bc and
Xiaomin Liu*b
aDepartment of Respiratory Medicine, The First Hospital, Jilin University, Changchun 130021, P. R. China
bState Key Laboratory of Luminescence and Applications, Changchun Institute of Optics, Fine Mechanics and Physics, Chinese Academy of Sciences, Changchun 130033, P. R. China. E-mail: xmliu@ciomp.ac.cn
cVan't Hoff Institute for Molecular Sciences, University of Amsterdam, Science Park 904, 1098 XH Amsterdam, The Netherlands
First published on 3rd July 2017
Abstract
Intensive investigations have been devoted to lanthanide-doped upconversion nanoparticles (UCNPs), which have shown great potential in applications such as biomedical imaging and therapy. Recently, various polymer-coated UCNPs, as novel bioprobes or nanocarriers, have been developed and their cellular uptake behavior and cytotoxicity have been widely studied. However, the interactions between the UCNPs and subcellular organelles are poorly understand, which restrict their applications in biomedicine. Herein, we engineered UCNPs with different surface charges (positive, negative, and neutral) and studied the dependence between cytotoxicity, internalization, and subcellular localization in normal and cancer cell lines. It was observed that UCNPs with positive or neutral charges entered most of the studied cell lines, whereas UCNPs with negative charges internalize mostly inside the cancer cell lines. Moreover, upon entering into the cells, these UCNPs are localized in different cell compartments, e.g. the cytoplasm, mitochondria or lysosomes, depending on their surface charges and incubation time with the cells. It is revealed that the cytotoxicity of the differently charged UCNPs towards the studied cell lines significantly depends on localization in the mitochondria rather than in the lysosomes or cytoplasm. The corresponding changes in the mitochondria structure were visualized showing the likelihood of cell death. These results have enriched our knowledge on the cytotoxicity of UCNPs in organelle and sheds light on the design of organelle-targeted UCNPs in tumor imaging and therapy.
1. Introduction
Progress in the design and manipulation of materials at the nano-scale has opened fascinating avenues for functional materials and constituted the basis for a broad range of applications, especially in biomedicine.1,2 In particular, nanoparticles are believed to have the potential to assist in the delivery of imaging contrast agents, therapeutic drugs or genetic materials to targeted lesion tissues and make the delivery procedure more precise, more effective, and less harmful to healthy tissues and organs.3–6 In recent years, inorganic nanomaterials such as silica nanoparticles,7,8 magnetic nanoparticles,9–11 carbon materials,12–14 metallic nanoparticles (like silver, gold),15–17 and up-conversion nanoparticles18–24 have been widely utilized for imaging and drug delivery in the laboratory. Particularly, lanthanide ion (Ln3+, such as Er3+, Tm3+, and Ho3+)-doped up-conversion nanoparticles (UCNPs), which are able to emit high-energy photons under excitation with near-infrared (NIR) light, have attracted significant interest in a variety of biological applications.25 The up-conversion light emitted by UCNPs is considered to be non-blinking and non-bleaching, less scattered, and capable of deep tissue penetration because excitation in the NIR region is within the optical transparency window of biological tissues. Besides imaging, UCNPs show great potential as nanocarriers for drug delivery, photodynamic therapy (PDT), gene therapy, immunotherapy and chemotherapy.26Note that the effective curative effect always requires drugs and therapeutics acting at specific subcellular organelles. For example, practical gene therapy has been excluded from the clinic for the past decade primarily due to the non-ideal intracellular processing of nanoparticle carriers and inefficient delivery of intact genes into the nucleus of target cells. Similarly, effective PDT critically depends upon the specific delivery of the nano-photosensitizer to the subcellular organelles. Furthermore, diseases associated with specific organelles may only become amenable to treatment upon the development of nanoparticle carriers that are capable of efficiently localizing therapeutics in the subcellular compartment of interest.27 Therefore, understanding the interactions between nanoparticles and the subcellular organelles are becoming a critical issue.
From the reports on both in vitro and in vivo investigations for a wide-range organic and inorganic nanoparticles over the past few decades, it turns out that the size, shape and surface coating properties of the colloidal particles play an important role in modulating their interactions with cells. Various UCNPs synthesis methods and surface modification strategies including silica coating, polymer coating, ligand oxidation, electrostatic layer by layer assembly and ligand exchange have been developed over the last several years to improve their water-dispersibility and bio-compatibility.28 For nanoparticles with the same size and shape, it has been shown that their surface charged properties play an important role in cellular internalization either in vitro or in vivo,29 for example, a surface charge of less than 15 mV has the smallest phagocytosis and long cycle performance.30 As far as UCNPs are concerned, the most common interactions, electrostatic interactions, between the charged UCNPs and the negatively charged plasma membrane of cells greatly affect the cytotoxicity and uptake efficiency of UCNPs. Many reports have demonstrated that positively charged UCNPs are clearly observed in cells following incubation, which is significantly higher than their neutral and negative counterparts.31–33 However, systematic studies are lacking on the intracellular uptake, especially the subcellular dynamic distribution and the relationships between the cytotoxicity and subcellular localization, which are the basis for unravelling the interactions between UCNPs and their biological environment.
Herein, we report our study on the surface charge dependent cell internalization, subcellular localization and cytotoxicity profile of UNCPs in healthy cells vs. malignant cells. Mouse microglia (BV-2) cells and human umbilical vein endothelial (HUVEC) cells were selected as non-transformed (normal) cells, whereas human alveolar adenocarcinoma (A549) and cholangiocarcinoma (QBC939) cells were selected as transformed (cancer) cell lines. Our results showed that positively charged UCNPs internalized in all cells, localizing preferentially in the lysosomes, followed by translocation to the mitochondria only in the QBC939 cells and subsequently becoming toxic to these cells. In contrast, UCNPs with a negative charge were internalized only in the QBC939 cells but not too much in the A549 cells, thus exhibiting toxicity only to the QBC939 cells. The UCNPs with neutral charge were not toxic to either normal cells or cancer cells, as these nanoparticles were primarily localized in the cytoplasm of the cells. The cytotoxicity of the differently charged UCNPs to the four different cell lines significantly depends on localization in the mitochondria rather than in the lysosomes or cytoplasm. This study provides a fundamental understanding of the surface charge of UCNPs on subcellular localization and toxicity, and provides a solid foundation for the biomedical application of up-conversion nanoparticles.
2. Experimental
2.1. Synthesis and surface modification of UCNPs
NaYF4: Yb3+, Er3+ UCNPs were synthesized using a typical procedure. 0.78 mmol of YCl3, 0.20 mmol of YbCl3 and 0.02 mmol of ErCl3 were placed into a 100 mL flask. Afterwards 6 mL of oleic acid (OA) and 15 mL of 1-octadecene (ODE) were added. This solution was heated to 100 °C for 10 min and then to 156 °C for 30 min, and subsequently allowed to cool to room temperature. Then, a solution of 4 mmol of NH4F (0.1482 g) and 2.5 mmol of NaOH (0.1 g) in 10 mL of methanol was added into the flask and the mixture was kept at 55 °C for 30 min. After evaporation of the methanol solvent, the solution was heated to 300 °C under an argon atmosphere for 1.5 h and cooled down to room temperature. The nanocrystals were precipitated with 10 mL of acetone, collected via centrifugation and redispersed in 10 mL of hexane.Preparation of the positively and negatively charged UCNPs: a two-step ligand exchange method was performed to render the hydrophobic UCNPs hydrophilic for further biological applications. In a typical process 10 mL of the hexane solution of the oleate-capped UCNPs was added to a glass bottle containing 10 mL of deionized water and was stirred vigorously for 2 h at room temperature while maintaining the pH around 4 by adding 0.1 M HCl solution (6%). After the reaction was completed, the oleic acid was removed from the aqueous solution by extracting the mixture three times with diethyl ether. The bare UCNPs were collected via centrifugation and washed two times with acetone before being redispersed in water. To this solution of bare UCNPs the ligand materials were added. Because of the high surface chemical activity, the ligand molecules can readily react with the dangling bonds on the ligand-free UCNPs and form a coordination bond. In the present study, the polymeric molecules PAA and PAAm were utilized to functionalize the UCNPs by mixing them directly with the bare UCNPs and washing afterwards with water for at least three times in order to remove the excess ligand materials. Preparation of the neutrally charged UNCPs: 2 mg of PEG-b-PCL and 0.5 mg of the UCNPs were mixed with 2 mL of tetrahydrofuran (THF), followed by the addition of 2 mL of H2O. The solution was then transferred to a bag and purified using dialysis (cut-off MW = 3500). Subsequently, the as-prepared micelles were purified via centrifugation to remove the excess free polymer, then filtered with a 0.22 mm filter.
2.2. Characterization of the UCNPs
Transmission electron microscopy (TEM) images were recorded on a JEOL-2010 microscope operated at an acceleration voltage of 200 kV. The TEM samples were prepared by dropping the sample solutions (5 mg mL−1 in water) onto a 300-mesh carbon coated copper grid (3 mm in diameter) followed by evaporation of the solvent. The Fourier transform infrared (FTIR) spectra of the bare UCNPs and UCNPs coated with PAA and PAAm were recorded on a Bruker Vertex 70 FTIR spectrometer. The up-conversion luminescence spectra were recorded on a Hitachi F-4500 fluorescence spectrophotometer equipped with a commercial CW NIR laser (980 nm). The hydrodynamic diameter and zeta potential of the UCNPs were determined using a Malvern Zetasizer Nano ZS system.2.3. Confocal imaging
Cells were cultured in standard RPMI-1640 medium with a supplement of 10% fetal bovine serum (FBS), 1% penicillin and 1% streptomycin, and incubated in a humidified atmosphere at 37 °C regulated with 5% CO2. The UCNPs were sterilized in an autoclave at 121 °C for 30 min before being added to the cell culture. Cell uptake experiments were performed by incubating the cell lines for 24 h with the various UCNPs at 37 °C and washed with PBS three times. Cells were fixed using 4% paraformaldehyde and stained with DAPI (0.1 mg mL−1) for 10 min. The samples were washed three times with PBS, followed by observation with a Nikon confocal microscope. For investigation of the intracellular distribution dynamics, the cells were incubated with the various UCNPs at 37 °C and washed three times with PBS. We used 300 nM MitoTracker Green for 40 min to stain the mitochondrion, 500 nM LysoTracker Red for 1 h to stain the lysosome at 37 °C following by gently washing three times with PBS. Up-conversion fluorescence and subcellular organelle imaging were then performed using a Nikon confocal microscope. For the JC-1 assay, the cells were incubated with the positively charged UCNPs for 24 h and washed thrice with PBS. The cells were then stained using JC-1, washed and used for imaging on a Nikon confocal microscope. The fluorescence of the J-monomer and J-aggregate can be detected simultaneously under excitation at 488 nm.2.4. Investigation of cellular uptake and cytotoxicity
Mouse microglia (BV-2) cells, human umbilical vein endothelial (HUVEC) cells, human alveolar adenocarcinoma (A549) cells and cholangiocarcinoma (QBC939) cells were obtained from the cell bank of the college of basic medical sciences, Jilin University. Cells growing in the log phase were seeded into a 96-well cell-culture plate and then incubated for 24 h at 37 °C under 5% CO2. RPMI 1640 solutions of the UCNPs (100 mL per well) at concentrations of 60 mg mL−1 were added to the wells of the treatment group. The cells were incubated for 24 h at 37 °C under 5% CO2. Subsequently, an evaluation of the cytotoxicity of the UCNPs was performed using a standard MTT assay method to the researched dependence between cytotoxicity and subcellular localization of the researched UCNPs and their biocompatibility. Briefly, the culture medium was removed and the cultures were washed twice with PBS. 100 μL of cell culture medium and 10 μL of the MTT reagent were added to each well and incubated at 37 °C for 4 h. When the purple precipitate of the formazan crystals was clearly visible under a microscope, 150 μL of dimethyl sulfoxide (DMSO) was added to each well and incubated in the dark for 2 h at room temperature until the purple crystals were dissolved completely. The absorbance was measured on a microplate reader (Bio-Rad) at 490 nm. The cell viability was expressed as a percentage of the control. The results are reported as the average ± SD of three samples.3. Results and discussion
3.1. Synthesis and characterization of the polymer-coated UCNPs
The original oleic acid-coated UCNPs were synthesized according to a literature procedure.34 The surface modification strategy of the UCNPs comprised of NaYF4 co-doped with 2% Er3+ and 20% Yb3+ ions was then executed as illustrated in Fig. 1. To change the surface charge properties of the UCNPs, firstly the bare UCNPs were obtained by removing the oleic acid on the UCNPs surface using a solution of HCl. Then, the UCNPs were functionalized with poly(acrylic acid) (PAA) and poly(allylamine) (PAAm), endowing the UCNPs with a negatively charged UCNPs(−) and positively charged UCNPs(+), respectively. Besides, the UCNPs were modified with poly(ethylene glycol)-block-poly(caprolactone) (PEG-b-PCL) to endow the UCNPs with neutrally charged UCNPs(0). Fig. 2 shows the TEM images of the bare UCNPs and UCNPs modified with the different polymers. The bare UCNPs exhibit monodispersion with an average diameter of 21 ± 3 nm, as can be seen in Fig. 2a.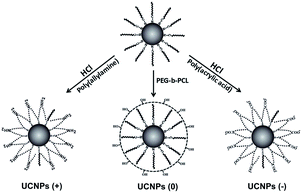 | ||
| Fig. 1 Surface modification of the original oleic acid-coated UCNPs with positive, neutral and negative charges, respectively. | ||
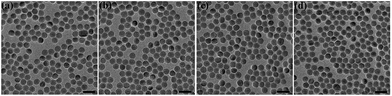 | ||
| Fig. 2 TEM characterization of the NaYF4: Yb3+, Er3+ UCNPs with a series of surface coatings. (a) Ligand-free UCNPs, (b) UCNP-PEG, (c) UCNP-PAA and (d) UCNP-PAAm. The scale bar is 50 nm. | ||
When compared with the bare UCNPs, no changes were observed in the diameter, monodispersity and morphology of the UCNPs with positive, negative and neutral charges, as shown in Fig. 2b–d, respectively. As can be seen in Fig. S1,† the up-conversion luminescence spectra of the UCNPs with different ligands remain similar, indicating that the surface coatings have a negligible influence on the up-conversion mechanism of the UCNPs. The up-conversion luminescence of the UCNPs can be split into two segments, a green region around 540 nm and a red region around 650 nm, as indicated in the inserted picture.
Fig. S2† reveals FTIR spectra of the nanoparticles. The band at 1724 cm−1 was attributed to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration mode of carboxylate anions, which are considered as evidence for the polyacrylic acid coating around the UCNPs. The band at 1565 cm−1 was assigned to the characteristic N–H bending vibrations, affirming the presence of PAAm around the UCNPs, indicating that the UCNPs were successfully modified with the polymer molecules, respectively. Fig. S3† reports on the hydrodynamic diameter of the UCNPs coated with the different ligands. When compared with the results obtained from the TEM analysis, the hydrodynamic diameters of the UCNPs show a marked increase, ca. 37 nm for the bare UCNPs, 78 nm for the UCNPs with PAAm, 106 nm for the UCNPs with PAA and 141 nm for the UCNPs with PEG-b-PCL, respectively. The results are in agreement with the chain length differences among the ligand materials. It was well known that macropinocytosis is an actin-based process that allows the large particles to enter the cells via large vesicles over 1 μm.29 Therefore, these three UCNPs modified with PAA, PAAm and PEG-b-PCL, respectively, can be easily internalized and delivered into the cells. It should be noted that the full width at half maximum (FWHM) of the distribution of the mean hydrodynamic diameter of the three kinds of UCNPs are almost the same as that observed for the bare UCNPs, ca. 28.59 nm, indicating that there was no aggregation after surface modification. The properties of these UCNPs with different surface charges are very important for its internalization in cells. The surface charges of the UCNPs were assessed using Zeta-potential analysis (Fig. S4†), where the negative, neutral and positively charges were confirmed for the UCNPs modified with PAA [UCNPs(−)], UCNPs modified with PEG-b-PCl [UCNPs(0)] and UCNPs modified with PAAm [UCNPs(+)], respectively. The dramatic differences in the Zeta-potentials also imply the successful surface functionalization of the UCNPs. Due to the modification with water-soluble and biocompatible ligand molecules, the UCNPs show excellent stability (over several months) without any agglomeration in deionized water, which is beneficial for their biomedical applications.
