Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Possible target-related proteins of stress-resistant rats suggested by label-free proteomic analysis
Jiao Chena,
Zhenzhen Wanga,
Shuai Zhanga,
Qidi Aib,
Shifeng Chua and
Nai-hong Chen *abc
*abc
aState Key Laboratory of Bioactive Substances and Functions of Natural Medicines, Institute of Materia Medica & Neuroscience Center, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100050, China. E-mail: chennh@imm.ac.cn; Fax: +86 1063165177; Tel: +86 10 63165177
bHunan University of Chinese Medicine, Changsha, Hunan 410208, China
cGuangzhou University of Chinese Medicine, Guangzhou 510000, China
First published on 22nd August 2017
Abstract
Stress plays a crucial role in the development of major depressive disorder, but the molecular mechanism underlying the susceptibility vs. resilience to stress remains unclear. To better understand these mechanisms, we used chronic unpredictable mild stress to develop a depressive rat model. We categorized them into stress resistant rats and stress sensitive rats by their performance in behavioral tests, including forced swim test and sucrose preference test. Brain regions were dissected, and prefrontal cortex (PFC) proteins extracted from stress resistant and stress sensitive rats were analyzed using label-free liquid chromatography-tandem mass spectrometry. Three hundred and four proteins were up-regulated and 323 proteins were down-regulated among the 1482 different proteins from stress resistant rats compared with that of the stress sensitive rats. Western blotting, immunohistochemical staining and electron microscopy were used to validate the results of the proteomic analysis. Some proteins differentially expressed in stress resistant and sensitive rats were found to be associated with several neurobiological processes, particularly with neurotransmission regulation. The results provide possible novel insights into the molecular mechanisms for stress resilience.
Introduction
Major depression disorder is a disabling disease that greatly influences public health. Both genetic and environmental factors, such as stress, contribute to the development of depression.1 However, some people are resilient to stress, even when they are exposed to extraordinary levels of stress and trauma.2,3 Resilience refers to the capacity of an individual to avoid negative social, psychological and biological consequences of extreme stress and maintain normal psychological and physical functioning even in stressful conditions.4 To find new therapeutic targets for depression, it is important to understand the neural and molecular mechanisms that mediate stress resilience.A large amount of literatures have implicated prefrontal cortex (PFC) in the pathophysiology of depression.5 Chronic stress can lead to dendritic atrophy of PFC neurons6 and PFC volume atrophy in MDD patients.7 Moreover, our previous study has demonstrated that gap junction dysfunction in PFC leads to depression.8 In addition, optogenetic activation of mPFC (medial prefrontal cortex) has an antidepressant-like effect in mice that are exposed to chronic social defeat.9 All this evidence indicates that PFC plays an important role in regulating stress susceptibility. However, there was no systematic research to determine molecular mechanisms of the PFC involved in stress resilient vs. vulnerability. To further accurately explore the molecular mechanisms associated with stress resilience, a quantitative proteomic analysis of PFC proteins from stress-resistant and stress-sensitive rats was performed using a label-free liquid chromatography-mass spectrometry (LC-MS) approach. Furthermore, western blotting, immunohistochemical staining and electron microscopy were used to validate the results of the proteomic analysis.
Materials and methods
Animals
Male Sprague-Dawley rats (Vital River Laboratories, Beijing, China) were housed under a 12 h light/12 h dark cycle at constant temperature (22 °C) with free access to food and water except when the animals were subjected to the stressors. Furthermore, 40 rats, weighing between 230 and 250 g at the beginning of the experiment were investigated with the test of chronic unpredictable mild stress. All experiments were performed in accordance with the guidelines established by the National Institutes of Health for the care and use of laboratory animals and were approved by the Animal Care Committee of the Peking Union Medical College and Chinese Academy of Medical Sciences.Chronic unpredictable mild stress (CUMS)
CUMS was used to develop the depressive rat model and this regimen consisted of once or twice daily exposure to different stressors including food deprivation, water deprivation, crowding, isolation, soiled cage, 4 h immobilization, 1 h shaker stress (160 rpm), 1 min tail pinch, cage tilt 45 degree overnight, reversal of the light/dark cycle, overnight light and swim stress at 4 °C. Control animals were housed under standard conditions, while the stressed rats were subjected to the CUMS for 8 weeks and then behavioral tests were used to test the depressive like behaviors (Table 1).| Day | Stressors |
|---|---|
| Monday | Restraint 4 h; 5 min hot water swim at 45 °C |
| Tuesday | Food deprivation 24 h; soiled cage |
| Wednesday | Isolation 24 h (one rat per cage); light on overlight |
| Thursday | Isolation 24 h; reversal of the light/dark cycle (light off 12 h and light on overnight) |
| Friday | Tail pinch 1 min; crowding overnight (8 rats per cage) |
| Saturday | Shaker stress 1 h (160 rpm); cage tilt 45 degree overnight |
| Sunday | 5 min swim stress at 4 °C; 24 h water deprivation |
Behavior tests
Sucrose preference test (SPT) was conducted after CUMS. Briefly, rats were habituated to 1% sucrose solution for 48 h, followed by a 4 h of water deprivation and 1 h exposure to two identical bottles filled with either sucrose solution or water. Sucrose preference was defined as the ratio of the volume of sucrose vs. total volume of sucrose and water consumed during the 1 h test.For the forced swim test (FST), rats were placed in a plastic cylinder (40 cm deep, 20 cm in diameter) filled with water at 23–25 °C up to a height of 25 cm from the base, and forced to swim for 15 min in the first session. After 24 h, the rats were reintroduced into the same cylinder, and their 5 min swim session was recorded. After each swim session, the rats were removed from the cylinder, dried with paper towels, and returned to their home cages. Water in the cylinder was renewed between subjects. The rest time was recorded as the depressive index when the rats floated in water without any struggle.
LC-MS analysis
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300× rpm for 3 min at 4 °C. The supernatant was collected, and the protein concentration was determined using the Qubit fluorescent protein quantification kit (Invitrogen) according to the manufacturer's instructions.
300× rpm for 3 min at 4 °C. The supernatant was collected, and the protein concentration was determined using the Qubit fluorescent protein quantification kit (Invitrogen) according to the manufacturer's instructions.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 trypsin to protein ratio at 37 °C for 12 h. Digested fractions were collected and stored at −80 °C without further treatment until the MS analysis.
50 trypsin to protein ratio at 37 °C for 12 h. Digested fractions were collected and stored at −80 °C without further treatment until the MS analysis.Western blotting
PFC region was dissected and homogenized in lysis buffer. Protein concentrations were determined by bicinchoninic acid protein assay. A total of 30 μg of proteins for each sample was separated by SDS-PAGE and then transferred to PVDF membrane (Millipore, Temecula, CA). The membrane was blocked with 3% BSA and incubated with primary antibody overnight at 4 °C (anti-complexin 2, RD, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000; anti-β-actin, sigma, 1
1000; anti-β-actin, sigma, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), followed by horseradishperoxidase (HRP)-conjugated secondary antibody (1
000), followed by horseradishperoxidase (HRP)-conjugated secondary antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000, KPL, Gaithersburg, MD). The protein bands were detected using enhanced chemiluminescence. Densitometric analysis of immunoreactivity for each protein was conducted using Gel-Pro Analyzer software (Media Cybernetics).
5000, KPL, Gaithersburg, MD). The protein bands were detected using enhanced chemiluminescence. Densitometric analysis of immunoreactivity for each protein was conducted using Gel-Pro Analyzer software (Media Cybernetics).
Electron microscopy (EM)
Processing and EM were conducted as previously described.8 Briefly, anesthetized animals were perfused, and the brains were trimmed to produce sections. They were postfixed with 2.5% glutaraldehyde for 2 h, washed with 0.1 M PBS and then exposed to 1% osmium tetraoxide for 2 h. After washing several times, the tissues were dehydrated with gradient alcohol and embedded in Epon resin. Randomly selected ultrathin sections were stained with uranyl acetate and lead citrate and were observed using a transmission electron microscope (H-7650, HITACHI, Tokyo, Japan).Immunohistochemistry
For immunohistochemistry, the sections were slide-mounted and boiled in citric acid, followed by treatment with 3% H2O2 for 15 min to quench the endogenous peroxidase activity. Sections were incubated in 10% normal donkey serum and 0.3% Triton X-100 for 30 min and then incubated overnight at 4 °C in primary antibody for complexin 2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, rabbit anti-complexin 2, RD). After secondary antibody incubation, sections were developed using Vector ABC kit and DAB kit. Images were taken with an upright microscope (PM20-35, OLYMPUS, Japan).
200, rabbit anti-complexin 2, RD). After secondary antibody incubation, sections were developed using Vector ABC kit and DAB kit. Images were taken with an upright microscope (PM20-35, OLYMPUS, Japan).
Statistical analysis
Data were expressed as the mean ± SEM. Differences among experimental groups were determined by one-way ANOVA. The level of statistical significance was set at p < 0.05. Bioinformatics analysis including the Uniprot knowledgebase and Gene Ontology (GO) database and networks were performed using the STRING 10 and PANTHER databases.Results
Stress resistant and stress sensitive rats were distinguished by their performance in behavioral tests
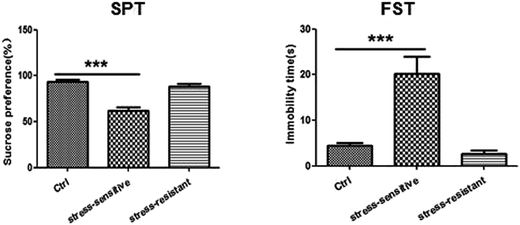
Our experimental workflow is illustrated in Fig. 1. After 8 weeks of stress, the animals were separated into two sub-populations based on their performance in SPT and FST as stress resistant and stress sensitive rats. The sucrose preference test showed that the stress resistant rats had more sucrose water than the stress sensitive rats. There was no significant difference among the groups in total fluid consumption for the 1 h test (data not show). In addition, the forced swim test demonstrated that the stress resistant rats spent less time in an immobile state than the stress sensitive rats (Fig. 2).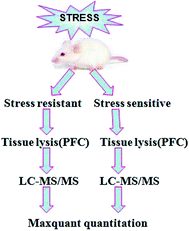 | ||
| Fig. 1 Experimental workflow. The process of the sample preparation, data acquisition, and data analysis is shown in the flowchart. | ||
Proteins identification and their functions in label-free analysis
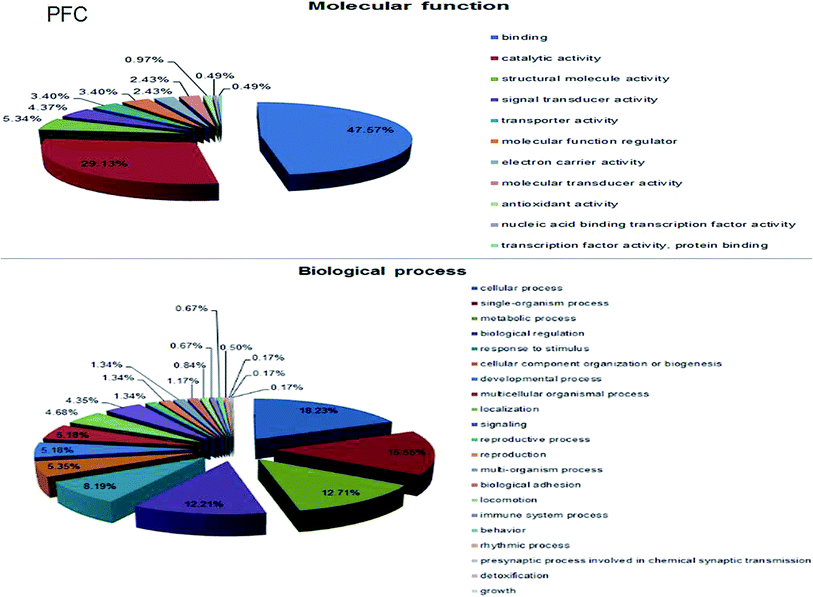
A total of 4160 proteins were identified in the samples from the PFC of the stress resistant and stress sensitive rats. Compared with stress sensitive rats, in stress resistant rats, a total of 1482 proteins showed a significant change in expression, out of which 304 proteins were up-regulated and 323 proteins were down-regulated. In addition, some proteins could not be detected in either stress resistant rats or stress sensitive rats (Fig. 3). To further understand the changes observed in the PFC between the stress resistant rats and stress sensitive rats, differentially expressed proteins were analyzed in terms of two ontology categories – “Molecular Function” and “Biological Process” – based on their annotations in GO domains. The proportional representation of various subcategories in each GO domain, with each value expressed as a percentage of the total, is shown in Fig. 4. Our results indicated that the differentially expressed proteins in stress resistant rats and stress sensitive rats were associated with diverse molecular functions and biological processes. The molecular functions of these proteins included binding, catalytic activity, structural molecule activity, signal transducer activity, transporter activity, molecular function regulator, electron carrier activity, molecular transducer activity, antioxidant activity, nucleic acid binding transcription factor activity and transcription factor activity. The biological processes mainly included the cellular process, single-organism process, metabolic process, biological regulation, response to stimulus, cellular component organization, developmental process, multicellular organismal process, localization and signaling. We also found that the proteins complexin 2, UFD1L, PFDN5, Septin11, ATP8A2, MANF, PCDH10 were involved in the stress resistant phenotype. The STRING knowledge database permits the evaluation of protein–protein interactions. We applied STRING 10 to our list of differentially expressed proteins. The data indicated close connectivity between complexin 2, Ufd1l, Pfdn5, and clasp1 (Fig. 5.). These proteins are associated with synaptic and cytoskeletal regulations. Several types of proteins, such as Manf, Septin11, Pcdh10, Atp8a2, were involved in other processes (Table 2).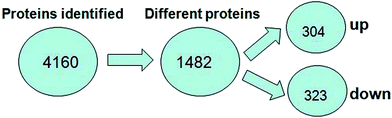 | ||
| Fig. 3 Number of proteins identified in the proteomic analysis. The number of up-regulated and down-regulated proteins in stress resistant rats is shown compared with that of stress sensitive rats. | ||
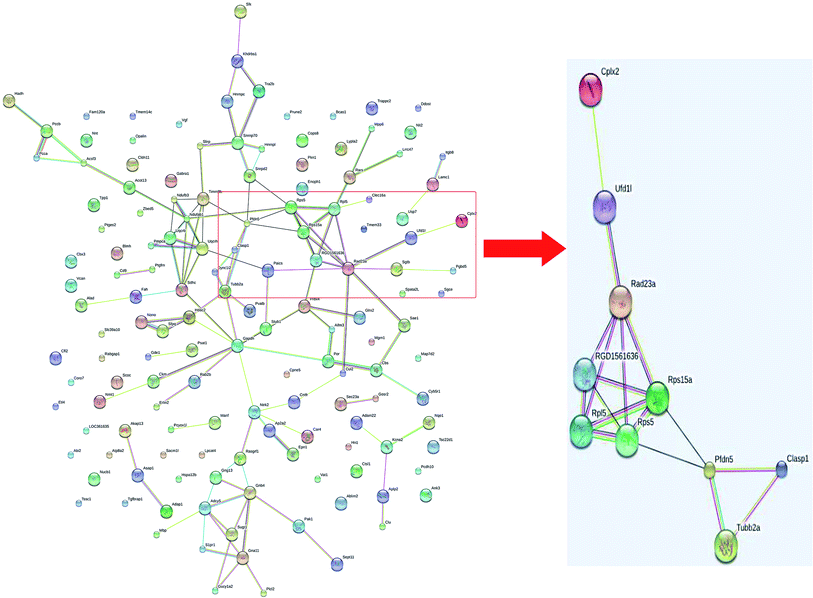 | ||
| Fig. 5 Interactions of the proteins differentially expressed between stress resistant rats and stress sensitive rats. | ||
| No. | Protein names | Gene names | p-Value | Location | Molecular function | Biological process |
|---|---|---|---|---|---|---|
| 1 | Complexin 2 | Cplx2 | <0.05 | Plasma membrane | Syntaxin-1 binding calcium-dependent protein | Vesicle docking involved in exocytosis |
| 2 | Phospholipid-transporting ATPase | Atp8A2 | <0.05 | Plasma membrane | Magnesium ion binding; ATP binding | Metabolic process |
| 3 | Protein Pcdh10 | Pcdh10 | <0.05 | Plasma membrane | Calcium ion binding | Cell adhesion |
| 4 | Mesencephalic astrocyte derived neurotrophy | Manf | <0.05 | Cytoplasm | Growth factor activity | Response to unfolded protein |
| 5 | Ubiquitin fusion degradation protein 1 | Ufd1l | <0.05 | Cytoplasm | K48-linked polyubiquitin binding | Proteasome mediated ubiquitin dependent protein catabolic |
| 6 | Septin-11 | Sept-11 | <0.05 | Cytoplasm | Nucleotide binding | Cell division; cell cycle |
| 7 | Prefoldin-5 | Pfdn5 | <0.05 | Cytoplasm | Unfolded protein binding | Negative regulation of transcription |
Validation of proteomic results
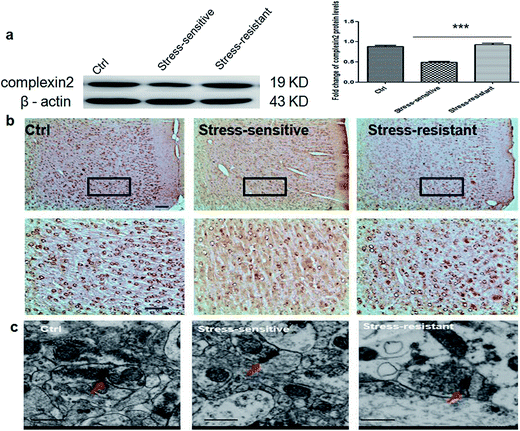
The expression of complexin 2 was validated using western blotting and immunohistochemistry. The result showed that the content of complexin 2 was significantly higher in the stress resistant rats compared to that of the stress sensitive rats. Furthermore, we also observed the presynaptic vesicles by electron microscopy. The result showed that the number of vesicles in the stress resistant rats was higher than that in the stress sensitive rats (Fig. 6).Discussion
In the present study, depressive-like behavioral tests were used to distinguish stress resistant rats and stress sensitive rats after CUMS exposure. The results showed that the stress resistant rats consumed more sucrose water than stress sensitive rats. Moreover, the stress resistant rats spent less time in immobile state than the stress sensitive rats. To search possible molecular mechanisms underlying this stress resistant phenotype, protein expression profiles of PFC from stress resistant and stress sensitive rats were investigated using proteomic analysis. The data showed that a certain amount of proteins exhibited changes in the stress resistant rats compared to that in the stress sensitive rats. In addition, the altered proteins belonged to several functional groups, suggesting that different cellular processes were involved in the stress resilience phenotype.Our data showed that the level of Atp8a2 was significantly higher in the stress resistant rats than in the stress sensitive rats. It was reported that Atp8a2 possesses phosphatidylserine translocase activity and is involved in the localization of phosphatidylserine to the inner leaflet of the plasma membrane.10 Altered phosphatidylserine distribution in neurons could affect the release of synaptic vesicles and transduction of action potentials along nerve fibers. Atp8a2 is specifically expressed in the nervous system. The mutation of the Atp8a2 gene causes axon degeneration and neurodegenerative diseases.11 Disruption of the Atp8a2 gene is associated with a severe neurological phenotype, including mental retardation, hypotonia and axonal degeneration.12 These data indicated that stress resistant symptoms in this animal model was associated with Atp8a2 increase in the PFC.
The level of ubiquitin fusion degradation 1-like (UFD1L) protein was found to have increased in the stress resistant rats. The ubiquitin-recognition protein UFD1L facilitates the clearance of misfolded proteins through the endoplasmic reticulum associated degradation pathway.13 Deficiency of this protein might lead to cell death or aberrant differentiation.14 In addition, UFD1L single nucleotide polymorphisms (SNP) have also been associated with schizophrenia.15 UFD1L polymorphism rs5992403 contributes to cognitive impairment in patients with schizophrenia. The UFD1L protein change herein raises the possibility that the stress resistant phenotype may be linked with UFD1L and the pathways associated with it.
We also observed that septin11 was higher in stress resistant rats. Septins are an evolutionarily group of GTP-binding and filament-forming proteins. They are known to interact with components of the cytoskeleton, such as actin and tubulin. In addition, the level of other proteins, CLASP1 and TUBB2a, involved in cytoskeleton regulation, was higher in stress resistant rats. These protein level alterations suggest modification of the microfilament structure, which in turn is strongly related to synaptic plasticity. Indeed, synaptic and cytoskeletal plasticity are involved in the development of psychiatric diseases, such as depression.16,17 Stress or antidepressant medication leads to alteration in the expression of the cytoskeletal proteins. Elevation can be found in the level of tubulin, which is the main component of the cytoskeletal system.18,19 Acute or chronic antidepressant treatment can affect the microtubular dynamics.20 Our results suggested cytoskeletal enhancement in the stress resistant phenotype. Moreover, we identified several proteins associated with cytoskeletal rearrangement, which were involved in the stress resistant behaviors. In addition, prefoldin5 (PFDN5) was also increased in the stress resistant rats. Prefoldin is a cochaperone that cooperates with the chaperonin containing TCP1 (CCT). It is known for its functional relevance in the cytoplasmic folding of actin and tubulin monomers during the cytoskeleton assembly.21,22 Deficiency in PFDN5 gene results in progressive neurodegeneration and reproductive abnormalities.23 The results suggested that PFDN5 might be involved in the stress resistant phenotype.
PCDH10, as a calcium-dependent adhesion molecule, is mainly expressed in the olfactory bulb, most parts of the limbic system and the cerebellum.24 It is reported that PCDH10 deficient mice have defects in axon pathways through ventral telencephalon. PCDH10 is essential for patterning of the putative guidance cues for thalamocortical projections.25 In addition, male mice lacking a single copy of PCDH10 exhibit abnormal social approach and social communication behaviors, as well as altered structure and function of amygdale synaptic connections, and PCDH10 also plays a role in the pathophysiology of autism disorder.26 Our data showed that PCDH10 was increased in the stress resistant rats, which indicated that PCDH10 was also involved in stress resilience phenotype.
Mesencephalic astrocyte-derived neurotrophic factor (Manf), is widely expressed in both neuronal and non-neuronal tissues. In the central nervous system (CNS), Manf protein expression is detected in several brain areas including the olfactory bulb, cerebral cortex, hippocampus, hypothalamus, striatum, substantia nigra (SN), and spinal cord. In the SN, Manf is detected in dopamine neurons.27 Increasing evidence indicates that Manf, when applied as extracellular proteins or delivered by viral vectors, can protect and repair midbrain dopamine neurons in vivo.28 Moreover, evidence shows that Manf is involved in the protection of existing neurons, in the regeneration of neurons and migration of newly formed neurons to the injury site in brain ischemia.29 Furthermore, as Manf localizes in the endoplasmic reticulum (ER), evidence suggests that Manf is important for the maintenance of ER homeostasis.30 Our data showed that Manf was increased in stress resilience rats compared with stress sensitive rats. This reminded us that the effect of Manf in neuron protection and ER homeostasis maintenance might be involved in the stress resilience phenotype.
Complexin 2 is a small, cytosolic protein that binds to the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) complex to regulate synaptic vesicle exocytosis.31 It is found mainly in the presynaptic terminal of olfactory bulb, striatum, hippocampus, thalamus, and cerebellum.32 Complexin 2 plays a role in neurotransmitter release.33 Phosphorylation of complexin by PKA regulates spontaneous neurotransmitter release and structural synaptic plasticity.34 Moreover, Jorquera believes that complexin 2 regulates both spontaneous and evoked neurotransmitter release by modulating the timing and properties of synaptotagmin activity.35 In addition, there is a growing body of evidence to support the idea that disturbed synaptic transmission contributes to the pathophysiology of mood disorders.36,37 Post-mortem studies have reported reduced complexin expression levels in cases of bipolar disorder and major depression.38 Herein, the expression of complexin 2 was increased in the stress resilience rats than stress sensitive rats. It seems likely that upregulation of complexin 2 contributes to the stress resilience phenotype by facilitating SNARE complex function and synaptic transmission at the synapse.
Conclusion
In summary, label-free LC-MS/MS quantification analysis was used, for the first time, to explore differentially expressed proteins in stress resistant rats compared with that in stress sensitive rats, and we found significant differences in the proteome profiles between stress resistant phenotype and stress sensitive phenotype. In total, 627 proteins were found to either increase or decrease in stress resistant rats compared with those in stress sensitive rats. Most importantly, 7 differentially expressed proteins were highly associated with neural biological processes. Thus, our findings provide a new insight into proteomic profiles of stress resistant phenotypes. The identification of these differentially expressed proteins will provide potential support to further research and eventually elucidate molecular mechanisms in the stress resistant phenotype.Acknowledgements
This study was supported by the National Natural Science Foundation of China Grants (81274122, 81202507, U1402221, 81573636), the National Mega-project for Innovative Drugs (2012ZX09301002-004, 2012ZX09301002-001), the Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT) (IRT1007), the Specialized Research Fund for the Doctoral Program of Higher Education of China (20121106130001), Beijing Natural Science Foundation (7131013, 7142115), Beijing Key Laboratory of New Drug Mechanisms and Pharmacological Evaluation Study (BZ0150). The State Key Laboratory Fund Open Project (GTZK201610), China Postdoctoral Science Foundation (2013M540066).References
- K. S. Kendler, L. M. Karkowski and C. A. Prescott, Am. J. Psychiatry, 1999, 156, 837–841 CrossRef CAS PubMed.
- H. Yu, D. D. Wang, Y. Wang, T. Liu, F. S. Lee and Z. Y. Chen, J. Neurosci., 2012, 32, 4092–4101 CrossRef CAS PubMed.
- V. Krishnan, M. H. Han, D. L. Graham, O. Berton, W. Renthal and S. J. Russo, Cell, 2007, 131, 391–404 CrossRef CAS PubMed.
- S. J. Russo, J. W. Murrough, M. H. Han, D. S. Charney and E. J. Nestler, Nat. Neurosci., 2012, 15, 1475–1484 CrossRef CAS PubMed.
- A. F. Arnsten, Nat. Rev. Neurosci., 2009, 10, 410–422 CrossRef CAS PubMed.
- E. B. Bloss, W. G. Janssen, D. T. Ohm, F. J. Yuk, S. Wadsworth and K. M. Saardi, J. Neurosci., 2011, 31, 7831–7839 CrossRef CAS PubMed.
- G. Rajkowska, J. J. Miguel-Hidalgo, J. Wei, G. Dilley, S. D. Pittman and H. Y. Meltzer, Biol. Psychiatry, 1999, 45, 1085–1098 CrossRef CAS PubMed.
- J. D. Sun, Y. Liu and Y. H. Yuan, Neuropsychopharmacology, 2012, 37(5), 1305–1320 CrossRef CAS PubMed.
- H. E. Covington, M. K. Lobo, I. Maze, V. Vialou, J. M. Hyman and S. Zaman, J. Neurosci., 2010, 30, 16082–16090 CrossRef CAS PubMed.
- X. Zhu, R. T. Libby, W. N. de Vries, R. S. Smith, D. L. Wright, R. T. Bronson, K. L. Seburn and S. W. John, PLoS Genet., 2012, 8, e1002853 CAS.
- V. A. van der Mark, P. J. Ronald, E. Oude and C. P. Coen, Int. J. Mol. Sci., 2013, 14, 7897–7922 CrossRef PubMed.
- P. Cacciagli, M. R. Haddad, C. Mignon-Ravix, B. El-Waly, A. Moncla, C. Missirian, B. Chabrol and L. Villard, Eur. J. Hum. Genet., 2010, 18, 1360–1363 Search PubMed.
- M. Chen, G. J. Gutierrez and Z. A. Ronai, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(22), 9119–9124 CrossRef CAS PubMed.
- H. H. Meyer, J. G. Shorter, J. Seemann, D. Pappin and G. Warren, EMBO J., 2000, 19, 2181–2192 CrossRef CAS PubMed.
- V. K. Ota, S. I. Belangero, A. Gadelha, F. T. Bellucco, D. M. Christofolini, T. I. Mancini, A. K. Ribeiro-dos-Santos, S. E. Santos, J. J. Mari, R. A. Bressan, M. I. Melaragno and M. A. Smith, J. Psychiatr. Res., 2010, 44, 1113–1115 CrossRef PubMed.
- E. Messaoudi, T. Kanhema, J. Soulé, A. Tiron, G. Dagyte, B. da Silva and C. Bramham, J. Neurosci., 2007, 27, 10445–10455 CrossRef CAS PubMed.
- Y. Li, A. Pehrson, J. Waller, E. Dale, C. Sanchez and M. Gulinello, Front. Neurosci., 2015, 9, 279 Search PubMed.
- C. Piubelli, M. Vighini, A. A. Mathe, E. Domenici and L. Carboni, Int. J. Neuropsychopharmacol., 2011, 14, 796–833 CrossRef CAS PubMed.
- M. Bianchi, C. Heidbreder and F. Crespi, Synapses, 2003, 49, 188–194 CrossRef CAS PubMed.
- C. Yang, G. Wang, H. Wang, Z. Liu and X. Wang, Behav. Brain Res., 2009, 205, 518–524 CrossRef PubMed.
- I. E. Vainberg, S. A. Lewis and H. Rommelaere, Cell, 1998, 93, 863–873 CrossRef CAS PubMed.
- G. Millan-Zambrano and S. Chavez, Open Biol., 2014, 4, 140085 CrossRef PubMed.
- Y. Lee, R. S. Smith and W. Jordan, J. Biol. Chem., 2011, 286, 726–736 CrossRef CAS PubMed.
- M. Frank and R. Kemler, Curr. Opin. Cell Biol., 2002, 14, 557–562 CrossRef CAS PubMed.
- M. Uemura, S. Nakao, S. T. Suzuki, M. Takeichi and S. Hirano, Nat. Neurosci., 2007, 10(9), 1151–1159 CrossRef CAS PubMed.
- H. Schoch, A. S. Kreibich, S. L. Ferri, R. S. White, D. Bohorquez, A. Banerjee, R. G. Port and H. C. Dow, Biol. Psychiatry, 2017, 81(3), 193–202 CrossRef CAS PubMed.
- P. Lindholm, J. Peranen, J. O. Andressoo, N. Kalkkinen and Z. Kokaia, Mol. Cell. Neurosci., 2008, 39, 356–371 CrossRef CAS PubMed.
- P. Lindholm, M. H. Voutilainen, J. Lauren, J. Peranen and V. M. Leppanen, Nature, 2007, 448, 73–77 CrossRef CAS PubMed.
- Y. Q. Yu, L. C. Liu, F. C. Wang, Y. Liang and D. Q. Cha, J. Cereb. Blood Flow Metab., 2010, 30, 79–91 CrossRef CAS PubMed.
- M. Lindahl, M. Saarma and P. Lindholm, Neurobiol. Dis., 2017, 97, 90–102 CrossRef CAS PubMed.
- H. T. McMahon, M. Missler, C. Li and T. C. Sudhof, Cell, 1995, 83, 111–119 CrossRef CAS PubMed.
- W. Freeman and A. J. Morton, Brain Res. Bull., 2004, 63, 33–44 CrossRef CAS PubMed.
- D. Glynn, H. E. Gibson, M. K. Harte, K. Reim, S. Jones, G. P. Reynolds and A. J. Morton, Hum. Mol. Genet., 2010, 19, 3402–3412 CrossRef CAS PubMed.
- R. W. Cho, L. K. Buhl, D. Volfson, A. Tran, F. Li, Y. Akbergenova and J. T. Littleton, Neuron, 2015, 88, 749–761 CrossRef CAS PubMed.
- R. A. Jorquera, S. Huntwork-Rodriguez, Y. Akbergenova, R. W. Cho and J. T. Littleton, J. Neurosci., 2012, 32, 18234–18245 CrossRef CAS PubMed.
- J. T. Coyle and R. S. Duman, Neuron, 2003, 38, 157–160 CrossRef CAS PubMed.
- P. Schloss and F. A. Henn, Pharmacol. Ther., 2004, 102, 47–60 CrossRef CAS PubMed.
- K. Sawada, C. E. Young, A. M. Barr, K. Longworth, S. Takahashi, V. Arango, J. J. Mann, A. J. Dwork, P. Falkai, A. G. Phillips and W. G. Honer, Mol. Psychiatry, 2002, 7, 484–492 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |