Open Access Article
Open Access ArticleFluorescent detection of point mutation via ligase reaction assisted by quantum dots and magnetic nanoparticle-based probes
S. Heidari Sharafdarkolaeia,
M. Motovali-Bashi a and
P. Gill
a and
P. Gill *bc
*bc
aDepartment of Genetics, Faculty of Sciences, University of Isfahan, Isfahan, Iran
bNanomedicine Group, Immunogenetics Research Center, Mazandaran University of Medical Sciences, Sari, Iran
cThalassemia Research Center, Mazandaran University of Medical Sciences, Sari, Iran. E-mail: pooriagill@yahoo.com; p.gill@mazums.ac.ir
First published on 12th May 2017
Abstract
A nanodiagnostic genotyping method was presented for point mutation detection directly in human genomic DNA based on ligase reaction coupled with quantum dots and magnetic nanoparticle-based probes. For this purpose, allele-specific probes, including a biotin-labeled common probe and two biotin-labeled allele-specific probes were designed for mutant and wild alleles of human beta globin gene (IVS-II-I G → A point mutation). When genomic DNA carried the mutation site, the common probe and allele-specific probe were ligated to form exponential amplified biotin-labeled fluorescence ligation products. These ligated products were captured by streptavidin-coated magnetic nanoparticles at one end and then attached to a QD 605-streptavidin conjugate at the other end to be detected fluorescently. Thereafter, the genotypes were identified conveniently according to the fluorescence color of quantum dots using a rotor-gene 6000Q real-time rotary analyzer. The results demonstrated the sensitivity and specificity percentages of this nanomolecular mutation detection method were 85.45% and 95.77% respectively. In addition, this method could be a high throughput and high sensitivity detection system that represents suitable non-PCR based nanodiagnostics for detection of other point mutations.
Introduction
Gene point mutations present important biomarkers for genetic diseases. However, existing point mutation detection methods suffer from low sensitivity, specificity and tedious assay processes.1 A major goal of mutation diagnostic methods is the ability to detect low abundant mutations with enough sensitivity to detect the mutant DNA without generating false positives or false negatives. Nucleic acid amplification methods fall into three categories: target, probe and signal amplification systems.2 The majority of diagnostic methods rely on PCR amplification of target sequences.3 The main disadvantages of PCR based methods although highly sensitive are amplification errors due to mis-priming, limited accuracy of discriminating single nucleotide variations, and limited multiplexing capability.1,4–6 Although a number of alternative PCR-free methods have been introduced, ligation-based techniques are the most widely used for diagnostic methods due to their exceptional specificity on base discrimination using ligase enzyme and robust multiplexing capabilities.1,7The ligase detection reaction (LDR) is a probe amplification method that uses two adjacent probes and the thermostable ligase.8 LDR employs two-allele specific probes known as discriminating probes, which differ in the base at their 3′-end and one common probe. Only if the discriminating primer fully complement to the target sequence, the ligase joins this primer to common one to form ligation products. Common or discriminating primer or both of them can be labelled, which is used for detection step.9–14 LDR has certain advantages over PCR based methods. LDR amplifies the probe molecule instead of producing fragments of DNA through polymerization of nucleotides so; few steps are involved in the amplification cycle, thereby reducing the time needed to complete the entire process. In addition, PCR requires high-quality DNA because it involves the amplification of large chains of nucleotides, whereas LDR does not, because the target sequence is usually small (36 to 60 nucleotides). Ligase reactions have exceptional specificity on base discrimination because of high fidelity thermostable ligase enzyme.1,7 The use of a thermostable ligase minimizes target independent ligation since the ligation reaction can be performed at or near the melting temperature (Tm) of the oligonucleotide probes.
Colloidal semiconductor nanocrystals, which are also called quantum dots (QDs) as a popular kind of fluorescent material that have several unique photophysical properties. Quantum dots have been found to be superior to traditional organic dyes as fluorescent markers with numerous advantageous features, such as longer lifetime, high extinction coefficient, high quantum yields, photostability, negligible phototoxicity and broad absorption spectrum but narrow emission spectra that is tuneable by changing the crystallite size.1,15–27
Quantum dots also have a brighter emission and a higher signal to noise ratio compared with organic dyes. The brightness of quantum dots compared to organic dyes are 10–20 times brighter.28 Quantum dots are stable fluorophores due to its inorganic composition, which reduces the effect of photo bleaching compared to organic dyes. The fluorescence time for quantum dots is about 10 to 40 ns, which is longer than the fluorescence of a few nanoseconds of organic dyes.29,30 The large Stokes shift (difference between peak absorption and peak emission wavelengths) reduces autofluorescence, which increases sensitivity.31 Lastly, the multicolour property of quantum dots allows the use of many probes to track several targets simultaneously.1,15–25,28 Nanoparticle-enhanced biosensors have been extensively researched. Of all nanoparticles, magnetic nanoparticles are proving to be a versatile tool, an excellent case in point being in DNA bioassays, where magnetic nanoparticles were often used for optimization of the hybridization and separation of target DNA. The efficient attachment of biomolecules to the surface of magnetic nanoparticles and bio recognition site interacting with the analyte is critical parameters in successful separation that determines the test sensitivity which offer promise here because their high surface to volume ratio provides a large number of sites available for molecular immobilization and high specific interaction.32–34 The use of magnetic nanoparticles especially has been increasing rapidly because they are superparamagnetic, non-toxic and of a small size and in response to a magnetic field they become easily magnetized and demagnetized.
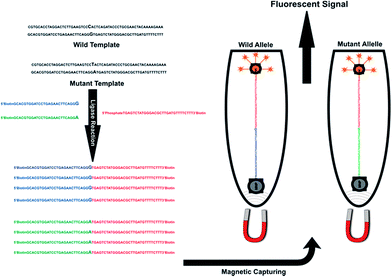
Here we describe a quantum dot-based LDR assay developed for the highly sensitive detection of single nucleotide polymorphisms (point mutations) in human genomic DNA. In this assay, the ligation was performed in a tube using a biotinylated common probe and an allele specific discriminating probe labelled with quantum dot. Schematic representation of the assay was shown in Fig. 1. According to a mutated site, LDR probes were designed for two alleles of a mutated site (Table 1). Among these oligomers, three label probes are needed for each biallelic marker. A 3′-biotin, 5′-phosphorylated common probe (common) and two 5′-biotin labelled allele-specific probes were designed which had the allelic base at the 3′-end (LPG probe for wild allele and LPA probe for mutant allele). Ligation reaction performed between two adjacent labelled probes when their hybridization was complementary to the template.
| Code | Sequences (5′ to 3′) | End modification |
|---|---|---|
| LPG | GCACGTGGATCCTGAGAACTTCAGGG | 5′ biotin |
| LPA | GCACGTGGATCCTGAGAACTTCAGGA | 5′ biotin |
| Common | TGAGTCTATGGGACGCTTGATGTTTTCTTT | 5′ phosphate, 3′ biotin |
Successful ligation of the two probes produces a biotin-tagged LDR product, which immobilized on streptavidin-conjugated magnetic nanoparticles (MNPs) through the biotin–streptavidin interaction. After re-moving the supernatant using magnetic separation, fluorescent signal of LDR products monitor by rotor-gene 6000TM real time rotary analyzer. It does not require amplification step and use biotin–streptavidin interaction for capturing of nanoparticles to ligation products, which was very fast and highly specific interaction. Separating and labelling steps do in one tube using this quick and easily biotin–streptavidin interaction.
Material and methods
Chemical and instruments
Rotor-Gene 6000TM real time rotary analyzer (Germany; https://www.qiagen.com) was used for signal detection of LDR product conjugated with QD. Qdot 605 ITK Streptavidin Conjugate Kit was purchased from Invitrogen (USA; https://www.thermofisher.com). A series of oligonucleotides were commercially synthesized by Bioneer (Korea; http://www.bioneer.com). Sequences of these oligos are listed in Table 1. We used Nano Drop system (Biochrome ltd, UK; http://www.biochrom.co.uk) for determination of DNA concentration. Streptavidin-coated magnetic nanoparticles were purchased from Chemicell (Germany; https://www.2bscientific.com) and TaqDNA ligase was obtained from New England Biolabs (USA; https://www.neb.com).Specimens and DNA extraction
All experiments were performed in compliance with the guideline “Biomedical research ethics review method involving people” (I.R. Iran), and approved by the ethical committee of Mazandaran University of Medical Science. Informed consents were obtained from human participants of this study. Fifty persons who their IVSII-1 (G → A) beta globin gene mutation investigated using ARMS–PCR method established in Mazandaran Thalassemia Research Center. Genomic DNA was isolated from leucocytes in peripheral blood of normal and patients subjects using phenol-chloroform extraction protocol. In addition, the genomic DNA concentration was assessed at 260 nm wavelength using UV-vis spectrophotometer.LDR amplification
LDR was performed using a Bio-Rad MJ mini thermal cycler. Extracted genomic DNA was used as LDR template. A set of 3 probes, including one common probe and two variant-specific probes were designed and ordered. All probes were labelled with biotin and attach to streptavidin coated magnetic nanoparticles and streptavidin coated quantum dots. Each LDR reaction contained 10 μL reaction volumes: 1 μL of 10× Taq ligase buffer; 4 units TaqDNA ligase; 20 μM each probes; template (20 ng genomic DNA). The conditions of LDR were the following: initial denaturation at 94 °C for 4 min, followed by 30 cycles of denaturation at 94 °C for 30 s, then 65 °C for 5 min.Coupling LDR ligated products
The proposed method employed MNPs to capture LDR ligated products through biotin–streptavidin interaction. The capturing process involved two major steps: (1) capturing ligated products using streptavidin-coated MNPs; (2) removal of non-specific absorption components on the surface of MNPs using washing buffer. Hence, 10 μl of LDR products with 2 μl of MNPs (1 mg ml−1) were incubated for 30 min at 37 °C. Then MNPs were absorbed using a magnetic plate and supernatant was discharged. Next, the MNPs were re-dispersed in incubation buffer and this washing procedure was repeated three times. Subsequently, 5 μl QD (0.2 μM) was added to the MNP–LDR mixture for 30 min at 37 °C for shaping MNP–LDR–QD nanocomplexes. Then, the nanocomplexes transferred to a magnetic plate, supernatant was discharged, and washing procedure was repeated three times again. Finally, deposited nanocomplexes were resuspended in 10 μl incubation buffer for fluorescent measurement.Fluorescent assay
Fluorescence ligation products on the surface of MNPs were detected by the rotor-gene 6000 real-time rotary analyzer and the corresponding signal were obtained which refer to QD 605 ITK Streptavidin in orange area.TEM characterization
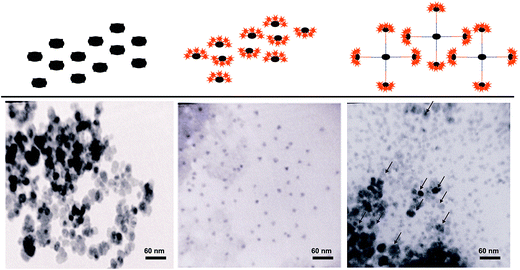
A few drops of each QDs, MNPs and ligation products sample were dispersed onto a 3 mm copper grid covered with a continuous carbon film and dried at room temperature. The TEM image of each sample was obtained on a CM30 transmission electron microscope (Philips EO, Netherlands) operating at 150 kV.Clinical performance of LDR–QD system
Fifty DNA samples were analyzed by LDR–QD system. True positive (TP), true negative (TN), false positive (FP) and false negative (FN) were determined by comparing the results of routine test (ARMS–PCR) and LDR–QD method. In this study, TP means routine detection test reported samples with IVS II-I (G–A) mutation and LDR–QD test confirmed it; TN means routine detection test reported samples without IVS II-I (G–A) mutation and LDR–QD test confirmed it; FP means detection test didn't report samples with IVS II-I (G–A) mutation but LDR–QD test reported samples with IVS II-I (G–A) mutation and FN means routine detection test reported samples with IVS II-I (G–A) mutation but LDR–QD test reported samples without IVS II-I (G–A) mutation. The sensitivity and specificity percentages were determined as follows: % sensitivity = TP/TP + FN, and % specificity = TN/TN + FP. The sensitivity of LDR–QD test were referred to the ability of the test for identifying those samples with IVS II-I (G–A) mutation, correctly. The specificity of LDR–QD test was referred to the ability of the test for identifying those samples without IVS II-I (G–A) mutation, correctly.35,36Results
TEM
To investigate the formation of the MNP–QD conjugate, TEM images were obtained from MNPs and QDs before and after ligation detection reaction (LDR). TEM micrographs showed that MNPs surrounded by QDs, which indicated the formation of the MNP–QD conjugates (Fig. 2).QD–LDR signals
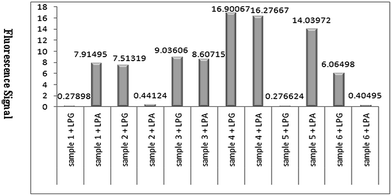
In the presence of a mismatch between two adjacent probes and template, ligation was not occurred. During LDR process, 3′ and 5′-biotin-LPA labelled ligation product was amplified for mutated template and 3′ and 5′-LPG-labeled ligation product was amplified for wild type template. These ligation products were attached to the QD streptavidin conjugate through biotin–streptavidin interaction and captured by streptavidin coated MNPs. The signal of QD was determined by detection of the fluorescence signal using rotor-gene 6000 real-time rotary analyzer in wavelength of 605 nm (orange area) (Fig. 3).QD–LDR genotyping
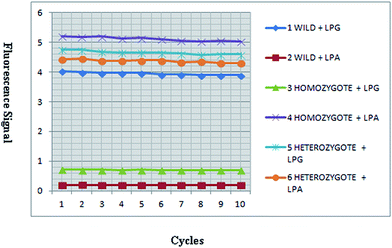
Genotypes were determined by detecting the fluorescence signals of QDs attached to ligation products (Fig. 3). Two LDR reactions were performed in separated tubes which was used wild allele specific probe (LPG) at one tube and mutant allele specific (LPA) at the other one. Both tubes exhibited fluorescence signal in heterozygote mutant sample, the tube that included of LPG exhibited signal in normal sample and the tube that consist of LPA had signal in homozygote mutant sample (Fig. 4).QD–LDR clinical performance
The clinical performance of the test was determined by comparing the results of LDR–QD test and ARMS. Table 2 summarized reliability values of LDR–QD test for diagnosis of β-thalassemia. In this study, among 50 persons with and without IVSII-1 (G–A) mutation, the false negative and positive results were 2 and 8 alleles respectively; however, the true positive and negative results were 47 and 43 alleles respectively. Moreover, the comparison between the results of the routine tests with the LDR–QD results indicated the sensitivity and specificity of this assay were 85.45% and 95.77% respectively (Table 2). Our results suggested that LDR–QD system could be suitable for detection of IVSII-1 (G–A) mutation.| LDR–QD test | ARMS test | Clinical performances | ||
|---|---|---|---|---|
| Mutant allele | Wilde Allele | % sensitivity | % specificity | |
| Mutant allele | 47 | 2 | 85.45 | 95.77 |
| Wilde allele | 8 | 43 | ||
High sensitivity and specificity of this assay for point mutation detection was attributed to the high-efficiency ligase based LDR and the signal amplification ability of QD-labelled probe.
Discussion
Beta thalassemia is considered the most common autosomal single gene disorder worldwide that can be found in more than 60 countries with a carrier population of up to 150–200 million. So far, close to 200 different mutations in the β-globin gene have been reported that associated with the onset of the disease37,38 the frequency and geographic distribution of beta thalassemia mutations is useful to establish a program for carrier screening, genetic counselling, prenatal diagnosis and for physicians to establish specific therapeutic approaches for patients with beta thalassemia.In this report, we described a nanodiagnostic method for beta-thalassemia point mutation using quantum dot-LDR system that could detect directly point mutations in human genomic DNA. Combining ligase detection reaction (LDR) and magnetic nanoparticle technique used for increased sensitivity, and employing fluorescence ligation products allowed detection analysis. The method has been demonstrated as convenient point mutation detection in human genome with the genotyping of the IVSII-1 (G → A) point mutation of β-globin gene associated with thalassemia disease. Although LDR was highly specific in base recognition, but it had very limited sensitivity. Consequently, LDR was usually combined with PCR that exponentially amplified the ligation products to a detectable level. The combined PCR–LDR process significantly improves the assay sensitivity but suffers from the complications of PCR.1,39 To our knowledge, this is the first report to demonstrate a practical biological application of LDR-quantum dot PCR-free system.
Streptavidin–biotin interaction was specifically chosen for this assay. This binding interaction was quick, reliable and strong.17,40 The success of the system depend on three components: (1) accuracy of the allele discrimination reaction by thermal DNA ligase, which has been well-established; (2) specificity biotin–streptavidin interaction between magnetic nanoparticles and quantum dots with ligation products; (3) the sensitive detection resulting from LDR product coupled with quantum dot and enrichment of streptavidin-coated magnetic nanoparticle.
The assay was able to differentiate between the homogeneous mutant, the heterozygous mutant and the wild type by analyzing the sample using QD-labelled mutant and wild type-specific probes. The method is simple, cost-effective, and paves a new way for point mutation detection as well as molecular diagnosis.41 While there have been studies such as those described by Meng and Battistella, which have reported the use of ligase detection system for point mutation detection, this study is unique in its integration of quantum dot and magnetic nanoparticles with LDR assay (Table 3).
| Detection method | Mutation(s) | Mechanism | Number of probes | Fluorescent molecule(s) | Time (hours) | Disadvantages | Ref. no. |
|---|---|---|---|---|---|---|---|
| Allele-specific real-time PCR | Codon 13 (G → A) of KRAS | Microfluidic cartridge containing preloaded primers and probes which enables the parallel detection of point mutation by allele-specific real-time PCR | 4 | FAM | 4 | Fabrication of the microfluidic system is expensive, a T–T, G–T, C–T, or C–A 3′-terminal mismatch error peone of allele specific PCR amplification | 42 |
| High resolution melt analysis | Fr 8/9 (+G), IVSI-1 (G > A), IVSI-5 (G > C), IVSI-110 (G > A), and CD44 (−C) | Is the quantitative analysis![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) of the melt curve of a DNA fragment following amplification by PCR of the melt curve of a DNA fragment following amplification by PCR |
2 | EvaGreen | 4 | Depends strongly on good PCR, instruments and dyes. Heterozygote detection does not depend on the type of single base substitution or the variant position within the PCR product. Small insertions and deletions may be somewhat more difficult to detect than substitutions | 43 |
| Ligase chain reaction | CD17 (A → T) | Two pairs of probe hybridization, ligation and signal detection | 6 | FAM and ROX | 3 | Prone to false-positive amplification | 44 |
| Gapped-ligase chain reaction | Codon12 and 13 (G → A) | PCR products use as template and DNA polymerase fills in a gap between annealed probes which are subsequently joined by DNA ligase | 4 | FAM, ROX and JOE | More than 4 | Need of polymerase, nucleotides and extension steps | 45 |
| Ligase detection reaction | IVS-I-1 (G → A), IVS-I-6 (T → C), IVS-I-110 (G → A), codon 39 (C → T), IVS-II-1 (G → A) | PCR amplification, probe hybridization and ligation to PCR products, signal detection | 5 | Cy5 | More than 4 | Combined PCR–LDR process suffers from the complications of PCR | 46 |
| Nano based ligase detection reaction | IVSII-1 (G → A) | Probe hybridization and ligation, separation by MNPs and quantum dot signal detection | 3 | QD | 3 | Providing non-PCR-based fluorescent analyzer | This study |
While there have been studies which have reported the use of ligation reaction for point mutation detection, this study is unique in its integration of both quantum dots and magnetic nanoparticles in LDR–PCR free system.47–49
Meng and Battistella described a convenient genotyping method capable of detecting point mutations directly in human genomic DNA based on the combination of ligase chain reaction (LCR) and microbead-enrichment technique. LCR probes, including a biotin-labelled common probe and two fluorescence-labelled allele-specific probes, were designed for two alleles of a mutated site. When genomic DNA carried the mutated site, the common probe and allele-specific probe were ligated to form exponential amplified biotin-labelled fluorescence ligation products. Streptavidin-coated microbeads enriched these ligated products, and genotypes were identified conveniently according to the fluorescence colour of microbeads using fluorescent microscopy. The method had been demonstrated as convenient point mutation detection in human genome with the genotyping of the CD17 (A → T) point mutation of β-globin gene associated with thalassemia disease.44
In the LCR, thermostable ligase lacking blunt-end ligation activity should be used to prevent the generation of background signals produced by the blunt-end ligation of the complementary probe pair duplexes.50 In a manner analogous to PCR, the ligated products could be exponentially amplified because the ligated probes serve as a template for the ligation reaction of the complementary probe pair in the following cycle. However, despite the improved efficiency, the LCR was prone to false-positive amplification. Because non-specifically ligated probes could also be used as a template so, the LCR was not as popular.51
Battistella et al. developed a genotyping assay based on LDR system that followed by a universal PCR (U-PCR) of genomic DNA-templated LDR product. The assay was designed to detect 7 prevalent mutations in the beta globin gene.46 Although this strategy had sufficiently high sensitivity and specificity to differentiate multiple targets but the combined PCR–LDR process suffer from the complications of PCR.
Several point mutation detection methods have been developed such as restriction fragment length polymorphisms (RFLP),52 temporal temperature gradient gel electrophoresis (TGGE),53 denaturing gradient gel electrophoresis (DGGE),54,55 reverse dot blot hybridization, amplification refractory mutation system (ARMS),56 high resolution melting (HRM).35 However, these technologies have some shortcomings. The main disadvantages of PCR based methods although highly sensitive are amplification errors due to mispriming, limited accuracy of discriminating single nucleotide variations, post-PCR processing steps such as gel electrophoresis and limited multiplexing capability.
To overcome these problems we presented a novel SNP assay that for the first time exploited MNPs and QDs coupled with LDR technique, which is removed amplification and post-PCR processing steps. Unique optical properties of QDs increase multiplexing capability of the assay. High throughput magnetic separation and high precision QD signal detection improve the sensitivity and selectivity of this assay.
Conclusion
This nanodiagnostic ligation assay was developed for accurately SNP/point mutation detection directly in human genomic DNA. This method did not require amplification step and use very fast and highly specific biotin–streptavidin interaction. Separating and labelling steps do in one tube using this quick and easily biotin–streptavidin interaction. Quantum dots have a broader excitation spectra and a narrow more sharply defined emission peak, which was not presented in organic fluorophores. Due to these properties, a single light source can be used to excite multicolour quantum dots simultaneously without signal overlap and allows the use of many probes to track several targets simultaneously, too. The large Stokes shift (difference between peak absorption and peak emission wavelengths) reduces autofluorescence, which increases sensitivity. In future using two different types of quantum dots signal can be detected in one tube, which reduces sample volume, processing times. Finally, this method matches the golden rules for the development of diagnostic applications. These include (1) low costs, (2) multiplexing capabilities, (3) short processing times, (4) low sample consumption, (5) easy adaptability to new formats and targets, (6) easily standardized and programmed.Acknowledgements
This study was supported partially by University of Isfahan and Vice Chancellor of Research and Technology, Mazandaran University of Medical Sciences. The authors thank also Central Research Laboratory, Mazandaran university of Medical Sciences. Also, the authors thank from Iran Nanotechnology Initiative (INI).Notes and references
- Y. Song, Y. Zhang and T. H. Wang, Small, 2013, 9, 1096–1105 CrossRef CAS PubMed.
- P. Gill, M. Forouzandeh, N. Eshraghi, M. Ghalami, M. Safa and M.-R. Noori-Daloii, Mol. Cell. Probes, 2008, 22, 103–109 CrossRef CAS PubMed.
- H. Sun, Y. Yang, L. Yang, B. Su, G. Jiang, K. Fei and D. Lu, BioMed Res. Int., 2014, 2014, 407537 Search PubMed.
- J. Ragoussis, Annu. Rev. Genomics Hum. Genet., 2009, 10, 117–133 CrossRef CAS PubMed.
- P. Y. Kwok and X. Chen, Curr. Issues Mol. Biol., 2003, 5, 43–60 CAS.
- Z. Tsuchihashi and N. C. Dracopoli, Pharmacogenomics J., 2002, 2, 103–110 CrossRef CAS PubMed.
- W. Cao, Trends Biotechnol., 2004, 22, 38–44 CrossRef CAS PubMed.
- F. Barany, Proc. Natl. Acad. Sci. U. S. A., 1991, 88, 189–193 CrossRef CAS.
- S. Watanabe, K. Hagihara, K. Tsukagoshi and M. Hashimoto, Anal. Chem., 2014, 86, 900–906 CrossRef CAS PubMed.
- X. L. Wang, S. G. Xie, L. Zhang, W. X. Yang, X. Wang and H. Z. Jin, World J. Gastroenterol., 2008, 14, 120–124 CrossRef CAS PubMed.
- R. Sinville, J. Coyne, R. J. Meagher, Y. W. Cheng, F. Barany, A. Barron and S. A. Soper, Electrophoresis, 2008, 29, 4751–4760 CrossRef CAS PubMed.
- A. E. Dent, C. T. Yohn, P. A. Zimmerman, J. Vulule, J. W. Kazura and A. M. Moormann, Am. J. Trop. Med. Hyg., 2007, 77, 250–255 CAS.
- P. Yi, W. Lu, J. Guo, Q. Liu, Z. Chen, J. Han and L. Li, Cell Biochem. Biophys., 2011, 61, 629–636 CrossRef CAS PubMed.
- X. T. Gregg, R. Kralovics and J. T. Prchal, Am. J. Hematol., 2000, 63, 184–191 CrossRef CAS PubMed.
- H. Zhou, J. Liu, J. J. Xu and H. Y. Chen, Anal. Chem., 2011, 83, 8320–8328 CrossRef CAS PubMed.
- H. Zhang, X. Fu, L. Liu, Z. Zhu and K. Yang, Anal. Biochem., 2012, 426, 30–39 CrossRef CAS PubMed.
- H. C. Yeh, Y. P. Ho, M. Shih Ie and T. H. Wang, Nucleic Acids Res., 2006, 34, e35 CrossRef PubMed.
- T. Jamieson, R. Bakhshi, D. Petrova, R. Pocock, M. Imani and A. M. Seifalian, Biomaterials, 2007, 28, 4717–4732 CrossRef CAS PubMed.
- W. R. Algar, A. J. Tavares and U. J. Krull, Anal. Chim. Acta, 2010, 673, 1–25 CrossRef CAS PubMed.
- E. Abbasi, T. Kafshdooz, M. Bakhtiary, N. Nikzamir, M. Nikzamir, M. Mohammadian and A. Akbarzadeh, Artif. Cells, Nanomed., Biotechnol., 2016, 44, 885–891 CAS.
- E. Petryayeva, W. R. Algar and I. L. Medintz, Appl. Spectrosc., 2013, 67, 215–252 CrossRef CAS PubMed.
- H. Xu, M. Y. Sha, E. Y. Wong, J. Uphoff, Y. Xu, J. A. Treadway, A. Truong, E. O'Brien, S. Asquith, M. Stubbins, N. K. Spurr, E. H. Lai and W. Mahoney, Nucleic Acids Res., 2003, 31, e43 CrossRef PubMed.
- X. He and N. Ma, Anal. Chem., 2014, 86, 3676–3681 CrossRef CAS PubMed.
- S. Su, J. Fan, B. Xue, L. Yuwen, X. Liu, D. Pan, C. Fan and L. Wang, ACS Appl. Mater. Interfaces, 2014, 6, 1152–1157 CAS.
- C. Y. Zhang, H. C. Yeh, M. T. Kuroki and T. H. Wang, Nat. Mater., 2005, 4, 826–831 CrossRef CAS PubMed.
- D. Diaz-Diestra, B. Thapa, J. Beltran-Huarac, B. R. Weiner and G. Morell, Biosens. Bioelectron., 2017, 87, 693–700 CrossRef CAS PubMed.
- Y. Tan, Y. Wang, M. Li, X. Ye, T. Wu and C. Li, Biosens. Bioelectron., 2017, 91, 741–746 CrossRef CAS PubMed.
- X. Gao, L. Yang, J. A. Petros, F. F. Marshall, J. W. Simons and S. Nie, Curr. Opin. Biotechnol., 2005, 16, 63–72 CrossRef CAS PubMed.
- A. P. Alivisatos, W. Gu and C. Larabell, Annu. Rev. Biomed. Eng., 2005, 7, 55–76 CrossRef CAS PubMed.
- W. C. Chan, D. J. Maxwell, X. Gao, R. E. Bailey, M. Han and S. Nie, Curr. Opin. Biotechnol., 2002, 13, 40–46 CrossRef CAS PubMed.
- A. Fu, W. Gu, C. Larabell and A. P. Alivisatos, Curr. Opin. Neurobiol., 2005, 15, 568–575 CrossRef CAS PubMed.
- E. Palecek and M. Fojta, Talanta, 2007, 74, 276–290 CrossRef CAS PubMed.
- X. Zhang, D. B. Reeves, I. M. Perreard, W. C. Kett, K. E. Griswold, B. Gimi and J. B. Weaver, Biosens. Bioelectron., 2013, 50, 441–446 CrossRef CAS PubMed.
- M. K. Pal, M. Rashid and M. Bisht, Biosens. Bioelectron., 2015, 73, 146–152 CrossRef CAS PubMed.
- M. Zafari, P. Gill, M. Kowsaryan, A. Alipour and A. Banihashemi, J. Matern.-Fetal Neonat. Med., 2015, 1–6 CrossRef PubMed.
- A. Rafati and P. Gill, Microchim. Acta, 2015, 182, 523–530 CrossRef CAS.
- M. R. Mahdavi, H. Karami, M. T. Akbari, H. Jalali and P. Roshan, Case Reports in Hematology, 2013, 2013, 906292 CrossRef PubMed.
- S. Hajihoseini, M. Motovali-Bashi, M. A. Honardoost and N. Alerasool, Iran. J. Public Health, 2015, 44, 380–387 Search PubMed.
- M. Hashimoto, F. Barany, F. Xu and S. A. Soper, Analyst, 2007, 132, 913–921 RSC.
- K. Sato, K. Hosokawa and M. Maeda, J. Am. Chem. Soc., 2003, 125, 8102–8103 CrossRef CAS PubMed.
- Z. Yuan, Y. Zhou, S. Gao, Y. Cheng and Z. Li, ACS Appl. Mater. Interfaces, 2014, 6, 6181–6185 CAS.
- O. Strohmeier, S. Laßmann, B. Riedel, D. Mark, G. Roth, M. Werner, R. Zengerle and F. von Stetten, Microchim. Acta, 2014, 181, 1681–1688 CrossRef CAS.
- M. Ramezanzadeh, M. Salehi and R. Salehi, Adv. Biomed. Res., 2016, 5, 71 CrossRef PubMed.
- X. Meng, X. Yang, K. Wang, Q. Guo, Y. Tan, Q. Mo and X. Xu, Talanta, 2010, 80, 1725–1729 CrossRef CAS PubMed.
- S. Jenner and D. Techel, Tumour Biol., 2015, 36, 6361–6368 CrossRef CAS PubMed.
- S. Battistella, F. Damin, M. Chiari, K. Delgrosso, S. Surrey, P. Fortina, M. Ferrari and L. Cremonesi, Clin. Chem., 2008, 54, 1657–1663 CAS.
- Y. Sun, X. Lu, F. Su, L. Wang, C. Liu, X. Duan and Z. Li, Biosens. Bioelectron., 2015, 74, 705–710 CrossRef CAS PubMed.
- M. B. Wabuyele, H. Farquar, W. Stryjewski, R. P. Hammer, S. A. Soper, Y. W. Cheng and F. Barany, J. Am. Chem. Soc., 2003, 125, 6937–6945 CrossRef CAS PubMed.
- Y. Cheng, Q. Du, L. Wang, H. Jia and Z. Li, Anal. Chem., 2012, 84, 3739–3744 CrossRef CAS PubMed.
- G. W. Shin, B. Chung and G. Y. Jung, Electrophoresis, 2014, 35, 1004–1016 CrossRef CAS PubMed.
- J. P. Schouten, C. J. McElgunn, R. Waaijer, D. Zwijnenburg, F. Diepvens and G. Pals, Nucleic Acids Res., 2002, 30, e57 CrossRef PubMed.
- J. Li, Zhongguo Yixue Kexueyuan Xuebao, 1992, 14, 42–45 CAS.
- R. V. Shaji, E. S. Edison, B. Poonkuzhali, A. Srivastava and M. Chandy, Clin. Chem., 2003, 49, 777–781 CAS.
- M. Losekoot, R. Fodde, C. L. Harteveld, H. van Heeren, P. C. Giordano and L. F. Bernini, Br. J. Haematol., 1990, 76, 269–274 CrossRef CAS PubMed.
- A. M. Dozy and Y. W. Kan, Clin. Genet., 1994, 45, 221–227 CrossRef CAS PubMed.
- J. A. Tan, J. S. Tay, L. I. Lin, S. K. Kham, J. N. Chia, T. M. Chin, N. B. Aziz and H. B. Wong, Prenatal Diagn., 1994, 14, 1077–1082 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2017 |