O stretching vibration mode of carboxylate anions, which are considered as evidence for the polyacrylic acid coating around the UCNPs. The band at 1565 cm−1 was assigned to the characteristic N–H bending vibrations, affirming the presence of PAAm around the UCNPs, indicating that the UCNPs were successfully modified with the polymer molecules, respectively. Fig. S3† reports on the hydrodynamic diameter of the UCNPs coated with the different ligands. When compared with the results obtained from the TEM analysis, the hydrodynamic diameters of the UCNPs show a marked increase, ca. 37 nm for the bare UCNPs, 78 nm for the UCNPs with PAAm, 106 nm for the UCNPs with PAA and 141 nm for the UCNPs with PEG-b-PCL, respectively. The results are in agreement with the chain length differences among the ligand materials. It was well known that macropinocytosis is an actin-based process that allows the large particles to enter the cells via large vesicles over 1 μm.29 Therefore, these three UCNPs modified with PAA, PAAm and PEG-b-PCL, respectively, can be easily internalized and delivered into the cells. It should be noted that the full width at half maximum (FWHM) of the distribution of the mean hydrodynamic diameter of the three kinds of UCNPs are almost the same as that observed for the bare UCNPs, ca. 28.59 nm, indicating that there was no aggregation after surface modification. The properties of these UCNPs with different surface charges are very important for its internalization in cells. The surface charges of the UCNPs were assessed using Zeta-potential analysis (Fig. S4†), where the negative, neutral and positively charges were confirmed for the UCNPs modified with PAA [UCNPs(−)], UCNPs modified with PEG-b-PCl [UCNPs(0)] and UCNPs modified with PAAm [UCNPs(+)], respectively. The dramatic differences in the Zeta-potentials also imply the successful surface functionalization of the UCNPs. Due to the modification with water-soluble and biocompatible ligand molecules, the UCNPs show excellent stability (over several months) without any agglomeration in deionized water, which is beneficial for their biomedical applications.
3.2. Surface-charge-dependent cell internalization
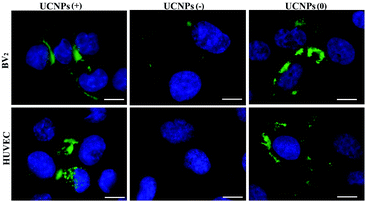
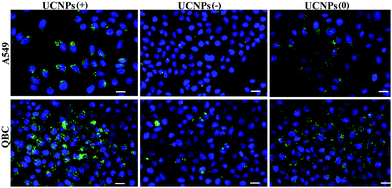
Confocal microscopy was employed to study the cellular uptake and intracellular localization of the polymer-coated UCNPs, where the UCNPs(−), UCNPs(+) and UCNPs(0) (1.0 mM) were incubated with two transformed carcinoma cell lines (A549 and QBC939) and two normal cell lines (BV-2 and HUVEC), respectively. We started with the internalization pattern in the two normal cells (Fig. 3). After incubation for 24 h, it was shown that the negatively charged carboxylated nanoparticles UCNPs(−) were minimally up-taken by the BV-2 cells and were not uptaken in the HUVEC cell line. However, both the positively charged UCNPs(+) and neutrally charged UCNPs(0) were up-taken by both cell lines, respectively. The intracellular fluorescence pattern of the UCNPs(0) was slightly diffused and a slightly further away from the cell nucleus, being different from the punctuated fluorescence pattern of UCNPs(+), which was mainly located around the cell nucleus. This difference seems to indicate a unique nanoparticle surface-charge dependent internalization mechanism with different intracellular compartmentalization among the nanoparticles and the two normal cell lines.Internalization experiments were also performed with two cancer cells (A549 and QBC939), which revealed that very few of the UCNPs(−) were only up-taken by the QBC939 cells but hardly by the A549 cells (Fig. 4). However, both cell lines were able to uptake the UCNPs(+). A higher degree of internalization and pronounced fluorescence of the UCNPs(+) in the QBC939 cells. In contrast, the diffused intracellular fluorescence images of the UCNPs(0) in both the A549 and QBC939 cells were observed, as shown in Fig. 4. Taken together, the above results suggest that the surface charge properties of the polymer-coated UCNPs dictate their differential internalization and localization in normal and cancer cells (Fig. S3 and S4,† respectively).
3.3. The intracellular distribution dynamics of the polymeric UCNPs
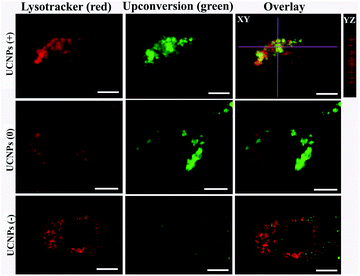
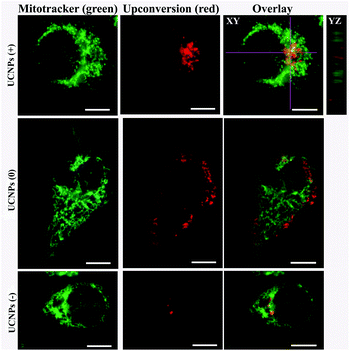
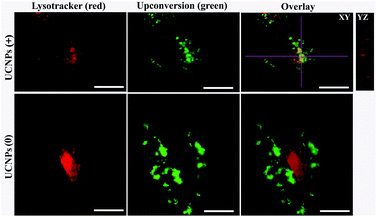
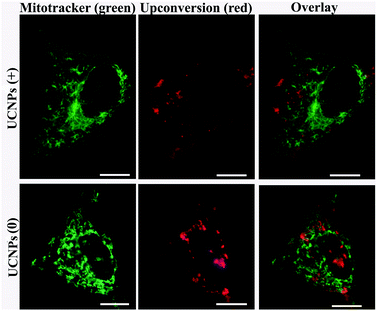
In order to corroborate that after internalization some of the polymer-coated UCNPs localized in the sub-cellular compartments, we treated the cells for 24 h with the UCNPs(−), UCNPs(+) and UCNPs(0) followed by treatment with mitotracker (a mitochondrion-specific dye) and lysotracker (a lysosome-specific dye), respectively. Mitotracker is a green fluorescent dye that specifically stains the mitochondria. Hence, the potential co-localization between the UCNPs (red) and mitotracker (green in mitochondria) should yield a yellow/orange overlap when the fluorescence images are merged. Similarly, lysotracker is a red fluorescent dye, which stains the lysosomes, a yellow/orange overlap of the fluorescence images should be observed when the UCNPs are localized with the lysotracker in the lysosomes. For the QBC939 cancer cells, as can be seen from Fig. 5, the results showed that the UCNPs(+) present a strong green punctuated luminescence pattern and are predominantly co-localized with lysotracker in the cells, as determined by the strong yellow color. In contrast, the UCNPs(0) showed a diffused distribution without localization in the lysosome of these cells. Little internalization and lysosome co-localization of the UCNPs(−) was observed in the QBC cells. For the mitochondrion, strong aggregation and a punctuated red luminescence pattern of the UCNPs(+), few small luminescence dot pattern of the UCNPs(−) and strong diffused luminescence pattern of the UCNPs(0) were observed in the QBC939 cells, respectively. Interestingly, the red signals originate from the UCNPs(+) partly co-localized with the green mitotracker dye giving a strong yellow color. However, the up-conversion emission of the UCNPs(0) and UCNPs(−) have negligible overlap with the mitochondrion (Fig. 6). These results indicate that the internalized UCNPs(+) are located in both the lysosomes and mitochondria in the QBC939 cancer cells.We further studied the internalization and organelle localization process of the UCNPs(+). The UCNPs(+) particles were initially internalized at 2 h (Fig. 7a and b). After continuous incubation for 24 h, more UCNPs(+) particles entered the cell and located in the subcellular structures including the cytoplasm, lysosomes and mitochondria (Fig. 7a–h). The amount and distribution of the internalized UCNPs(+) particles varied with time. The UCNPs(+) particles gradually trafficked to the lysosomes (at 2, 8 and 12 h, Fig. 7a, c and e). Within 24 h, most of the UCNPs(+) particles gradually entered and filled the lysosomes, as evidenced by the stronger and stronger yellow color (Fig. 7g). The lysosomes act as the waste disposal system of the cell by digesting unwanted materials in the cytoplasm, both from outside of the cell and obsolete components inside the cell. Therefore, when the UCNPs entered the cells, they were preferentially up-taken by the lysosomes. Whereas, at 8 h, the red emission of the UCNPs(+) presented a diffused pattern and have no obvious overlap with green mitotracker (Fig. 7b and d). At 12 h, some of UCNPs(+) particles appeared in the mitochondria, where they began to reside in a punctuated luminescence pattern (Fig. 7f). The number of UCNPs(+) particles in the mitochondria increased gradually. At 24 h, the UCNPs(+) particles exhibited an aggregated state and a strong yellow color was clearly seen in the mitochondria, suggesting that the UCNPs(+) particles were in the mitochondria (Fig. 7h). Based on these results it can be speculated that the UCNPs(+) particles in the QBC939 cells trafficked from the lysosomes to the cytoplasm and then to the mitochondria as internalization proceeded.
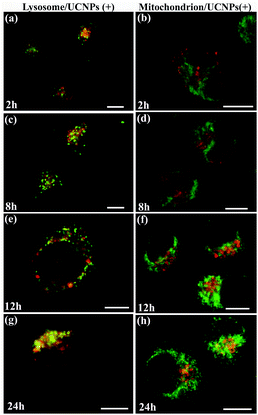 | ||
| Fig. 7 The time-dependent internalization and subcellular co-localization of UCNPs(+) in the lysosome and mitochondrion of QBC939 cells. | ||
In the case of normal HUVEC cells (Fig. 8), the distribution of UCNPs(+) inside the cell co-localized partly with the lysotracker marker. Meanwhile, when the UCNPs(0) were used, a diffused distribution of the internalized nanoparticles was observed with minimal co-localization with the lysosome. As far as the mitochondrion was concerned (Fig. 9), the up-taken positive and neutral charged UCNPs in the HUVEC cells did not show co-localization with the mitochondria. These results demonstrate that in HUVEC cells, the UCNPs(+) partly localized in the lysosome, while neutral UCNPs(0) primarily localized in the cytosol. The subcellular distribution of the differently charged UCNPs in the A549 and BV-2 cell lines were also investigated as shown in Fig. S5–S8,† which present very similar localization patterns with those observed for the HUVEC cell line. In both the A549 and BV-2 cell lines, the UCNPs(+) partly localized in the lysosome but hardly co-localized with the mitochondria. The fluorescence pattern of the UCNPs (0) occurs in the cell cytoplasm and do not overlap with either the lysosome or with the mitochondria. Mitochondrial and lysosomal co-localization experiments were not performed in the HUVEC and BV-2 cell lines for the UCNPs(−) due to the fact that the UCNPs(−) were not up-taken by both cells lines. All these results led to the conclusion that the surface charge of the UCNPs dictates the nanoparticles' subcellular localization in cancer and normal cells.
3.4. The intracellular-distribution-dependent cytotoxicity of the UCNPs with different charges
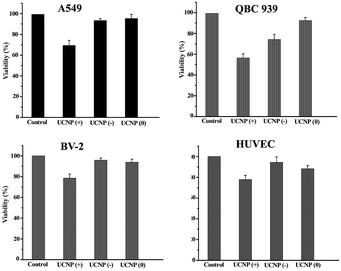
In order to answer how the surface-charge-dependent internalization and intracellular (mitochondria vs. lysosomal or cytoplasmic) localization of the polymeric UCNPs affects the nanoparticle's cytotoxicity, cell viability (MTT) assays were performed. Interestingly, the UCNPs(+) exhibited various degrees of toxicity to the QBC939, A549, BV-2 and HUVEC cell lines studied (Fig. 10). Notably, the UCNPs(+) were more toxic to the QBC939 cell line when compared to the others, since only QBC939 exhibited mitochondria co-localization. Interestingly, although both the A549 and QBC939 cells belong to cancer cell lines, the UCNPs(+) demonstrated a negligible cytotoxicity to the A549 cells when compared to the QBC939 cell line. The difference in cytotoxicity of the UCNPs(+) between QBC939 cells and A549 cells was possibly derived from the difference in the nature between the two cell lines, for example, cholangiocarcinoma is a kind of higher malignant cancer that is slowly growing but highly metastasizing than lung cancer. From the intracellular localization pattern of the UCNPs(+) in the A549, BV-2 and HUVEC cell lines, it can be seen that some UCNPs(+) co-localized with the lysosomes, whereas hardly co-localized with mitochondria. Therefore, the UCNPs(+) exhibited little or slight cytotoxicity to these cells, which testify that the cytotoxicity of the UCNPs(+) depends more whether they are localized in the mitochondria rather than in the lysosomes. UCNPs(0) did not exhibit any toxicity to the QBC939, A549, BV-2 and HUVEC cell lines studied, despite most of these nanoparticles are localized in the cytoplasm. UCNPs(−) had different degrees of toxicity, depending on the nanoparticle's intracellular localization. For instance, the UCNPs(−) nanoparticles were not toxic to the HUVEC, BV-2 or A549 cell lines, whereas they present a little toxicity to the QBC939 cancer cells. This can be rationalized by the minimal uptake of UCNPs(−) by normal cells, as opposed to the QBC939 cancer cells, which exhibited enhanced nanoparticle uptake.The ICP analysis of Y3+ concentrations in the studied cell lines treated with the different charged UCNPs (shown in Table S1†) was consistent with the toxicity results. The main conclusions from the study are, (i) the UCNPs(+) are much more toxic to all the cell lines studied than the UCNPs(−) or UCNPs(0); (ii) the QBC939 cancer cells are the most sensitive to the surface charge of the nanoparticles among all the cell lines studied; (iii) the localization of the UCNPs(+) in mitochondria, as opposed to the cytoplasm or lysosome, leads to much greater cytotoxicity.
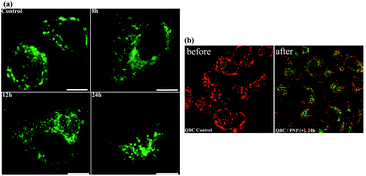
It is well-known that the mitochondria take part in oxidative phosphorylation and function as the energy factory of cells. Mitochondria in the cell can form a highly interconnected dynamic tubular network and undergo frequent fusion and fission mediated by fusion–fission proteins, which maintains the mitochondrial morphology and function. Once apoptosis perturbing the balance is triggered, the mitochondrial tubular network begins to fragment. These changes are essential for the downstream events of apoptosis including cytochrome c release and the caspase cascade. The fusion–fission proteins influence the course of apoptosis via controlling the mitochondrial morphology and ultrastructure.35 Based on this, an examination was performed on the mitochondria morphology using confocal microscopy to check whether mitochondrial fission and fusion play a role in the regulation of the response to UCNPs(+). As illustrated in Fig. 11a, mitochondria in the cell exhibited fragmented and short tubular shapes without nanoparticles. After incubation with the UCNPs(+) for 8 h, the mitochondria elongated to form a thinner and longer tubule network, and obviously long tubule-like mitochondria were observed in most of the cells. However, in the cells cultured with the UCNPs(+) for 12 h, some of the mitochondria were reversed to small punctate forms and almost all of the mitochondria became dots after 24 h of treatment. The mitochondrion-specific dye JC-1 was used to detect changes in the mitochondrial membrane potential. Apoptotic cells mainly show green fluorescence, while healthy cells mainly show red fluorescence, making JC-1 suitable for the detection of mitochondrial damage.36 After 24 h of exposure of the cells to the UCNPs(+), the JC-1 results show that the UCNPs(+) induce mitochondrial damage in the QBC939 cells. The QBC939 cells show a decreased ratio of red to green fluorescence (Fig. 11b) and exhibit impaired mitochondrial membranes that may disrupt cellular metabolism, release reactive oxygen species from the mitochondria and even cause apoptosis. Thus, the UCNPs(+) in the QBC939 cells have indeed significantly influenced cellular metabolism via the mitochondrial pathway. We can, from these observations, come to the conclusion that an intracellular migration process of the UCNPs(+) occurred in which they migrated to the lysosomes and subsequently moved to the mitochondria. Their mitochondrial storage impairs the mitochondrial structures and disrupts the production of ATP energy molecules, and thus, the main reason for their cellular toxicity.
4. Conclusions
In summary, NaYF4: Yb3+, Er3+ UCNPs with different surface charge properties have been studied to understand their internalization, subcellular localization and cytotoxicity profile employing two cancer cell lines and two normal cell lines. It can be concluded that the internalization and subcellular localization of UCNPs play a key role in their cytotoxicity profile, exhibiting significant toxicity when they are localized in the mitochondria of cancer cells. Whereas minimal toxicity was observed when they are localized into the cytoplasm. Such a subcellular migration and distribution study enriches our understanding on how up-conversion nanoparticles interact with cells and the origin of their cytotoxicity, which sheds new light on the design of novel up-conversion nanoparticles for biomedical applications. In addition, the UCNPs(+) exhibit a passive targeting to the mitochondrion of QBC939 cells and may serve as an efficient drug-carrier for the treatment of cholangiocarcinoma.Acknowledgements
This work was financially supported by the NSF of China (61575194, 11474278, 11674316, 11604331 and 11374297) and the Project of Science and Technology Agency, Jilin Province (20170520113JH, 20170520112JH and 20170519002JH).Notes and references
- E. K. Lim, T. Kim, S. Paik, S. Haam, Y. M. Huh and K. Lee, Chem. Rev., 2015, 115, 327 CrossRef CAS PubMed.
- D. E. Lee, H. Koo, I. C. Sun, J. H. Ryu, K. Kim and I. C. Kwon, Chem. Soc. Rev., 2012, 41, 2656 RSC.
- W. Dong, Y. Li, D. Niu, Z. Ma, J. Gu, Y. Chen, W. Zhao, X. Liu, C. Liu and J. Shi, Adv. Mater., 2011, 23, 5392 CrossRef CAS PubMed.
- Z. Liu, X. Sun, N. Nakayama-Ratchford and H. Dai, ACS Nano, 2007, 1, 50 CrossRef CAS PubMed.
- A. A. Bhirde, V. Patel, J. Gavard, G. Zhang, A. A. Sousa, A. Masedunskas, R. D. Leapman, R. Weigert, J. S. Gutkind and J. F. Rusling, ACS Nano, 2009, 3, 307 CrossRef CAS PubMed.
- R. S. Chang, M. S. Suh, S. Kim, G. Shim, S. Lee, S. S. Han, K. E. Lee, H. Jeon, H. G. Choi, Y. Choi, C. W. Kim and Y. K. Oh, Biomaterials, 2011, 32, 9785 CrossRef CAS PubMed.
- J. Lu, M. Liong, Z. Li, J. I. Zink and F. Tamanoi, Small, 2010, 6, 1794 CrossRef CAS PubMed.
- J. M. Rosenholm, A. Meinander, E. Peuhu, R. Niemi, J. E. Eriksson, C. Sahlgren and M. Linden, ACS Nano, 2009, 3, 197 CrossRef CAS PubMed.
- H. Lee, E. Lee, D. K. Kim, N. K. Jang, Y. Y. Jeong and S. Jon, J. Am. Chem. Soc., 2006, 128, 7383 CrossRef CAS PubMed.
- N. Lee, H. Kim, S. H. Choi, M. Park, D. Kim, H. C. Kim, Y. Choi, S. Lin, B. H. Kim, H. S. Jung, H. Ki, K. S. Park, W. K. Moon and T. Hyeon, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 2662 CrossRef CAS PubMed.
- J. Huang, L. Bu, J. Xie, K. Chen, Z. Cheng, X. Li and X. Chen, ACS Nano, 2010, 4, 7151 CrossRef CAS PubMed.
- S. Y. Hong, G. Tobias, K. T. Al-Jamal, B. Ballesteros, H. Ali-Boucetta, S. Lozano-Perez, P. D. Nellist, R. B. Sim, C. Finucane, S. J. Mather, M. L. H. Green, K. Kostarelos and B. G. Davis, Nat. Mater., 2010, 9, 485 CrossRef CAS PubMed.
- X. J. Liang, H. Meng, Y. Wang, H. He, J. Meng, J. Lu, P. C. Wang, Y. Zhao, X. Gao, B. Sun, C. Chen, G. Xing, D. Shen, M. M. Gottesman, Y. Wu, J. J. Yin and L. Jia, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 7449 CrossRef CAS PubMed.
- J. T. Robinson, K. Welsher, S. M. Tabakman, S. P. Sherlock, H. Wang, R. Luong and a. H. Dai, Nano Res., 2010, 3, 779 CrossRef CAS PubMed.
- D. Kim, Y. Y. Jeong and S. Jon, ACS Nano, 2010, 4, 3689 CrossRef CAS PubMed.
- I. C. Sun, D. K. Eun, H. Koo, C. Y. Ko, H. S. Kim, D. K. Yi, K. Choi, I. C. Kwon, K. Kim and C. H. Ahn, Angew. Chem., Int. Ed., 2011, 50, 9348 CrossRef CAS PubMed.
- S. Lee, E. J. Cha, K. Park, S. Y. Lee, J. K. Hong, I. C. Sun, S. Y. Kim, K. Choi, I. C. Kwon, K. Kim and C. H. Ahn, Angew. Chem., Int. Ed., 2008, 47, 2804 CrossRef CAS PubMed.
- Y. I. Park, K. T. Lee, Y. D. Suh and T. Hyeon, Chem. Soc. Rev., 2015, 44, 1302 RSC.
- G. Chen, H. Qiu, P. N. Prasad and X. Chen, Chem. Rev., 2014, 114, 5161 CrossRef CAS PubMed.
- W. Fan, W. Bu and J. Shi, Adv. Mater., 2016, 28, 3987 CrossRef CAS PubMed.
- N. M. Idris, M. K. G. Jayakumar, A. Bansal and Y. Zhang, Chem. Soc. Rev., 2015, 44, 1449 RSC.
- K. Liu, X. M. Liu, Q. H. Zeng, Y. L. Zhang, X. G. Kong, Y. H. Wang, F. Cao, S. A. G. Lambrechts, M. C. G. Aalders and H. Zhang, ACS Nano, 2012, 6, 4054 CrossRef CAS PubMed.
- F. Wang and X. G. Liu, Acc. Chem. Res., 2014, 47, 1378 CrossRef CAS PubMed.
- C. Yao, P. Y. Wang, X. M. Li, X. Y. Hu, L. Y. Wang and F. Zhang, Adv. Mater., 2016, 28, 9341 CrossRef CAS PubMed.
- L. Prodi, E. Rampazzo, F. Rastrelli, A. Speghinic and N. Zaccheronia, Chem. Soc. Rev., 2015, 44, 4922 RSC.
- J. Zhou, Q. Liu, W. Feng, Y. Sun and F. Y. Li, Chem. Rev., 2015, 115, 395 CrossRef CAS PubMed.
- J. G. Huang, T. Leshuk and F. X. Gua, Nano Today, 2011, 6, 478 CrossRef CAS.
- S. L. Gai, C. X. Li, P. P. Yang and J. Lin, Chem. Rev., 2014, 114, 2343 CrossRef CAS PubMed.
- J. Nam, N. Won, J. Bang, H. Jin, J. H. Park, S. W. Jung, S. H. Jung, Y. G. Park and S. J. Kim, Adv. Drug Delivery Rev., 2013, 65, 622 CrossRef CAS PubMed.
- N. Kamaly and O. C. Farokhzad, Nat. Rev. Mater., 2016, 12, 738 CAS.
- M. K. Tsang, C. F. Chan, K. L. Wong and J. H. Hao, J. Lumin., 2015, 157, 172 CrossRef CAS.
- C. F. Chan, M. K. Tsang, H. G. Li, R. F. Lan, F. L. Chadbourne, W. L. Chan, G. L. Law, S. L. Cobb, J. H. Hao, W. T. Wong and K. L. Wong, J. Mater. Chem. B, 2014, 2, 44 RSC.
- Y. Gu, X. Qiao, J. Zhang, Y. Y. Sun, Y. M. Tao and S. X. Qiao, Chem. Res. Chin. Univ., 2017, 32(3), 474 CrossRef.
- Y. Wang, K. Liu, X. M. Liu, K. Dohnalov, T. Gregorkiewicz, X. G. Kong, M. C. G. Aalders, W. J. Buma and H. Zhang, J. Phys. Chem. Lett., 2011, 2, 2083 CrossRef CAS.
- Z. Q. Fan, H. M. Yu, N. Cui, X. G. Kong, X. M. Liu, Y. L. Chang, Y. Wu, L. K. Sun and G. Y. Wang, Exp. Cell Res., 2015, 335, 68 CrossRef CAS PubMed.
- L. M. Wang, Y. Liu, W. Li, X. M. Jiang, Y. L. Ji, X. C. Wu, L. G. Xu, Y. Qiu, K. Zhao, T. T. Wei, Y. F. Li, Y. L. Zhao and C. Y. Chen, Nano Lett., 2011, 11, 772 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra04487a |
| This journal is © The Royal Society of Chemistry 2017 |