Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Pathogen inactivation of cruciferous phytoalexins: detoxification reactions, enzymes and inhibitors
M. Soledade C. Pedras
 * and
Abbas Abdoli
* and
Abbas Abdoli

Department of Chemistry, University of Saskatchewan, 110 Science Place, Saskatoon, SK S7N 5C9, Canada. E-mail: s.pedras@usask.ca; Fax: +1-306-966-4730; Tel: +1-306-966-4772
First published on 2nd May 2017
Abstract
Phytoalexins (elicited antimicrobial plant defenses) that are resistant to pathogen degradation provide the producing plants with higher disease resistance levels, and, conversely, their transformation by pathogens makes plants more susceptible to diseases. In this context, the transformation of cruciferous phytoalexins carried out by fungal pathogens is of great concern due to the global significance of cruciferous crops. This review covers the detoxification pathways of cruciferous phytoalexins, the corresponding detoxifying enzymes and their natural and synthetic inhibitors. Paldoxins (inhibitors of fungal detoxifying enzymes) are examined as a potentially sustainable strategy to control plant pathogenic fungi. As a spin-off of the reviewed work, some of the biotransformation reactions could be applied to selectively convert more complex molecules to novel or known products.
1. Introduction
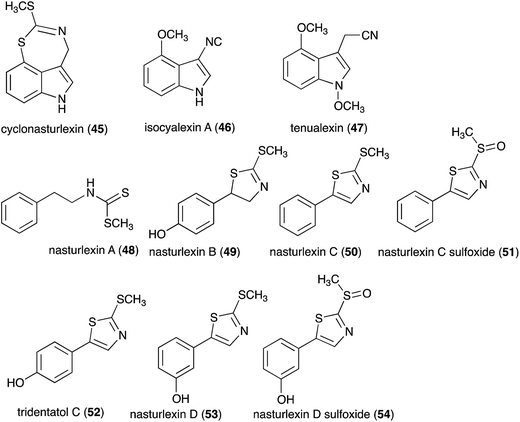
Pathogens and plants have co-evolved for centuries in an arms race that continues to escalate.1,2 While plants can recognize pathogen signals and prevent invasion using various defense mechanisms, in certain circumstances pathogens are able to evade these mechanisms. Plant defense mechanisms involve numerous biosynthetic pathways that produce a great diversity of secondary metabolites (natural products) with specific roles. Some plant defense mechanisms are constitutive and others are induced, i.e. expressed as a response to challenges posed by pathogens, pests, or environmental factors. Induced mechanisms provide the plant with an enhanced level of resistance against a range of invaders such as fungal pathogens.3 Phytoalexins are antimicrobial plant metabolites part of the elicited defense responses caused by biotic and abiotic stress, thus are not synthesized by constitutive metabolic pathways.4 The chemical structures of phytoalexins have diverse biosynthetic origins, and are usually related within the same plant family. For example, the majority of the cruciferous phytoalexins reported to date are derived from (S)-tryptophan,4–7 but a new structural group derived from (S)-phenylalanine has been recently reported.8,9 Cruciferous phytoalexins display a wide range of antifungal activities against different phytopathogenic fungal species. Crucifers are economically important crops that include sources of edible and industrial oils (canola, mustard, rapeseed), vegetables (cabbage, cauliflower, broccoli, turnip, rutabaga) and condiments (horseradish, mustard, wasabi).Between 1986, when the first cruciferous phytoalexins were reported, and 2011, the chemical structures, syntheses and antimicrobial activities of 44 cruciferous phytoalexins were reported.4 Since the last comprehensive review of cruciferous phytoalexins was published in 2011 (Fig. 1),4 ten new phytoalexins have been reported: cyclonasturlexin (45),5 isocyalexin A (46),10 tenualexin (47),7 nasturlexins 48–51, 53, 54 (ref. 8 and 9) and tridentatol C (52)8 (Fig. 2). Isocyalexin A (46) was the first isocyanide (or isonitrile, –NC) reported from the plant kingdom, although cyanide (or nitriles, –CN) containing metabolites are common in plants. For example, tenualexin (47), isolated from wall rocket,7 appears to be the most potent antifungal compound among the nitrile containing phytoalexins (indolyl-3-acetonitrile (38), caulilexin C (39) and arvelexin (40)).11 Interestingly, watercress is the only crucifer species reported that biosynthesizes phytoalexins using two independent pathways, initiated from the primary precursors amino acids (S)-tryptophan and (S)-phenylalanine. To date, only (S)-phenylalanine derived phytoalexins, the nasturlexins 48–51, 53, 54 and tridentatol C (52), but no indolyl phytoalexins, were reported from winter cress and upland cress. Nitrile containing phytoalexins such as tenualexin (47) are likely biosynthesized from the corresponding indolyl acetaldoximes, while cyclonasturlexin (45) is biosynthetically derived from brassinin (1) via the indolyl glucosinolate glucobrassicin.5 Considering the large number of crucifer species (more than 3700 species) and the relatively low number of species (<50) analyzed for phytoalexin production, it is likely that phytoalexin structures deriving from phenylalanine/tyrosine and their homologues will match the variety of functional groups present in those derived from (S)-tryptophan.
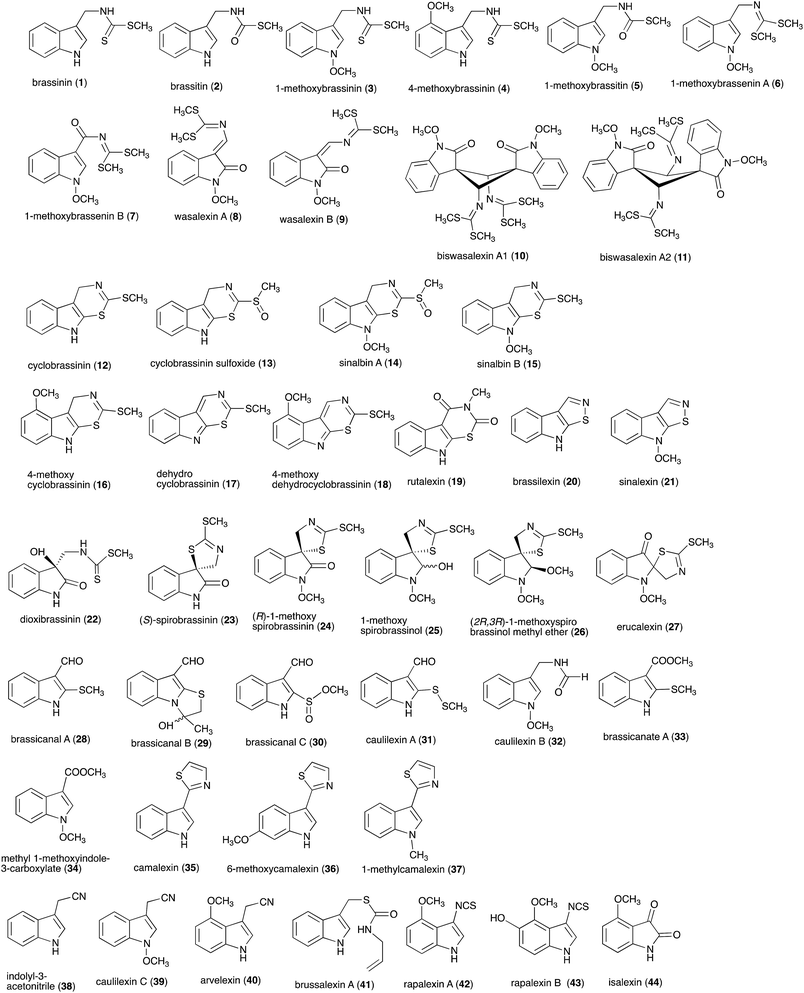 | ||
| Fig. 1 Chemical structures and names of cruciferous phytoalexins 1–44 reported up to 2011.4 | ||
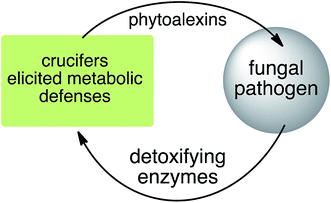
Phytoalexins resistant to pathogen detoxyfication provide the producing plants with higher disease resistance levels, and, conversely, their transformations by pathogens make plants more susceptible to diseases. Our group has pioneered work on the transformation of crucifer phytoalexins by plant pathogens and is one of the very few working in the field. We have been investigating the metabolic transformations of various crucifer phytoalexins by fungal plant pathogens since 1991 (ref. 12) with the goal of developing inhibitors of these reactions as crop protection agents. An overview of the catabolic pathways of phytoalexins, their products, enzymes and inhibitors is reported.
2. Biotransformation and detoxification of phytoalexins
Plant fungal pathogens are able to elude plant defense mechanisms using a variety of strategies that include production of enzymes with diverse catalytic activities.13 For example, some fungi carry out effective detoxification of metabolites such as phytoalexins using detoxifying enzymes that exhaust these valuable defenses (Fig. 3).14 Fungal pathogens of crucifers such as Alternaria brassicicola (Schwein.) Wiltshire, Botrytis cinerea Pers. Fr. (teleomorph Botryotinia fuckeliana (de Bary) Whetzel), Leptosphaeria maculans (Desm.) Ces. et Not. [asexual stage Phoma lingam (Tode ex Fr.) Desm.] (canola and mustard virulent isolates), L. biglobosa, Rhizoctonia solani Kuhn and Sclerotinia sclerotiorum (Lib.) de Bary were shown to detoxify cruciferous phytoalexins using different reaction types.4 The type of reaction carried out (e.g. oxidation, reduction, hydrolysis) depends on both the fungal species and the chemical structure of the phytoalexin.2.1 Brassinin and cyclobrassinin
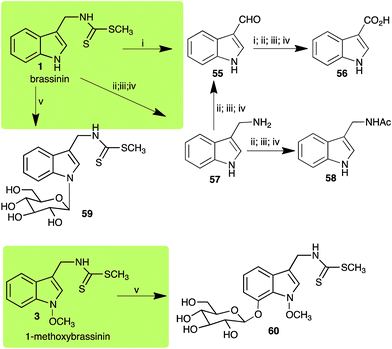
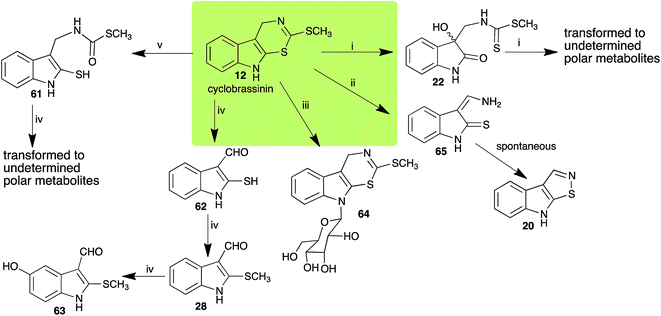
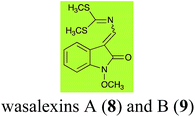
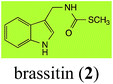
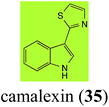
Brassinin (1), 1-methoxybrassinin (3) and cyclobrassinin (12), the first cruciferous phytoalexins reported,15 are produced mainly by Brassica species.4,16 Brassinins 1, 3 and 4 are the first naturally occurring dithiocarbamates reported from plants; coincidentally, synthetic dithiocarbamates have been used as pesticides for decades.17 As summarized in Scheme 1, brassinin (1) was detoxified by several fungal species (A. brassicicola,18 L. biglobosa,19 L. maculans canola virulent isolates,12 S. sclerotiorum20 and B. cinerea21) to metabolites 55–59. The transformation of brassinin (1) to indole-3-carboxaldehyde (55) is the only enzymatic transformation of a dithiocarbamate group to an aldehyde group12 reported to date. Similarly, the metabolism of cyclobrassinin (12) by different fungal species (A. brassicicola,22 L. biglobosa,23,24 L. maculans canola virulent isolates,23,24 R. solani,25 S. sclerotiorum20) was found to yield different products (Scheme 2). Surprisingly, some of these intermediate products were shown to be phytoalexins, which were further metabolized to nontoxic products. To date, S. sclerotiorum is the only fungal pathogen reported to use glucosylation for detoxification of phytoalexins.Brassinin detoxifying enzymes produced by A. brassicicola (brassinin hydrolase, BHAb), L. maculans mustard virulent isolates (brassinin hydrolase, BHLm)26 and L. maculans canola virulent isolates (brassinin oxidase, BOLm)27 were isolated from wild type fungal isolates, purified to homogeneity and characterized. BOLm was the first phytoalexin detoxifying enzyme purified and characterized, a milestone achievement. The gene for brassinin glucosyl transferase (BGTSs) from S. sclerotiorum was cloned and expressed in Saccharomyces cerevisiae. The purified recombinant enzyme was able to glucosylate brassinin effectively.28 BOLm, BHLm and BHAb are elicited detoxifying enzymes, induced by stress, not produced under normal culture conditions. Importantly, it was established that BHAb and BHLm have different physical properties and substrate specificities, although both enzymes catalyze the hydrolysis of brassinin (1) to the corresponding amine (57).26 More recently, cyclobrassinin hydrolase was isolated from A. brassicicola (CHAb), purified to homogeneity and shown to have different substrate specificity and characteristics from those of BHAb.29 Unexpectedly, it was discovered that the cruciferous phytoalexins cyclobrassinin (12), camalexin (35) and wasalexins A (8) and B (9), inhibited BOLm activity,27 whereas cyclobrassinin (12) inhibited BHAb and BHLm,26 and camalexin (35), dioxibrassinin (22) and brassilexin (20) inhibited CHAb (Table 1).29 These discoveries suggest that phytoalexin blends produced in crucifers have multiple functions, including inhibition of specific fungal detoxifying enzymes, in addition to the role of inhibiting the growth of microbial pathogens.
| Inhibitors (#) | Enzymea: % inhibition | Inhibitors (#) | Enzymea: % inhibition |
|---|---|---|---|
| a BOLm: brassinin oxidase from Leptosphaeria maculans (canola virulent isolates); BHAb: brassinin hydrolase from Alternaria brassicicola; BHLm: brassinin hydrolase from L. maculans (brown mustard virulent isolates) CHAb: cyclobrassinin hydrolase from A. brassicicola. Inhibitors at 0.30 mM; brassinin (1) at 0.10 mM. | |||
 |
BOLm: 37 ± 8 |  |
BOLm: 16 ± 2 |
| BHAb: 26 ± 4 | CHAb: 93 ± 2 | ||
 |
BOLm: 14 ± 4 | 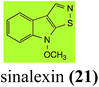 |
CHAb: 52 ± 2 |
 |
BHAb: 18 ± 2 | 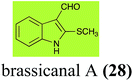 |
CHAb: 43 ± 3 |
 |
BOLm: 53 ± 4 | 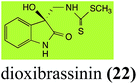 |
CHAb: 58 ± 5 |
| CHAb: 65 ± 5 | |||
 |
BOLm: 63 ± 5 | 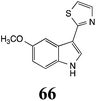 |
BOLm: 72 ± 1 |
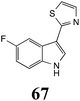 |
BOLm: 63 ± 2 | 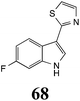 |
BOLm: 46 ± 2 |
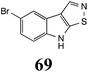 |
BOLm: 45 ± 2 | 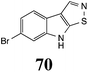 |
BOLm: 63 ± 4 |
 |
BOLm: 14 ± 6 | 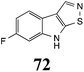 |
BOLm: 40 ± 5 |
 |
BOLm: 40 ± 2 | 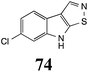 |
BOLm: 66 ± 7 |
 |
BOLm: 18 ± 4 | 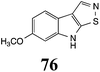 |
BOLm: 38 ± 4 |
 |
CHAb: 73 ± 5 | 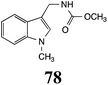 |
BHAb: 46 ± 3 |
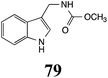 |
BHAb: 21 ± 4 | 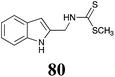 |
BOLm: 23 ± 6 |
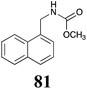 |
BHAb: 89 ± 2 | 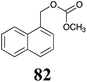 |
BHAb: 62 ± 4 |
 |
BOLm: 25 ± 7 |  |
BHAb: 52 ± 3 |
The design of selective inhibitors (coined paldoxins, phytoalexin detoxification inhibitors) of these fungal detoxifying enzymes was proposed as a strategy to protect crops that produce the corresponding phytoalexins.30 Selective inhibitors of such detoxifying enzymes, i.e. paldoxins, are less likely to affect non-targeted organisms, thus are anticipated to have lower impact on ecosystems and become a sustainable strategy to control plant pathogens. Paldoxins could prevent the depletion of the plants' natural defenses by the detoxifying enzymes of plant pathogens. By definition, the ideal paldoxins would not be toxic to living organisms, whether animal, plant or microbe.31
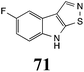
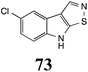
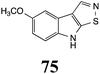
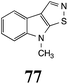
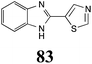
Initially, potential paldoxins of BOLm were designed by replacement of the dithiocarbamate group of brassinin (1) with various functional groups and of the indolyl moiety with naphthalenyl and phenyl moieties.32 In vitro screening of this synthetic library using purified BOLm indicated that none of the synthetic compounds were inhibitory to BOLm, but demonstrated for the first time that the phytoalexins camalexin (35) and cyclobrassinin (12) were competitive inhibitors of BOLm.27 Later on, compounds with scaffolds based on various phytoalexins (Table 1), including camalexin (35)33 and brassilexin (20),34 were shown to inhibit BOLm. 6-Chlorobrassilexin (74) and 5-methoxycamalexin (66) were determined to be the best competitive inhibitors of BOLm to date (Table 1).34 However, due to their substantial antifungal activity against L. maculans, these inhibitors are not considered paldoxins. This antifungal activity is not surprising considering that the designed inhibitor structures were based on phytoalexins. More recently, synthetic compounds that did not contain the indole nucleus and displayed low antifungal activity were found to inhibit BHAb,35 suggesting that such potential paldoxins could be applied to protect mustard crops against A. brassicicola.
| Phytoalexin (#) Scheme # | Fungal species: transformation product(s) (#) |
|---|---|
| 1-Methoxybrassinin (3) Scheme 1 | Sclerotinia sclerotiorum: 7-O-glucosyl-1-methoxybrassinin (60) |
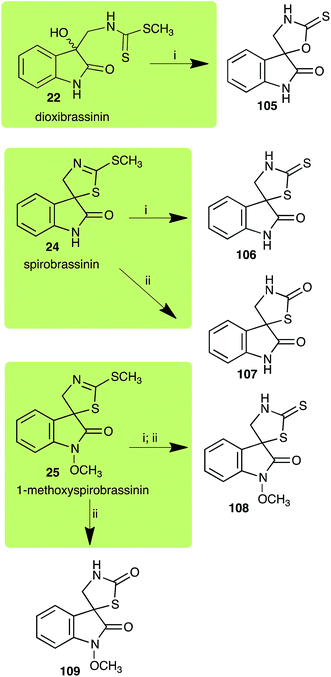
| 1-Methoxyspirobrassinin (25) Scheme 7 | Leptosphaeria maculans (brown mustard virulent isolates): 1-methoxy-2′-thioxospiro[indole-3,5′-thiazolidin]-2-one (108) |
| S. sclerotiorum: 1-methoxyspiro[indole-3,5′-thiazolidin]-2,2′-dione (109); 1-methoxy-2′-thioxospiro[indole-3,5′-thiazolidin]-2-one (108) | |
| 1-Methylcamalexin (37) Scheme 10 | Alternaria brassicicola: 1-methylindole-3-thiocarboxamide (118); 1-methylindole-3-carbonitrile (119); 1-methylindole-3-carboxylic acid (120) |
| Rhizoctonia solani: 2-(1-methyl-3-indolyl)-oxazoline (121); 1-methylindole-3-carboxamide (122); 1-methylindole-3-carbonitrile (123) | |
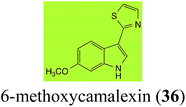
| 6-Methoxycamalexin (36) Scheme 11 | A. brassicicola: 6-methoxyindole-3-thiocarboxamide (124); 6-methoxyindole-3-carbonitrile (125); 6-methoxyindole-3-carboxylic acid (126) |
| S. sclerotiorum: 1-glucosyl-6-methoxycamalexin (127) | |
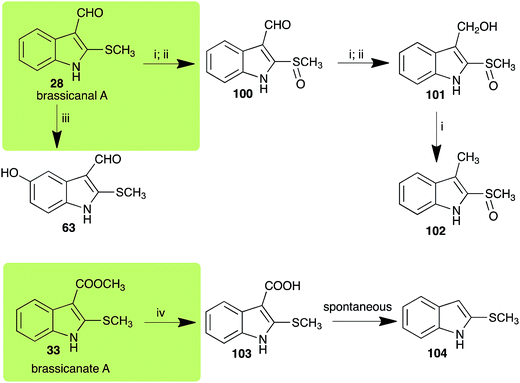
| Brassicanal A (28) Scheme 6 | L. maculans (canola virulent isolates): brassicanal A sulfoxide (100); 3-(hydroxymethyl)indole-2-methylsulfoxide (101); 3-methylindole-2-methylsulfoxide (102) |
| R. solani: 5-hydroxybrassicanal A (63) | |
| S. sclerotiorum: brassicanal A sulfoxide (100); 3-(hydroxymethyl)indole-2-methylsulfoxide (101) | |
| Brassicanate A (33) Scheme 6 | A. brassicicola: 2-methylsulfanylindole-3-carboxylic acid (103) |
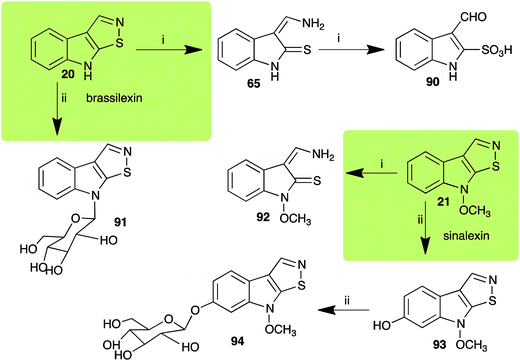
| Brassilexin (20) Scheme 4 | L. maculans (canola virulent isolates): 3-aminomethyleneindole-2-thione (65); 3-formylindole-2-sulfonic acid (90) |
| S. sclerotiorum: 1-glucosylbrassilexin (91) | |
| Brassinin (1) Scheme 1 | A. brassicicola: indole-3-methanamine (57); indole-3-carboxaldehyde (55); indole-3-carboxylic acid (56); N′-acetylindole-3-methanamine (58) |
| B. cinerea: indole-3-methanamine (57); indole-3-carboxaldehyde (55); indole-3-carboxylic acid (56); N′-acetylindole-3-methanamine (58) | |
| L. biglobosa: indole-3-methanamine (57); indole-3-carboxaldehyde (55); indole-3-carboxylic acid (56); N′-acetylindole-3-methanamine (58) | |
| L. maculans (canola virulent isolates): indole-3-carboxaldehyde (55); indole-3-carboxylic acid (56) | |
| L. maculans (brown mustard virulent isolates): indole-3-methanamine (57); indole-3-carboxaldehyde (55); indole-3-carboxylic acid (56); N′-acetylindole-3-methanamine (58) | |
| S. sclerotiorum: 1-glucosylbrassinin (59) | |
| Camalexin (35) Scheme 9 | A. brassicicola: indole-3-thiocarboxamide (114); indole-3-carbonitrile (115); indole-3-carboxylic acid (56) |
| B. cinerea: indole-3-thiocarboxamide (114); indole-3-carbonitrile (115); indole-3-carboxylic acid (56) | |
| R. solani: 5-hydroxycamalexin (111); 2-formamidophenyl-5-hydroxy-2′-thiazolylketone (112); 5-hydroxyindole-3-carbonitrile (113) | |
| S. sclerotiorum: 6-hydroxycamalexin (116); 6-O-glucosylcamalexin (117) | |
| Cyclobrassinin (12) Scheme 2 | A. brassicicola: S-methyl[(2-sulfanylindolyl-3)methyl]thiocarbamate (61) |
| L. maculans (canola virulent isolates): dioxibrassinin (22) | |
| L. biglobosa: 3-aminomethyleneindole-2-thione (65) | |
| R. solani: 2-sulfanylindole-3-carboxaldehyde (62); brassicanal A (28); 5-hydroxybrassicanal A (63) | |
| S. sclerotiorum: 1-glucosylcyclobrassinin (64) | |
| Dioxibrassinin (22) Scheme 7 | L. maculans (brown mustard virulent isolates): 2′-thioxospiro[indole-3,5′-oxazolidin]-2-one (105) |
| Erucalexin (27) Scheme 8 | L. maculans (canola virulent isolates): dihydroerucalexin (110) |
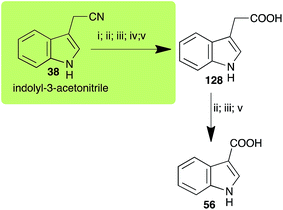
| Indolyl-3-acetonitrile (38) Scheme 12 | A. brassicicola: indolyl-3-acetic acid (128); indole-3-carboxylic acid (56) |
| L. maculans (canola virulent isolates): indolyl-3-acetic acid (128) | |
| L. maculans (brown mustard virulent isolates): indolyl-3-acetic acid (128) indole-3-carboxylic acid (56) | |
| R. solani: indolyl-3-acetic acid (128); indole-3-carboxylic acid (56) | |
| S. sclerotiorum: indolyl-3-acetic acid (128) | |
| Isalexin (44) Scheme 5 | A. brassicicola: 1,3-dihydro-3-hydroxy-4-methoxyindol-2-one (99) |
| Rutalexin (19) Scheme 5 | A. brassicicola: N-methyl 2-sulfanylindole-3-carboxamide (95); rutapyrone (96) |
| Sinalexin (21) Scheme 4 | L. maculans (canola virulent isolates): 3-aminomethylene-1-methoxyindoline-2-thione (92) |
| S. sclerotiorum: 7-hydroxysinalexin (93); 7-O-glucosylsinalexin (94) | |
| Spirobrassinin (24) Scheme 7 | L. maculans (brown mustard virulent isolates): 2′-thioxospiro[indole-3,5′-thiazolidin]-2-one (106) |
| S. sclerotiorum: spiro[indole-3,5′-thiazolidin]-2,2′-dione (107) | |
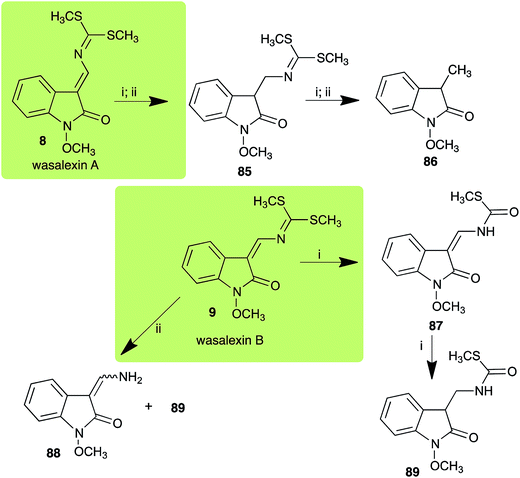
| Wasalexin A (8) Scheme 3 | L. maculans (canola virulent isolates): dihydrowasalexin (85); 3-methyl-1-methoxy-2-oxindole (86) |
| L. maculans (brown mustard virulent isolates): dihydrowasalexin (85); 3-methyl-1-methoxy-2-oxindole (86) | |
| Wasalexin B (9) Scheme 3 | L. maculans (canola virulent isolates): S-methyl 1-methoxy-3-aminomethylene-2-oxindole thiocarbamate (87); methyl 1-methoxy-3-aminomethyl-2-oxindole dithiocarbamate (89) |
| L. maculans (brown mustard virulent isolates): 3-(aminomethylene)-1,3-dihydro-1-methoxyindol-2-one (88); methyl 1-methoxy-3-aminomethyl-2-oxindole dithiocarbamate (89) |
2.2 Wasalexins A and B
Wasalexins A (8) and B (9) were isolated from the crucifers wasabi36 and salt cress,37 whereas only wasalexin A (8) was isolated from stinkweed.38 Wasalexins A (8) and B (9) were obtained by synthesis as a mixture of E (8) and Z (9) isomers in a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio.39 Although the more stable E isomer (8) could be separated by crystallization of the synthetic mixture, 9 could not be obtained as a single compound. For this reason, metabolic studies were conducted with 8 as a single compound, but investigation of the metabolism of 9 was conducted using a mixture of 8 and 9 (2
1 ratio.39 Although the more stable E isomer (8) could be separated by crystallization of the synthetic mixture, 9 could not be obtained as a single compound. For this reason, metabolic studies were conducted with 8 as a single compound, but investigation of the metabolism of 9 was conducted using a mixture of 8 and 9 (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio).40 It was determined that wasalexin A (8) detoxified by both canola and brown mustard virulent isolates of L. maculans via reduction of the double bond in the side chain of the oxoindole ring, as summarized in Scheme 3.40 Because both wasalexins A (8) and B (9) were detoxified by L. maculans canola and brown mustard isolates, a correlation between the production of wasalexins by the three plant species (all reported to be resistant to L. maculans canola virulent isolates) and their disease resistance to L. maculans could not be established. It is conceivable that in planta, additional metabolites are produced that inhibit the metabolism of wasalexins by L. maculans.
1 ratio).40 It was determined that wasalexin A (8) detoxified by both canola and brown mustard virulent isolates of L. maculans via reduction of the double bond in the side chain of the oxoindole ring, as summarized in Scheme 3.40 Because both wasalexins A (8) and B (9) were detoxified by L. maculans canola and brown mustard isolates, a correlation between the production of wasalexins by the three plant species (all reported to be resistant to L. maculans canola virulent isolates) and their disease resistance to L. maculans could not be established. It is conceivable that in planta, additional metabolites are produced that inhibit the metabolism of wasalexins by L. maculans.
2.3 Brassilexin and sinalexin
L. maculans canola virulent isolates detoxified brassilexin (20), via 3-aminomethyleneindole-2-thione (65), to the polar metabolite 3-formylindolyl-2-sulfonic acid (90, Scheme 4).41 Similar to brassilexin (20), sinalexin (21) was detoxified by reduction of the isothiazole ring to 3-aminomethylene-1-methoxyindole-2-thione (92), which decomposed spontaneously in aqueous solution.41 Similar to brassinins 1 and 3 and cyclobrassinin (12), S. sclerotiorum detoxified both brassilexin (20) and sinalexin (21) to the glucosyl derivatives 91 and 94, respectively.422.4 Rutalexin, isalexin and rapalexin A
The pathway for transformation of rutalexin (19), a rutabaga phytoalexin, by A. brassicicola has been reported recently to produce the hybrid metabolite rutapyrone (Scheme 5).43 In cultures of A. brassicicola rutalexin (19) was hydrolyzed and decarboxylated to the sulfanylamide 95, which in turn reacted with the least hindered epoxide carbon of phomapyrone G (98) to yield the adduct rutapyrone (96). Sulfanylamide 95 oxidized spontaneously to the disulfide 97 (the major product), which in solution oxidized spontaneously to several other products.43 The biotransformation of isalexin (44), another rutabaga phytoalexin, in cultures of A. brassicicola yielded several products that included 1,3-dihydro-3-hydroxy-4-methoxyindol-2-one (99).43 This structure was confirmed by comparison with a synthetic sample prepared by NaBH4 reduction of isalexin (44). In solution, metabolite 99 re-oxidized spontaneously to isalexin (44). By contrast, rapalexin A (42), a rutabaga phytoalexin as well, was stable in cultures of A. brassicicola.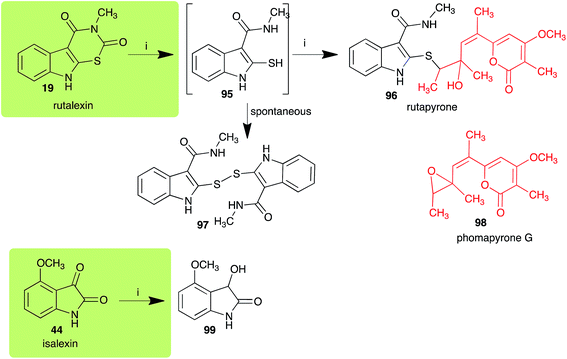 | ||
| Scheme 5 Detoxification of the cruciferous phytoalexins rutalexin (19) and isalexin (44) by a plant fungal pathogen: (i) Alternaria brassicicola. | ||
2.5 Brassicanal A and brassicanate A
Brassicanal A (28) was hydroxylated by R. solani25 to 63, and transformed by L. maculans (canola virulent isolates)44 and S. sclerotiorum42 to brassicanal A sulfoxide (100) and 3-hydroxymethylindole-2-methylsulfoxide (101); 101 was further transformed by L. maculans to 3-methylindole-2-methylsulfoxide (102), as shown in Scheme 6. The biotransformation products were significantly less inhibitory to R. solani, L. maculans and S. sclerotiorum, hence these transformations were considered detoxifications. The transformation of brassicanate A (33), another rutabaga phytoalexin, by A. brassicicola yielded 2-methylsulfanediylindole-3-carboxylic acid (103), which spontaneously decarboxylated in aqueous acidic solutions to yield the non-toxic product 104 (Scheme 6).432.6 Dioxibrassinin, brussalexin A, spirobrassinins and erucalexin
Dioxibrassinin (22) was metabolized by canola virulent isolates of L. maculans, but no biotransformation products were detected or isolated,23 whereas brown mustard virulent isolates detoxified dioxibrassinin (22) to 2′-thioxospiro[indoline-3,5′-oxazolidin]-2-one (105) (Scheme 7).45 By contrast, brussalexin A (41) was not stable in culture medium or in water, yielding indolyl-3-methanol that decomposed to yield mainly 3,3′-diindolylmethane.46Spirobrassinin (24) was not transformed by L. maculans canola virulent isolates but was transformed by brown mustard virulent isolates to metabolite 106 (Scheme 7).45 1-Methoxyspirobrassinin (25) was metabolized to the spirothiazolidinethione 108 by L. maculans (brown mustard virulent isolates) (Scheme 7).45 S. sclerotiorum transformed both spirobrassinin (24) and 1-methoxyspirobrassinin (25) to metabolites 107–109 (Scheme 7).42 Because S. sclerotiorum did not use glucosylating enzymes to transform 24 and 25, it was suggested that glucosylating enzymes were only used for detoxification of phytoalexins with strong antifungal activities against this fungus.42 Erucalexin (27) was detoxified by L. maculans (canola virulent isolates) by reduction to a diastereomeric mixture of 3-dihydroerucalexins (110) (Scheme 8). The relative configurations of the diastereomeric mixture of dihydroerucalexins were established by 1H NMR and NOE spectroscopy.46
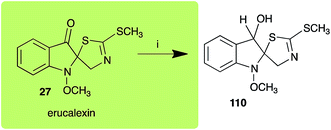 | ||
| Scheme 8 Detoxification of the cruciferous phytoalexin erucalexin (27) by a plant fungal pathogen: (i) Leptosphaeria maculans (canola virulent isolates). | ||
2.7 Camalexins
The phytoalexin camalexin (35) is perhaps the most studied of the cruciferous phytoalexins, not surprisingly because it is produced by a crucifer model species (A. thaliana) that had its genome published in 2000.47 Camalexin (35) is detoxified by A. brassicicola,48 B. cinerea,21 R. solani49 and S. sclerotiorum,42 but not by A. brassicae or L. maculans (canola virulent isolates).50 The products of transformation of camalexin (35) by each fungal species were different, except for the detoxification pathway of camalexin (35) in A. brassicicola and B. cinerea that occurred via identical intermediates (Scheme 9). In all cases the final products were found to be less toxic to each species than camalexin (35).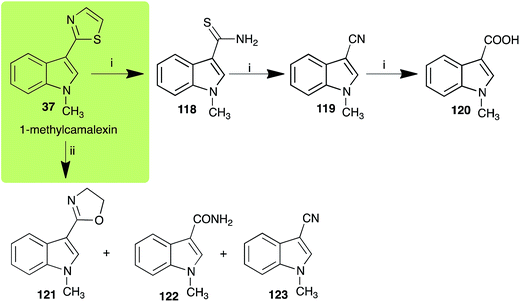 | ||
| Scheme 10 Detoxification of the cruciferous phytoalexin 1-methylcamalexin (37) by plant fungal pathogens: (i) Alternaria brassicicola; (ii) Rhizoctonia solani. | ||
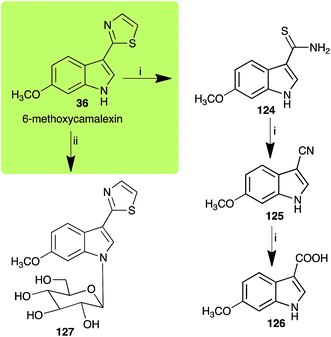 | ||
| Scheme 11 Detoxification of the cruciferous phytoalexin 6-methoxycamalexin (36) by plant fungal pathogens: (i) Alternaria brassicicola; (ii) Sclerotinia sclerotiorum. | ||
The rate of transformation of camalexin (35) by A. brassicicola48 was much slower than that observed by B. cinerea.21 Different transformation rates are consistent with the defensive role of camalexin (35) in A. thaliana, which displays resistance to A. brassicicola and susceptibility to B. cinerea. It is noteworthy that glucosylation of camalexin (35) was preceded by oxidation of C-6 (Scheme 9), whereas direct glucosylation of the indole nitrogen of the phytoalexins brassinin (1), cyclobrassinin (12) and brassilexin (20) by S. sclerotiorum was observed. A detailed pathway for transformation of camalexins by A. brassicicola was proposed based on analyses of the metabolism of synthetic isomers and bioisosteres of camalexin (35). It was suggested that enzymatic oxidation of the thiazole ring of camalexins 35, 36 and 37 occurs via intermediate(s) that after hydrolysis yield the corresponding thiocarboxamides 114, 118 and 124.48
2.8 Indolyl-3-acetonitrile
Indolyl-3-acetonitrile (38) was determined to be a phytoalexin in brown mustard (B. juncea), but a constitutive antimicrobial metabolite (phytoanticipin) in other Brassica species.51 Indolyl-3-acetonitrile (38) was metabolized by L. maculans (canola and brown mustard virulent isolates), R. solani, S. sclerotiorum52 and A. brassicicola.11 Acids 128 and 56, unlike the parent metabolite indolyl-3-acetonitrile (38), had no detectable inhibitory effects on any of the fungal species.3. Conclusion and prospects
As summarized in Table 2, the metabolism and detoxification pathways of more than twenty cruciferous phytoalexins by plant pathogenic fungal species have been reported. Because rapalexin A (42) and camalexin (35) are not metabolized by L. maculans, these phytoalexins are of great interest to improve the disease resistance of crucifers to this pathogen, one of the most destructive to oilseed crops. In addition, because camalexins 35–37 are transformed slowly by A. brassicicola, incorporation of the biosynthetic pathway of camalexins into cultivated Brassica species could have a great positive impact in obtaining resistance to both L. maculans and A. brassicicola. Incorporation of novel defense pathways in agriculturally important crops through plant breeding is an alternative that could decrease the unsustainable use of fungicides.To date, five fungal enzymes mediating the detoxification of brassinin (1) and cyclobrassinin (12) were purified and characterized: BOLm, BHLm, BHAb, CHAb and BGTSs. The enzymes involved in these detoxifications were shown to be inducible and display substrate specificity. Remarkably, it was established that some crucifer phytoalexins are competitive inhibitors of these detoxifying enzymes. This unprecedented conclusion suggested that phytoalexin blends, e.g. brassinin (1) and cyclobrassinin (12), produced by particular plant species in response to attack by a pathogen have multiple functions, including inhibition of fungal growth and inhibition of specific enzymes produced by pathogenic fungi to overcome the plants' defenses.
Finally, as a spin off of the work reviewed above, some of the biotransformation reactions could be applied to selectively convert complex molecules to novel or known products. For example, considering that the fungus S. sclerotiorum is easy to culture and carries out efficient and selective glucosylation of indolyl compounds containing diverse functional groups, including brassinin (1), brassilexin (20), sinalexin (21) and camalexin (35), this fungal species could be exploited for the selective glucosylation of more complex substrates.
Based on work carried out over more than two decades, a new strategy to control fungal pathogens of crucifers has emerged. That is, because specific crucifer fungal pathogens produce specific enzymes to detoxify cruciferous phytoalexins, it follows that selective inhibitors of these enzymes, coined paldoxins, could be designed and produced to prevent phytoalexin degradation in planta (Fig. 4). For example, considering that the detoxifications of brassinin (1) and cyclobrassinin (12) are carried out by specific enzymes (BOLm, BHLm, BHAb, CHAb), application of paldoxins of these enzymes to plants could enable the accumulation of 1 and 12 (and their derivatives) to levels necessary to inhibit pathogen growth. In addition, paldoxins of the enzymes that catalyze the detoxifications of brassilexin (20), sinalexin (21) and camalexins 35–37 could have similar application, yet these enzymes remain to be purified and characterized. Nonetheless, in examples where phytoalexins are detoxified by multiple parallel pathways, the paldoxin strategy is unlikely to have a positive impact in controlling plant fungal pathogens.
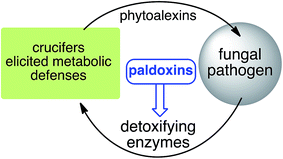 | ||
| Fig. 4 Proposed strategy to protect crucifer crops: paldoxins = phytoalexin detoxification inhibitors. | ||
It has been proposed that the ideal paldoxins need to be selective inhibitors that could allow accumulation of a plant's own defenses. Selective inhibitors are less likely to affect non-targeted organisms and thus are anticipated to have lower impact on cultivated ecosystems. As such, paldoxins cannot be toxic to living organisms, whether animal, plant or microbe. Furthermore, mixtures of paldoxins to act synergistically with the natural disease resistance factors of plants could be optimized to effectively protect crops with minimal environmental impact. In conclusion, further work needs to be done to advance the current knowledge on phytoalexin detoxification and design of inhibitors before field applications of this strategy can be adopted.
References
- R. Maor and K. Shirasu, The arms race continues: battle strategies between plants and fungal pathogens, Curr. Opin. Microbiol., 2005, 8, 399–404 CrossRef CAS PubMed.
- N. Pumplin and O. Voinnet, RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence, Nat. Rev. Microbiol., 2013, 11, 745–760 CrossRef CAS PubMed.
- C. M. J. Pieterse, C. Zamioudis, R. L. Berendsen, D. M. Weller, S. C. M. Van Wees and P. A. H. M. Bakker, Induced systemic resistance by beneficial microbes, Annu. Rev. Phytopathol., 2014, 52, 347–375 CrossRef CAS PubMed.
- M. S. C. Pedras, E. E. Yaya and E. Glawischnig, The phytoalexins from cultivated and wild crucifers: chemistry and biology, Nat. Prod. Rep., 2011, 28, 1381–1405 RSC.
- M. S. C. Pedras and Q. H. To, Unveiling the first indole-fused thiazepine: structure, synthesis and biosynthesis of cyclonasturlexin, a remarkable cruciferous phytoalexin, Chem. Commun., 2016, 52, 5880–5883 RSC.
- M. S. C. Pedras and E. E. Yaya, Dissecting metabolic puzzles through isotope feeding: a novel amino acid in the biosynthetic pathway of the cruciferous phytoalexins rapalexin A and isocyalexin A, Org. Biomol. Chem., 2013, 11, 1149–1166 CAS.
- M. S. C. Pedras and E. E. Yaya, Tenualexin, other phytoalexins and indole glucosinolates from wild cruciferous species, Chem. Biodiversity, 2014, 11, 910–918 CAS.
- M. S. C. Pedras and Q. H. To, Non-indolyl cruciferous phytoalexins: nasturlexins and tridentatols, a striking convergent evolution of defenses in terrestrial plants and marine animals?, Phytochemistry, 2015, 113, 57–63 CrossRef CAS PubMed.
- M. S. C. Pedras, M. Alavi and Q. H. To, Expanding the nasturlexin family: nasturlexins C and D and their sulfoxides are phytoalexins of the crucifers Barbarea vulgaris and B. verna, Phytochemistry, 2015, 118, 131–138 CrossRef CAS PubMed.
- M. S. C. Pedras and E. E. Yaya, The first isocyanide of plant origin expands functional group diversity in cruciferous phytoalexins: synthesis, structure and bioactivity of isocyalexin A, Org. Biomol. Chem., 2012, 10, 3613–3616 CAS.
- M. S. C. Pedras and S. Hossain, Interaction of cruciferous phytoanticipins with plant fungal pathogens: indole glucosinolates are not metabolized but the corresponding desulfo-derivatives and nitriles are, Phytochemistry, 2011, 72, 2308–2316 CrossRef CAS PubMed.
- M. S. C. Pedras and J. L. Taylor, Metabolic transformation of the phytoalexin brassinin by the blackleg fungus, J. Org. Chem., 1991, 56, 2619–2621 CrossRef CAS.
- S. Zeilinger, V. K. Gupta, T. E. S. Dahms, R. N. Silva, H. B. Singh, R. S. Upadhyay, E. V. Gomes, C. K. Tsui and S. C. Nayak, Friends or foes? Emerging insights from fungal interactions with plants, FEMS Microbiol. Rev., 2015, 40, 182–207 CrossRef PubMed.
- M. S. C. Pedras and P. W. K. Ahiahonu, Metabolism and detoxification of phytoalexins and analogs by phytopathogenic fungi, Phytochemistry, 2005, 66, 391–411 CrossRef CAS PubMed.
- M. Takasugi, N. Katsui and A. Shirata, Isolation of three novel sulphur-containing phytoalexins from the Chinese cabbage Brassica campestris L. ssp. pekinensis (cruciferae), J. Chem. Soc., Chem. Commun., 1986, 1077 RSC.
- M. S. C. Pedras, Metabolism and detoxification of phytoalexins from crucifers and application to the control of fungal plant pathogens, in Biotechnology of Crucifers, ed. S. K. Gupta, Springer, New York, Heidelberg, Dordrecht, London, 2013, ch. 13, pp. 151–171 Search PubMed.
- P. E. Russell, A century of fungicide evolution, J. Agric. Sci., 2005, 143, 11–25 CrossRef CAS.
- M. S. C. Pedras, P. B. Chumala, W. Jin, M. S. Islam and D. W. Hauck, The phytopathogenic fungus Alternaria brassicicola: phytotoxin production and phytoalexin elicitation, Phytochemistry, 2009, 70, 394–402 CrossRef CAS PubMed.
- M. S. C. Pedras and J. L. Taylor, Metabolism of the phytoalexin brassinin by the blackleg fungus, J. Nat. Prod., 1993, 56, 731–738 CrossRef CAS.
- M. S. C. Pedras, P. W. K. Ahiahonu and M. Hossain, Detoxification of the cruciferous phytoalexin brassinin in Sclerotinia sclerotiorum requires an inducible glucosyltransferase, Phytochemistry, 2004, 65, 2685–2694 CrossRef CAS PubMed.
- M. S. C. Pedras, S. Hossain and R. B. Snitynsky, Detoxification of cruciferous phytoalexins in Botrytis cinerea: spontaneous dimerization of a camalexin metabolite, Phytochemistry, 2011, 72, 199–206 CrossRef CAS PubMed.
- M. S. C. Pedras, A. Abdoli, P. B. Chumala, P. Saha and G. Schatte, Unprecedented spirocyclization of 3-methyleneindoline-2-thiones during hydrolysis of the phytoalexin cyclobrassinin, Bioorg. Med. Chem. Lett., 2013, 23, 484–487 CrossRef CAS PubMed.
- M. S. C. Pedras and F. I. Okanga, Strategies of fungal pathogens: detoxification of a cruciferous phytoalexin by mimicry, Chem. Commun., 1998, 67–68 RSC.
- M. S. C. Pedras and F. I. Okanga, A convenient synthesis of the cruciferous phytoalexins brassicanal A and brassilexin by mimicry of a fungal detoxification pathway, Chem. Commun., 1998, 1565–1566 RSC.
- M. S. C. Pedras and F. I. Okanga, Strategies of cruciferous pathogenic fungi: detoxification of the phytoalexin cyclobrassinin by mimicry, J. Agric. Food Chem., 1999, 47, 1196–1202 CrossRef CAS PubMed.
- M. S. C. Pedras, Z. Minic and V. K. Sarma-Mamillapalle, Substrate specificity and inhibition of brassinin hydrolases, detoxifying enzymes from the plant pathogens Leptosphaeria maculans and Alternaria brassicicola, FEBS J., 2009, 276, 7412–7428 CrossRef CAS PubMed.
- M. S. C. Pedras, Z. Minic and M. Jha, Brassinin oxidase, a fungal detoxifying enzyme to overcome a plant defense - purification, characterization and inhibition, FEBS J., 2008, 275, 3691–3705 CrossRef CAS PubMed.
- A. C. Sexton, Z. Minic, A. J. Cozijnsen, M. S. C. Pedras and B. J. Howlett, Cloning, purification and characterisation of brassinin glucosyltransferase, a phytoalexin-detoxifying enzyme from the plant pathogen Sclerotinia sclerotiorum, Fungal. Genet. Biol., 2009, 46, 201–209 CrossRef CAS PubMed.
- M. S. C. Pedras and Z. Minic, The phytoalexins brassilexin and camalexin inhibit cyclobrassinin hydrolase, a unique enzyme from the fungal pathogen Alternaria brassicicola, Bioorg. Med. Chem., 2014, 22, 459–467 CrossRef CAS PubMed.
- M. S. C. Pedras, M. Jha and P. W. K. Ahiahonu, The synthesis and biosynthesis of phytoalexins produced by cruciferous plants, Curr. Org. Chem., 2003, 7, 1635–1647 CrossRef CAS.
- M. S. C. Pedras, The chemical ecology of crucifers and their fungal pathogens: boosting plant defenses and inhibiting pathogen invasion, Chem. Rec., 2008, 8, 109–115 CrossRef CAS PubMed.
- M. S. C. Pedras and M. Jha, Toward the control of Leptosphaeria maculans: design, syntheses, biological activity, and metabolism of potential detoxification inhibitors of the crucifer phytoalexin brassinin, Bioorg. Med. Chem., 2006, 14, 4958–4979 CrossRef CAS PubMed.
- M. S. C. Pedras, Z. Minic and V. K. Sarma-Mamillapalle, Synthetic inhibitors of the fungal detoxifying enzyme brassinin oxidase based on the phytoalexin camalexin scaffold, J. Agric. Food Chem., 2009, 57, 2429–2435 CrossRef CAS PubMed.
- M. S. C. Pedras, Z. Minic, V. K. Sarma-Mamillapalle and M. Suchy, Discovery of inhibitors of brassinin oxidase based on the scaffolds of the phytoalexins brassilexin and wasalexin, Bioorg. Med. Chem., 2010, 18, 2456–2463 CrossRef CAS PubMed.
- M. S. C. Pedras, Z. Minic and S. Hossain, Discovery of inhibitors and substrates of brassinin hydrolase: probing selectivity with dithiocarbamate bioisosteres, Bioorg. Med. Chem., 2012, 20, 225–233 CrossRef CAS PubMed.
- M. S. C. Pedras, J. L. Sorensen, F. I. Okanga and I. L. Zaharia, Wasalexins A and B, new phytoalexins from wasabi: isolation, synthesis, and antifungal activity, Bioorg. Med. Chem. Lett., 1999, 9, 3015–3020 CrossRef CAS PubMed.
- M. S. C. Pedras and A. M. Adio, Phytoalexins and phytoanticipins from the wild crucifers Thellungiella halophila and Arabidopsis thaliana: Rapalexin A, wasalexins and camalexin, Phytochemistry, 2008, 69, 889–893 CrossRef CAS PubMed.
- M. S. C. Pedras, P. B. Chumala and M. Suchy, Phytoalexins from Thlaspi arvense, a wild crucifer resistant to virulent Leptosphaeria maculans: structures, syntheses and antifungal activity, Phytochemistry, 2003, 64, 949–956 CrossRef CAS PubMed.
- M. S. C. Pedras and M. Jha, Concise syntheses of the cruciferous phytoalexins brassilexin, sinalexin, wasalexins, and analogues: expanding the scope of the Vilsmeier formylation, J. Org. Chem., 2005, 70, 1828–1834 CrossRef CAS PubMed.
- M. S. C. Pedras and M. Suchy, Metabolism of the crucifer phytoalexins wasalexins A and B in the plant pathogenic fungus Leptosphaeria maculans, Org. Biomol. Chem., 2006, 4, 3526–3535 CAS.
- M. S. C. Pedras and M. Suchy, Detoxification pathways of the phytoalexins brassilexin and sinalexin in Leptosphaeria maculans: isolation and synthesis of the elusive intermediate 3-formylindolyl-2-sulfonic acid, Org. Biomol. Chem., 2005, 3, 2002–2007 CAS.
- M. S. C. Pedras and M. Hossain, Metabolism of crucifer phytoalexins in Sclerotinia sclerotiorum: detoxification of strongly antifungal compounds involves glucosylation, Org. Biomol. Chem., 2006, 4, 2581–2590 CAS.
- M. S. C. Pedras and A. Abdoli, Biotransformation of rutabaga phytoalexins by the fungus Alternaria brassicicola: unveiling the first hybrid metabolite derived from a phytoalexin and a fungal polyketide, Bioorg. Med. Chem., 2017, 25, 557–567 CrossRef CAS PubMed.
- M. S. C. Pedras and A. Q. Khan, Biotransformation of the brassica phytoalexin brassicanal A by the blackleg fungus, J. Agric. Food Chem., 1996, 44, 3403–3407 CrossRef CAS.
- M. S. C. Pedras and R. B. Snitynsky, Impact of cruciferous phytoalexins on the detoxification of brassilexin by the blackleg fungus pathogenic to brown mustard, Nat. Prod. Commun., 2010, 5, 883–888 CAS.
- M. S. C. Pedras and V. K. Sarma-Mamillapalle, The cruciferous phytoalexins rapalexin A, brussalexin A and erucalexin: chemistry and metabolism in Leptosphaeria maculans, Bioorg. Med. Chem., 2012, 20, 3991–3996 CrossRef CAS PubMed.
- The Arabidopsis genome initiative, Analysis of the genome sequence of the flowering plant Arabidopsis thaliana, Nature, 2000, 408, 796–815 CrossRef PubMed.
- M. S. C. Pedras and A. Abdoli, Metabolism of the phytoalexins camalexins, their bioisosteres and analogues in the plant pathogenic fungus Alternaria brassicicola, Bioorg. Med. Chem., 2013, 21, 4541–4549 CrossRef CAS PubMed.
- M. S. C. Pedras and A. Q. Khan, Biotransformation of the phytoalexin camalexin by the phytopathogen Rhizoctonia solani, Phytochemistry, 2000, 53, 59–69 CrossRef CAS PubMed.
- M. S. C. Pedras, A. Q. Khan and J. L. Taylor, The phytoalexin camalexin is not metabolized by Phoma lingam, Alternaria brassicae, or phytopathogenic bacteria, Plant Sci., 1998, 139, 1–8 CrossRef CAS.
- M. S. C. Pedras, C. M. Nycholat, S. Montaut, Y. Xu and A. Q. Khan, Chemical defenses of crucifers: elicitation and metabolism of phytoalexins and indole-3-acetonitrile in brown mustard and turnip, Phytochemistry, 2002, 59, 611–625 CrossRef CAS PubMed.
- M. S. C. Pedras and S. Montaut, Probing crucial metabolic pathways in fungal pathogens of crucifers: biotransformation of indole-3-acetaldoxime, 4-hydroxyphenylacetaldoxime, and their metabolites, Bioorg. Med. Chem., 2003, 11, 3115–3120 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |