Open Access Article
Open Access ArticleA novel, label-free fluorescence detection method for the formation of G-quadruplex DNA based on DNA-templated silver nanoclusters†
Jiaona Xu and
Chunying Wei *
*
Key Laboratory of Chemical Biology and Molecular Engineering of Ministry of Education, Institute of Molecular Science, Shanxi University, Taiyuan 030006, P. R. China. E-mail: weichuny@sxu.edu.cn
First published on 5th May 2017
Abstract
A novel, label-free fluorescence detection method was proposed for the formation of the G-quadruplex DNAs based on placing two DNA-Ag NCs together to light-up fluorescence. Herein, we designed two tailored DNA sequences noted as target DNA (abbreviated as T-DNA) and complementary DNA (abbreviated as C-DNA), respectively. T-DNA contains two cytosine-rich sequences at the 3′- and 5′-end responsible for Ag NCs synthesis, together with the guanine-rich (G-rich) DNA sequence in the middle segment. C-DNA was designed completely complementary to the G-rich segment of the T-DNA. Provided that the middle G-rich oligonucleotide of T-DNA folds into G-quadruplex in the presence of monovalent cations, the located at 3′ and 5′ terminuses two darkish DNA-Ag NCs approximate, leading to the enhanced fluorescence signal readout. Upon addition of the C-DNA, the G-quadruplex structure is unfolded and the two DNA-Ag NCs are separated, then the fluorescence signal is quenched. On the contrary, when the G-rich oligonucleotide with mutational bases failed to form the G-quadruplex, the fluorescence signal decreased clearly. This design allowed specific and general detection of the formation of diverse G-quadruplexes with easy operation and low cost. We believed that the proposed approach laid a foundation for the researchers in bioimaging, pharmacology, diagnostics, and other fields.
Introduction
During the past decades, DNA-templated silver nanoclusters (DNA-Ag NCs),1 as a new class of promising fluorescence nanomaterials, have been extensively used in biosensing2–5 and bioimaging.6,7 Compared with traditional luminescent probe,8,9 DNA-Ag NCs show advantages of stability, biocompatibility, ultra-small size, lower toxicity, and a strong fluorescence emission.10,11 Moreover, it has been demonstrated that the fluorescence emission and fluorescence quantum yield of Ag NCs could be regulated by modulating the sequence and length of DNA templates.Some G-rich DNA sequences can fold into four-stranded secondary structures, titled as G-quadruplex DNAs, which are formed by planar G-tetrads through Hoogsteen hydrogen bonding and interactions with monovalent cations.12 G-quadruplexes, widely located in plenty of important regulatory regions including telomeric DNA and oncogene promoters, play a significant role in regulation of genetic transcription, recombination, and replication.13 Thus, there is a need to develop techniques that allow the study of G-quadruplex, which puts the focus on the successful detection of the formation of the G-quadruplex DNAs in the first place. Nowadays, many kinds of methods have been developed for the detection of the formation of the G-quadruplex such as circular dichroism,14 electrochemistry,15 surface plasmon resonance,16 and fluorescence resonance energy transfer.17 However, they pose several challenges, such as the need for complex labeled processes, a high cost of operation, and sophisticated instrumentation.
Herein, based on the phenomenon that placing two darkish DNA-Ag NCs together can light-up fluorescence,18 we present a novel, label-free sensing platform to detect the formation of the G-quadruplex DNAs using DNA-Ag NCs as a fluorescence probe. As shown in Scheme 1, the DNA template (T-DNA) designed containing two components, the Ag NC-nucleation segment at the two terminuses and the G-rich oligonucleotide. In the presence of monovalent cations such as K+ and Na+, the G-rich oligonucleotide folded into G-quadruplex,19 leading to the two darkish DNA-Ag NCs approximated, thus the fluorescence intensity of DNA-Ag NCs rapidly increased. Upon addition of the complementary DNA (C-DNA) with the G-rich oligonucleotide, the G-quadruplex structure was opened and the two DNA-Ag NCs were separated, then the fluorescence of DNA-Ag NCs was quenched.
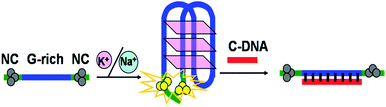 | ||
| Scheme 1 Schematic illustration of the assay strategy for the detection of the formation of the G-quadruplex DNAs. | ||
Experimental
Reagents and apparatus
Oligonucleotides used in this work were supplied by Shanghai Sangon Biotechnology Co. Ltd. (Shanghai, China), and the sequences of oligonucleotides were listed in Table S1.† Silver nitrate (AgNO3, 99.8%) and sodium borohydride (NaBH4, 98%) were purchased from Aladdin Bio-Chem Technology Co. Ltd. (Shanghai, China) and used without further purification. Phosphate buffer solution (PBS, pH 6.6) was used in all of the experiments. All solutions were prepared using Millipore Milli-Q water (18.2 MΩ cm).Fluorescence spectra were recorded at room temperature on a Fluoromax-4 Spectrofluorometer (Horiba Jobin Yvon Inc., France). The emission spectra were recorded with 5 nm slit widths for both excitation and emission. UV-vis absorbance measurements were performed on a Cary 50 UV-vis-NIR Spectrophotometer (Varian Inc., USA). All spectra were obtained by using a 1 cm path length quartz cell. The average size of DNA-Ag NCs was verified by transmission electron microscopy (TEM) using a JEOL JEM-2100 high resolution TEM instrument operated at 200 kV. A drop of clear aqueous solution was carefully placed on an ultrathin pure carbon film and dried at ambient condition for TEM characterization. Time-resolved fluorescence measurements were performed using FL920 Lifetime Fluorescence Spectrometer (Edinburgh Instruments, Livingston, UK) operating in the time-correlated single photon counting (TCSPC) mode with a semiconductor laser (405 nm) as the excitation source. For data analysis, commercial software by Edinburgh Instruments was used. The picosecond transient fluorescence decays were fitted with multi-exponential (n) function, 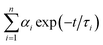 , where αi is weight percentages of the decay components with time constants of τi. The average excited state lifetime was expressed by the equation
, where αi is weight percentages of the decay components with time constants of τi. The average excited state lifetime was expressed by the equation 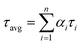 , when
, when  . The 1.5% agarose gel electrophoresis (AGE) analysis of the DNAs was carried out in 1× Tris–acetic–EDTA at a constant voltage of 110 V for about 40 min. The gel was scanned using the Alpha Imager (Protein Simple, USA). CD spectra were performed on a Chirascan Circular Dichroism Spectrometer (Applied Photophysics Ltd., Surrey, UK), and the spectrum was recorded from 220 to 320 nm at 1 nm intervals using a quartz cell of 1 mm optical path length and an instrument scanning speed of 120 nm min−1 at room temperature.
. The 1.5% agarose gel electrophoresis (AGE) analysis of the DNAs was carried out in 1× Tris–acetic–EDTA at a constant voltage of 110 V for about 40 min. The gel was scanned using the Alpha Imager (Protein Simple, USA). CD spectra were performed on a Chirascan Circular Dichroism Spectrometer (Applied Photophysics Ltd., Surrey, UK), and the spectrum was recorded from 220 to 320 nm at 1 nm intervals using a quartz cell of 1 mm optical path length and an instrument scanning speed of 120 nm min−1 at room temperature.
Preparation of DNA-Ag NCs
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6
6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and followed by the vigorous shaking of the solution for 30 s, the reaction mixture was incubated in the dark at 4 °C for one hour to form Ag NCs.
1) and followed by the vigorous shaking of the solution for 30 s, the reaction mixture was incubated in the dark at 4 °C for one hour to form Ag NCs.Results and discussion
The choice of the template DNA
Here we chose three different DNA sequences (TBA1, TBA2, and TBA3) as the templates (Table S1†) to form the T-DNA-Ag NCs, and their optical characterizations were shown in Fig. S1.† There was an apparent difference in the fluorescence intensity of three T-DNA-Ag NCs, and TBA2-based Ag NCs presented the strongest fluorescence, that is, there was the same Ag NC-nucleation segment in 5′- and 3′- terminus. As such, TBA2 was selected as the template to detect the formation of the G-quadruplex DNAs in the following experiments.The viability of the design
The veracity of the strategy was assessed first. The introduction of C-TBA2 (Table S1†) that can hybridize with G-rich segment (TBA) of sequence TBA2 resulted in the two DNA-Ag NCs to be separated, thus the fluorescence of DNA-Ag NCs would be quenched. TBA adopts an intramolecular chair type G-quadruplex structure in buffer containing K+ or Na+,20 although the structure is more stable for the former because Na+ has a low binding affinity for TBA.20,21 Here it was observed that the TBA2-Ag NCs showed the stronger fluorescence intensity in PBS buffer containing K+ than containing Na+ (Fig. S2†), suggesting the stable G-quadruplex structure brings the 5′-end and the 3′-end closer and gives the stronger emission. In order to improve the sensitivity of assay, we selected the buffer containing K+ to perform the following experiments.As we know that the concentration of K+ affects the stability of G-quadruplex,20,22 which might bring about the change of the fluorescence intensity of DNA-Ag NCs, meanwhile, the ionic strength also plays a key role in the formation of DNA-Ag NCs.23 In order to optimize the experimental conditions, different concentrations of K+ was added to the PBS buffer, and the F/F0 values were employed to evaluate the sensitivity of the assay. F is the fluorescence intensity of the TBA2-Ag NCs, and F0 is the fluorescence intensity of TBA2-Ag NCs after hybridization with the C-TBA2. Agarose gel electrophoresis (AGE) was used to examine whether DNA duplexes were formed after addition of C-DNA (Fig. S3†). Relative to that of TBA2-DNA (lanes 1), the slower migration band appeared in the TBA2-DNA/C-TBA2-DNAs (lanes 2), which indicated that the C-DNA could effectively hybridize to the corresponding T-DNA to form a duplex structure.
According to the results from Fig. 1 and S4,† the buffer containing 40 mM K+ gives the largest F/F0 value, thus it was chosen and used throughout experiments. It was found that the F/F0 value even reached about 16-fold in the condition of optimum buffer (Fig. 1A). There were two peaks at about 430 and 560 nm in the absorption spectra of TBA2-Ag NCs (Fig. 1B). Upon excitation at ∼560 nm, the obtained TBA2-Ag NCs showed a maximum emission at ∼620 nm (inset of Fig. 1B), while no fluorescence emission occurred upon excitation at ∼430 nm. The peak at ∼430 nm was corresponded to the absorption of Ag nanoparticles, only the peak at ∼560 nm was the characteristic absorption of the DNA-Ag NCs.3 So we chose 560 nm as the wavelength of excitation to measure the emission spectra of the TBA2-Ag NCs (Fig. 1C).
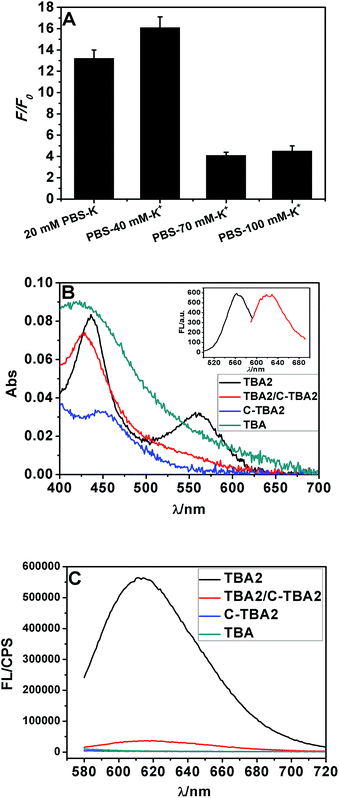 | ||
| Fig. 1 The Ag NCs synthesized with the sequences TBA2, C-TBA2, TBA and TBA2/C-TBA2 in Table S1† as the templates in phosphate buffer containing 40 mM K+. (A) The effect of ionic strength on the fluorescence intensity of Ag NCs. (B) UV-vis absorption spectra and (C) fluorescence emission spectra of TBA2-, TBA2/C-TBA2-, C-TBA2- and TBA-Ag NCs (λex = 560 nm). The inset showed the excitation (λmax = 560 nm) and emission spectra (λmax = 620 nm). | ||
Considering that Ag+ or Ag NCs may destabilize the duplex or the G-quadruplex to produce the G-rich and C-rich single strand in solution, and these sequences would be form the other DNA-Ag NCs that might affect our results, herein we separately measured the absorption and fluorescence spectra of the Ag NCs based on G-rich sequence be capable of folding the G-quadruplex (TBA) and their complementary C-rich sequence (C-TBA2). As shown in Fig. 1B and C, both C-TBA2- and TBA-Ag NCs fail to give other absorbance peaks except a plasma absorption band about 430 nm, expectedly, the fluorescence emission were not also observed upon excited at 560 nm. Thus we can conclude that the fluorescence DNA-Ag NCs was still formed at the CCCTTAATCCCC segment of TBA2, even if Ag+ or Ag NCs could destabilize the duplex DNA or G-quadruplex.
To confirm whether the formation of Ag NCs had an effect on the conformation of TBA G-quadruplex or not, CD spectrum of TBA2-AgNCs were measured. As shown in Fig. S5,† the CD spectrum presented a broad positive band centered at 275 nm and a negative band at 245 nm, and the positions of both peaks obviously deviated from those in TBA G-quadruplex itself, where a strong positive band at 295 nm and a weak negative band at 270 nm.20 We speculated that the CD spectrum of TBA2-AgNCs might be largely attributed to the conformation of the closed ssDNA-Ag NCs containing 12 bases and another 3 complementary base pairs (AAA/TTT) located at 3′- and 5′-terminus, that is, the characteristic peak at 295 nm of G-quadruplex were buried in this broad positive band, and the other negative peak at 270 nm was surpassed by the positive peak located at same position. It was reasonable that the 15 bases long sequences located at 3′- and 5′-end were responsible for the whole conformation of TBA2-AgNCs, because the TBA G-quadruplex itself contains only two tetrads.
Sensitivity of the detection system
To investigate the selectivity of the sensing platform and the possibility of detecting mutations, two-base mutational G-rich oligonucleotide segment of TBA2 (M-TBA2) was selected to study the specificity of the method under the optimal condition. It was found that both absorption peak and fluorescence intensity of M-TBA2-Ag NCs were lower than those of TBA2-Ag NCs (Fig. 2), which should result from the fact that mutational G-rich oligonucleotide cannot form G-quadruplex, leading to the two DNA-Ag NCs cannot approach. The results indicated that the proposed strategy had good selectivity for the detection of G-quadruplex DNAs formation.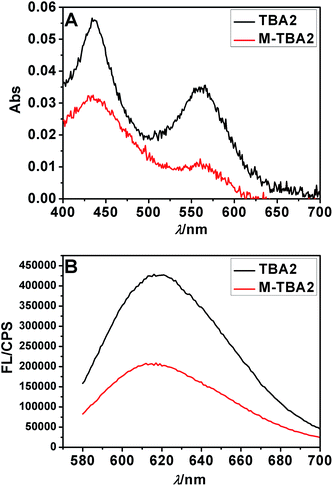 | ||
| Fig. 2 The Ag NCs synthesized with the sequences TBA2 and M-TBA2 in Table S1† as the templates. UV-vis absorption spectra (A) and fluorescence emission spectra (B). | ||
TEM and lifetime analysis
Fig. S6† showed the typical TEM images of the Ag NCs, and the histogram of cluster size distribution was given in insets. The results revealed that the as-prepared DNA-Ag NCs had an average size of 2.0 ± 0.3 nm and good dispersibility. There were no distinct changes in the average size of TBA2-, TBA2/C-TBA2-, and M-TBA2-Ag NCs, suggesting the Ag NCs were not aggregated into the large nanoparticles.To further elucidate the quenching mechanism, the time-correlated single photon counting (TCSPC) technique was used to measure the lifetimes of Ag NCs (Fig. S7†). The fluorescence transients of Ag NCs presented three exponential time constants as tabulated in Table S2.† According to the results in Table S2,† there were no obvious changes in the average fluorescence lifetimes of TBA2-, TBA2/C-TB2-, and M-TBA2-Ag NCs.
Versatility of the detection system
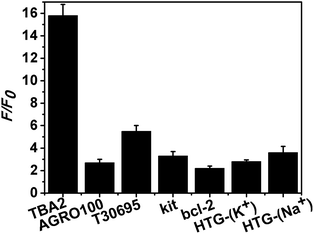
Thus far, we have demonstrated the feasibility of this novel method. In order to validate its general applicability in detecting the formation of the G-quadruplex DNAs, we further chose other G-rich oligonucleotides that can fold into G-quadruplexes, such as AGRO100,24 T30695,25 kit,26 bcl-2,27 and HTG.28 The G-rich segment of TBA2 was replaced by these G-rich sequences, respectively, and their sequences were given in Table S1.† As shown in Fig. S8,† after hybridization with the corresponding C-DNA (Fig. S3†), the absorption peak and the fluorescence intensity of the T-DNA-Ag NCs were obviously decreased. Besides, both the G-rich (O-DNA) and their complementary C-rich sequence (C-DNA) sequences did not affect the fluorescence of duplex-Ag NCs. Although C-kit- and C-bcl-2-Ag NCs presented a strong fluorescence because both C-rich sequences might be good templates to form DNA-Ag NCs, their maximum emission wavelengths were obviously different from those the kit- and bcl-2-AgNCs. Importantly, based on these results, we could conclude that Ag+ or Ag NCs failed to destabilize the duplex DNA because the same peak positions of duplex- and T-DNA-Ag NCs were observed for kit and bcl-2, meanwhile, the fluorescence intensity of duplex-Ag NCs was lower than that of either T-DNA or C-DNA. If Ag+ or Ag NCs destroyed the structure of duplex DNA, the produced C-rich and T-DNA single-stranded templates should synthesized the respective fluorescence Ag NCs. Due to the common contribution of the two kinds of Ag NCs, it would not only result in the blue shift of the peak position of duplex-Ag NCs, but also the fluorescence intensity of duplex-Ag NCs should be stronger than that of either T-DNA- or C-DNA-Ag NCs. In conclusion, this novel method had the specific ability for detection of the formation of G-quadruplex DNAs.According to the results from Fig. 3, the F/F0 value of TBA2 was larger than the others, which should be attributed to the structural difference of the G-quadruplex DNAs. It was reported that G-rich oligonucleotides of TBA2 can form chair-type intramolecular G-quadruplex in the presence of K+,20 resulting in the located at 5′ and 3′ termina two darkish DNA-Ag NCs more approximated than the other G-quadruplex structures including parallel-stranded (AGRO100,24 T30695,25 and kit,26), mixed parallel/antiparallel-stranded (bcl-2, HTG in buffer containing K+),27,28 and basket-type (HTG in buffer containing Na+) conformations.28 In addition, the F/F0 values of the kit- and AGRO100-Ag NCs were lower than that of the T30695-Ag NCs while these DNAs have parallel conformation, which might be due to the difference of DNA sequences and microenvironment that affects the fluorescence of Ag NCs. In the case of HTG G-quadruplex, the located 5′ and 3′ termina two darkish DNA-Ag NCs were closer in buffer containing Na+ (basket-type structure) than containing K+ (mixed parallel/antiparallel-stranded structure), expectedly, it was observed that the fluorescence intensity of the former was stronger than that of the latter. In short, the F/F0 value was closely related to the structure of G-quadruplex DNA, and the more closer the 5′ and 3′ termina was, the larger the F/F0 was.
Conclusions
In summary, a novel and label-free fluorescence detection method for the formation of the G-quadruplex DNAs was developed based on placing two darkish DNA-Ag NCs together light-up system. This novel approach presented a universal feasibility for variety of G-quadruplex structures whereas the chair-type structure of TBA gave the largest sensitivity among them, that is, the assay partially depended on the structure of G-quadruplex DNAs, the more closer the 5′ and 3′ termina was, the higher the sensitivity was. In addition, the ionic strength, DNA sequences and the stability of G-quadruplex structures also affect the sensitivity of assay. This simple and cost-effective design may provide a flexible strategy for the G-quadruplex DNAs based researches.Acknowledgements
We greatly appreciate financial support from the National Natural Science Foundation of China (21171108) and Shanxi Provincial Fund for Natural Sciences (201601D011026).Notes and references
- B. Han and E. Wang, Anal. Bioanal. Chem., 2012, 402, 129–138 CrossRef CAS PubMed.
- K. Zhang, K. Wang, X. Zhu, Y. Gao and M. Xie, Chem. Commun., 2014, 50, 14221–14224 RSC.
- K. Zhang, T. Ren, K. Wang, X. Zhu, H. Wu and M. Xie, Chem. Commun., 2014, 50, 13342–13345 RSC.
- K. Zhang, K. Wang, M. Xie, X. Zhu, L. Xu, R. Yang, B. Huang and X. Zhu, Biosens. Bioelectron., 2014, 52, 124–128 CrossRef CAS PubMed.
- J. Xu and C. Wei, Biosens. Bioelectron., 2017, 87, 422–427 CrossRef CAS PubMed.
- J. Yin, X. He, K. Wang, Z. Qing, X. Wu, H. Shi and X. Yang, Nanoscale, 2012, 4, 110–112 RSC.
- J. Yin, X. He, K. Wang, F. Xu, J. Shangguan, D. He and H. Shi, Anal. Chem., 2013, 85, 12011–12019 CrossRef CAS PubMed.
- Z. Guo, L. Murphy, V. Stein, W. A. Johnston, S. Alcala-Perez and K. Alexandrov, J. Am. Chem. Soc., 2016, 138, 10108–10111 CrossRef CAS PubMed.
- S. Lin, W. Gao, Z. Tian, C. Yang, L. Lu, J.-L. Mergny, C.-H. Leung and D.-L. Ma, Chem. Sci., 2015, 6, 4284–4290 RSC.
- I. Diez, M. I. Kanyuk, A. P. Demchenko, A. Walther, H. Jiang, O. Ikkala and R. H. Ras, Nanoscale, 2012, 4, 4434–4437 RSC.
- N. Enkin, E. Sharon, E. Golub and I. Willner, Nano Lett., 2014, 14, 4918–4922 CrossRef CAS PubMed.
- Y. Wu and R. M. Brosh, FEBS J., 2010, 277, 3470–3488 CrossRef CAS PubMed.
- S. Müller, S. Kumari, R. Rodriguez and S. Balasubramanian, Nat. Chem., 2010, 2, 1095–1098 CrossRef PubMed.
- P. L. T. Tran, J.-L. Mergny and P. Alberti, Nucleic Acids Res., 2011, 39, 3282–3294 CrossRef CAS PubMed.
- Y. Fan, X. Chen, J. Kong, C. Tung and Z. Gao, Angew. Chem., 2007, 119, 2097–2100 CrossRef.
- L. Tang, I. S. Chun, I. Wang, J. Li, X. Li and Y. Lu, Anal. Chem., 2013, 85, 9522–9527 CrossRef CAS PubMed.
- C. Lu, H. Yang, C. Zhu, X. Chen and G. Chen, Angew. Chem., 2009, 121, 4879–4881 CrossRef.
- B. Yin, J. Ma, H.-N. Le, S. Wang, Z. Xu and B. Ye, Chem. Commun., 2014, 50, 15991–15994 RSC.
- D. Sen and W. Gilbert, Nature, 1990, 344, 410–414 CrossRef CAS PubMed.
- S. Nagatoishi, Y. Tanaka and K. Tsumoto, Biochem. Biophys. Res. Commun., 2007, 352, 812–817 CrossRef CAS PubMed.
- B. I. Kankia and L. A. Marky, J. Am. Chem. Soc., 2001, 123, 10799–10804 CrossRef CAS PubMed.
- J.-L. Mergny, A.-T. Phan and L. Lacroix, FEBS Lett., 1998, 435, 74–78 CrossRef CAS PubMed.
- W. Chen, G. Lan and H.-T. Chang, Anal. Chem., 2011, 83, 9450–9455 CrossRef CAS PubMed.
- V. Đapić, P. J. Bates, J. O. Trent, A. Rodger, S. D. Thomas and D. M. Miller, Biochemistry, 2002, 41, 3676–3685 CrossRef.
- T. Li, E. Wang and S. Dong, J. Am. Chem. Soc., 2009, 131, 15082–15083 CrossRef CAS PubMed.
- G. R. Vandenbark, Y. Chen, E. Friday, K. Pavlik, B. Anthony, C. deCastro and R. E. Kaufman, Cell Growth Differ., 1996, 7, 1383–1392 CAS.
- J. Dai, T. S. Dexheimer, D. Chen, M. Carver, A. Ambrus, R. A. Jones and D. Yang, J. Am. Chem. Soc., 2006, 128, 1096–1098 CrossRef CAS PubMed.
- A. T. Phan, V. Kuryavyi, K. N. Luu and D. J. Patel, Nucleic Acids Res., 2007, 35, 6517–6525 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Additional figures and DNA sequences for detection. See DOI: 10.1039/c7ra02984e |
| This journal is © The Royal Society of Chemistry 2017 |