Open Access Article
Open Access ArticleThe modulation effect of the convexity of silicon topological nanostructures on the growth of mesenchymal stem cells†
Yang Zou‡
a,
Hongqing Feng‡a,
Han Ouyanga,
Yiming Jina,
Min Yua,
Zhuo Liuab and
Zhou Li *a
*a
aBeijing Institute of Nanoenergy and Nanosystems, Chinese Academy of Sciences, National Center for Nanoscience and Technology (NCNST), Beijing, 100083, P. R. China. E-mail: zli@binn.cas.cn
bSchool of Biological Science and Medical Engineering, Beihang University, Beijing 100191, China
First published on 17th March 2017
Abstract
A series of different topological nanostructures are fabricated on silicon wafer using metal-assisted chemical etching. The modulation effect of these nanostructures on the size, filopodia generation and growth orientation of the rat mesenchymal stem cells (MSCs) are studied. These topological nanostructures tend to induce the MSCs to have smaller size, but they generate much more filopodia compared to the flat silicon control. The modulation effects of these nanostructures are dependent on their surface convexity, as analysed by grey-level value histogram and fast Fourier transformation (FFT). A surface with a higher portion of convex area is better at supporting larger cell sizes. The wavenumber analysis by FFT further determines its effect on filopodia generation. In addition, the growth orientation of the cells are also guided by the surface convexity. On the porous and spongy surface, the cell filopodia extend and grow in avoidance of large sinking pits. On the columnar and spiny surface, the cell body and filopodia extend only on the tips of these nanostructures. Our study reveals that surface convexity is an important factor modulating cell behavior, and convexity analysis by image processing can work as a fast and simple evaluation standard to design topological nanostructures.
1. Introduction
Specific topological nanostructures have remarkable influences on cell behaviours because the physiological cellular micro-environment is in nanoscale. For example, osteocytes are isolated from each other in the skeletal system, but they are also connected through the bone matrix and form a network for material transportation and signal transmission.1,2 Also, natural extracellular matrix has abundant topology in nanoscale.3 The size of a single collagen is approximately 300 nm long and 1.5 nm wide.4 These extracellular molecules form dozens of micron filamentous fiber structures with a diameter of about 260–410 nm.5 In short, proper topological nanostructures are very critical for the physiological functions of cells and tissues.As a result, the design of artificial nanoscale patterns to modulate cell behaviours has attracted wide interest in the field of bionics. At present, major research on the topological nanostructures includes nanogratings, nanogrooves, nanoposts, nanopillars, nanopores and nanoconcave arrays.6–9 Various methods including soft lithography, laser-beam etching, electron-beam lithography, and hot embossing have been employed to prepare geometric patterns of different sizes on different materials.10–13 Both organic and inorganic nanostructured substrates have been developed, including polymers, carbon-based nanotubes and nanofibers, titanium oxide nanotubes, ZnO nanowires, and silicon-based nanostructures.14
The topological nanostructures affect mammalian cells of almost all types and modulate various cell behaviours.15–19 Surface material, energy, chemistry, rigidity, roughness, and structure size and height have all been reported to influence the growth and development of cells cultured on these surfaces.20–22 For example, for cardiomyocytes cultured on polystyrene or polyurethane substrate, the organization of the actin cytoskeleton, focal adhesion complexes, and beating rates were all regulated by the combinational effects of the surface topography and substrate stiffness.23 For mesenchymal stem cells (MSCs) cultured on porous silicon gradient, the spreading of rat and human MSCs were all suppressed on the large-pore-size region and increased towards the small-pore-size region (diameters less than 100 nm).24
For nanostructure-induced stem cell differentiation, many processes have been developed in recent years. Osteogenesis differentiation has been demonstrated in many kinds of topological structures, especially in TiO2 nanotubes.25,26 PDMS nanostructures with ordered ridges and grooves could also induce MSCs cultured on 400 nm pitch to differentiate into osteocytes.18 As on the grooved polystyrene nanostructures, however, the MSCs were driven to adipogenesis and myogenesis differentiation, but not osteogenesis differentiation.27 Also, the porous silicon nanostructures of about 200 nm diameter greatly enhanced both the osteogenesis and adipogenesis differentiation of MSCs.28 The 200 nm polystyrene nanopore-patterned surfaces significantly promoted the pancreatic differentiation of human embryonic cells and induced pluripotent stem cells, highly upregulating the expression of PDX1 and profoundly enhancing the production of insulin, glucagon, and somatostatin.29 Human MSCs cultured on a layer of single-wall carbon nanotube with a vertical height less than 100 nm were found to differentiate into neurogenic cells over the first week of culture.30
Apart from the above discoveries, however, a global and in-depth insight on the influence of various aspects of the topological nanostructures on the cells has not been achieved. Meanwhile, there have been few quick evaluation methods to determine whether a certain surface topology is beneficial to promoting specific cell behaviours. In this study, a low-cost, efficient, and easy-to-operate method, metal-assisted chemical etching, was applied on silicon wafers to prepare a series of silicon substrates with various topological structures in nanoscale. We cultured rat MSCs directly on these silicon topological nanostructures (STNS) to observe their influence on the cell spreading, filopodia generation and growth orientation. We found that the convexity of the nanostructures, as analysed by image processing, can quickly and qualitatively predict the influence of the topological structures on MSC growth and development.
2. Experimental details
2.1 STNS preparation
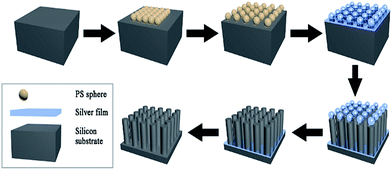
Metal-assisted chemical etching was applied to prepare STNS on silicon wafers.31,32 Hydrogen peroxide was used to oxidize silicon with silver as catalyst, and hydrofluoric acid was used to etch off the silicon dioxide. Silver cannot be deposited onto the silicon wafer where polystyrene (PS) spheres were previously adhered, so silicon wire-array structure substrates were formed beneath the PS spheres. The preparation process for STNS is shown in detail in Fig. 1. First, PS spheres were dragged with silicon wafer and closely attached onto the wafer. Second, the PS spheres were isolated from each other by 60 W plasma for 5 min. Third, a layer of silver film was deposited onto silicon wafers by magnetron sputtering. Fourth, the silicon wafer was etched with a chemical etchant composed of 8.76 ml H2O, 1.92 ml 30% HF and 0.27 ml 40% H2O2. Fifth, the PS spheres were washed away using acetone. Finally, the silver film was dissolved using nitric acid, and the materials were baked in the oven. Experimental conditions were modulated to obtain various STNS. As for the needle-like and pillar-like structures, a close-packed PS sphere monolayer was assembled on the Si wafer. Silver film was deposited by magnetron sputtering system for 2 min. Then, Si wafer was immersed into the etchant from 30 min to 2 h. As for the samples with porous structures, Si wafer with patterned photoresist was prepared by lithography. After depositing silver film, the wafer was immersed in the etchant for 30 min. As for the samples with pyramid structures, Si wafer was immersed in warm NaOH solution instead of etchant chemical. Nine different topology patterns were obtained in this study. The chemical etching reaction equations were as follows:| Si + 2H2O → SiO2 + 4H+ + 4e− | (1) |
| SiO2 + 6HF → H2SiF6 + 2H2O | (2) |
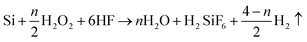 | (3) |
2.2 MSC extraction
Male SD rats weighing 150 g were sacrificed and wiped with ethanol. The hind legs were stripped and rinsed with phosphate buffer solution. The MSCs were blown out with Dulbecco minimum essential medium (DMEM) using a 10 ml syringe until the bone become white. The MSCs were repeatedly pipetted and filtered through a strainer, and then centrifuged at 1000 rpm for three minutes at room temperature. The MSC pellets were re-suspended with DMEM medium (10% FBS). Then, they were cultured in a 6-well plate which had been treated with poly-lysine the night before. All experiments were performed in compliance with the Experimental Safety Guidelines of Beijing Institute of Nanoenergy and Nanosystems, Chinese Academy of Sciences (BINN, CAS). The experiment had been approved by the Biosafety and Bioethics Committee of BINN.2.3 MSC culture on the STNS
Four STNS were chosen for the cell culture experiment: porous, spongy, columnar, and spiny. The silicon wafer without topology was set as the control group. The samples were placed into a 24-well plate and soaked with 75% ethanol overnight, and then dried before culture of MSCs. The seeded cells were diluted to 2 × 105 per well. The cells were cultured for 48 h, then fixed with an aldehyde fixative for 15 minutes, and immersed in chloroauric acid for 2 hours to deposit a layer of gold on the surface for SEM observation. Then, the cells were dehydrated with gradient ethanol (30%, 50%, 75%, 80%, 95%, and 100%, respectively). Dehydration in the first five gradients was done once for 15 minutes, and in the last gradient, twice for 30 minutes. The samples were air-dried overnight after dehydration in a super-clean bench. The cell morphologies were observed under scanning electron microscope (FE-SEM, Hitachi SU8020) for detailed analysis and photographic data recording.2.4 Image processing
The SEM images of the four STNS, porous, spongy, columnar, and spiny, were taken perpendicular to the surface and processed using Matlab. Images with the same magnification were chosen for processing. For each type of STNS, representative regions with area of 460 × 460 pixels were analysed. The grey-level values of each pixel in the images were obtained, and normalized to be within the range of 0–255. The number of pixels with grey-level values within each pair of consecutive integers were counted and calculated. The wavenumber of the convex and concave areas in the STNS were evaluated using two-dimensional fast Fourier transformation (FFT).2.5 Statistical analysis
For each of the STNS, more than 4 repeats were processed. The cell size and filopodia were measured using ImageJ. The statistical significance of differences between each group was analysed by t-test using SPSS.3. Results and discussion
3.1 STNS fabrication and morphology
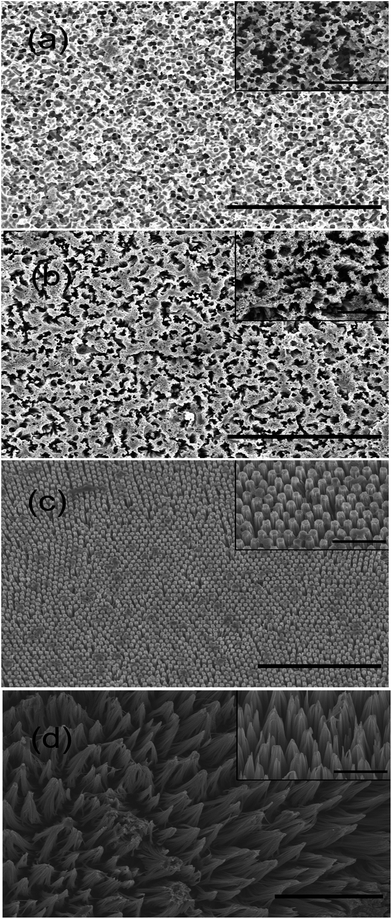
Nine typical STNS were produced using chemical etching (Fig. S1†): irregular porous, aperture array, spongy, long-columnar, spiny, short-columnar, fascicular, conical, and pyramid STNS. Among them, four typical STNS were chosen for cell culture experiment and more detailed analysis: porous, spongy, columnar (long), and spiny STNS (Fig. 2). They are representatives of two different morphology types. Porous and spongy STNS had consecutive convex surfaces with discrete concave areas. Columnar and spiny STNS had consecutive concave areas with discrete convex spots. On the porous STNS, there were many circular pores that were formed by etching on the surface. On the spongy STNS, the etched parts were more irregular and linked to form crooked gullies. On the columnar STNS, arrays of columns were distributed evenly on the surface. On the spiny STNS, the Si nanowires were congregated at the tips in clusters and formed scattered tip points on the surface.3.2 Image processing using histogram and FFT
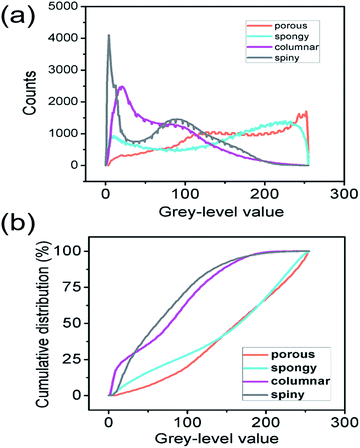
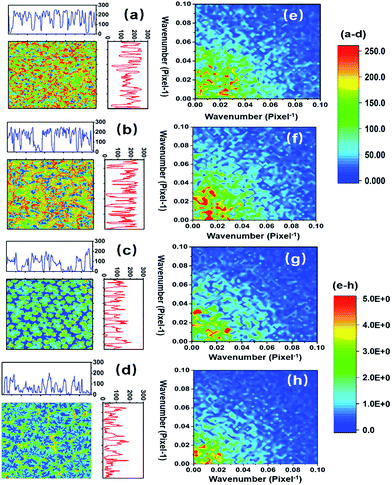
The numerical distribution of the grey-level values of each type of STNS is shown in Fig. 3. Fig. 3(a) is the smoothed line of the histogram of the four STNS types, showing the pixel counts for a certain grey-level value. For the porous and spongy STNS, which were convex area-dominated, the lines went upwards as the grey-level values increased. This meant that there were more pixels with a high grey-level value than those with a low grey-level value. For the other two, which were concave area-dominated, the lines went downwards as the grey-level values increased. This meant that there were more pixels with a low grey-level value than those with a high grey-level value. The cumulative distributions of the grey-level values of the four STNS types were also calculated and are represented in Fig. 3(b), which shows the percentage of pixels that are over a certain grey-level value. In the cumulative distribution, the porous and spongy, or the columnar and spiny, almost overlapped with each other, respectively, making it very difficult to distinguish them from each other.The surface convexity was further evaluated for the spatial distribution of the grey-level values as shown in Fig. 4. The one-dimensional grey-level value changes in a certain line and row are shown in Fig. 4(a–d). The overall two-dimensional changes were analysed using FFT. The wavenumbers per pixel in the X and Y axis are shown in Fig. 4(e–h). The wavenumber in the spatial domain revealed by FFT is equivalent to the frequency of temporal signals in the time domain, which reflects the fluctuation level of the signals.33,34 The porous and spongy STNS had wavenumber components in the range of 0–0.10, and the major components were in the range of 0–0.04. The columnar and spiny STNS had wavenumber components in the range of 0–0.08, and the major components were in the range of 0–0.03. Compared to the porous STNS, the wavenumbers of the spongy STNS had larger amplitudes in the range of 0–0.04, especially in the relatively higher region (0.03–0.04). In the comparison between columnar and spiny STNS, the wavenumbers of the columnar STNS had larger amplitudes in the range of 0–0.03, especially in the relatively higher frequency region (0.02–0.03). Therefore, the FFT analysis was an effective way to distinguish the surface morphology that had similar numerical grey-level value distribution. The spongy STNS had more fluctuations than the porous one, and the columnar STNS had more fluctuations than the spiny one.
3.3 MSCs size expansion and filopodia generation
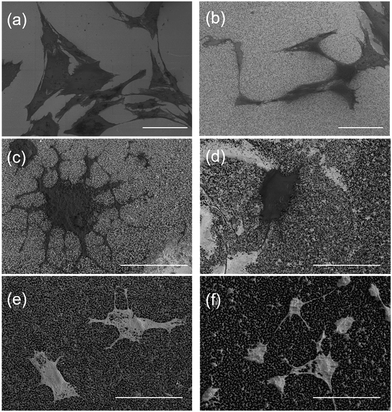
Cells grown on the STNS surfaces are shown in Fig. 5. Generally, the MSCs growing on the control Si wafer were large in size, but had few filopodia (Fig. 5(a)). Contrarily, the MSCs growing on the STNS were smaller, but had many filopodia (Fig. 5(b), porous STNS, as an example). The images of MSCs on the STNS with higher magnifications are shown in Fig. 5(c–f). The spread of the cells and the generation of the filopodia are clearly shown. More SEM images of MSCs growing on the four STNS types are shown in Fig. S2.† ImageJ was used to measure the cell sizes quantitatively. The results are shown in Fig. 6(a). For the porous, spongy, columnar and spiny STNS, the average cell size was 959.46 (±116.58) μm2, 575.78 (±164.74) μm2, 437.00 (±70.84) μm2, and 171.96 (±54.56) μm2, respectively.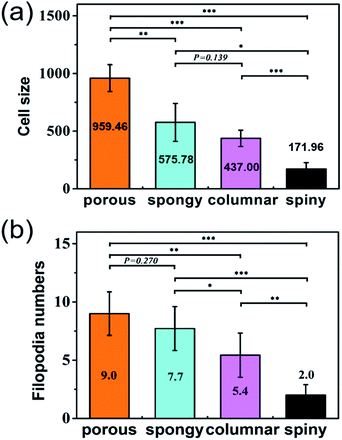 | ||
| Fig. 6 The cell size (a) and the numbers of filopodia (b) on the four kinds of STNS. The statistical analysis is marked in the figures (*p < 0.05; **p < 0.01; ***p < 0.001). | ||
The average cell size was largest on the porous STNS; it was significantly larger than any of the others. The average cell size on the spongy STNS was larger than that on the columnar STNS, but the difference was not statistically significant. The average cell size on the spiny STNS was the smallest, and the difference was significant with all the others. Meanwhile, the numbers of cellular filopodia in the experiment groups (Fig. 6(b)) were increased significantly compared to the control group. The cells growing on the porous STNS had 9.0 filopodia each on average, and those on the spongy STNS had 7.7 each. On columnar STNS, the average number of filopodia was 5.4, and on spiny STNS, it was only 2.
3.4 The filopodia growth preference
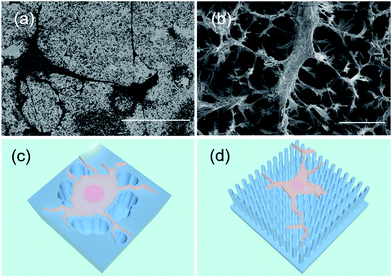
In addition, the preference of filopodia growth orientation on the nanostructures was revealed in this experiment. In different types of STNS, a similar phenomenon was observed: the cell filopodia tended to grow along the edges of the convex area but not into the concave area. They actively avoided the tufted and barrier morphology, and grew along the periphery of the pits and defects (Fig. 7(a)). On the concave columnar and spiny STNS, the filopodia extended on the tops of the pillars and bundle tips, and did not grow to the concave area (Fig. 7(b)). The schematic growth patterns of MSCs on the two surfaces are drawn in Fig. 7(c and d). On the convex STNS, the filopodia protruded from the cell body and extended on the raised surface between the defects and pits. On the concave STNS, the cell filopodia elongated on the projected structures, and the cell body grew and extended on the top of the nanotips. Because the raised area was continuous on the convex STNS, but discontinuous on the concave STNS, the cells grew larger on the convex STNS than on the concave STNS. The preference of the filopodia growth extension was consistent with the cell size spread because they both relied on the convex area.3.5 Discussion on the convexity modulation of MSC growth
Based on our experiment, it can be concluded that a consecutively convex surface with limited concave areas was beneficial for cell spread and growth, while a consecutively concave surface with limited convex areas was not. Cell geometry and size has important influence on cell fate and functions. When the cell adhesion area is small to a certain degree, apoptosis will happen.35 The cell size and shape also regulate stem cell differentiation through Rac1 and N-cadherin genes.36 As a result, to achieve successful cell culture and modulation by nanopatterning, a surface capable of supporting adequate cell adhesion and spread is critical. Meanwhile, the STNS have the ability to induce filopodia generation and growth, and the “convex” surface showed better ability than the “concave” one. Filopodia play important roles in cell sensing, migration, function and differentiation.37–39 For example, in neuron development, filopodia extend from the growth cone at the leading edge to form axons; filopodia-like projections have also been linked to dendrite creation when new synapses are formed in the brain.40 In macrophages, filopodia act as phagocytic tentacles and pull bound objects towards the cell for phagocytosis.41 MSCs cultured on the cell-adhesive, peptide-functionalized Au nanorods were found to have more filopodia growing and faster differentiation into adipocytes than on flat controls.42 Therefore, a surface able to modulate filopodia growth is very important for cell function and differentiation modulation, which is a fundamental aim of topological nanostructure design.In this study, we discovered that MSCs growing on the STNS had smaller cell size but greater filopodia generation. A similar phenomenon was observed in osteoprogenitor cells growing on arrays of nanopits with 120 nm diameters.43 The increased filopodia and decreased cell size may be caused by the different attachment forces between the topological nanostructures and cell membrane proteins, which also influence the focal adhesion formation and cytoskeletal organisation in the cells. LX-2 and HepG2 cells growing on arrays of silicon nanowire of 100 nm diameters were also found to have smaller size.44 The expression of type I collagen and R-actin was downregulated after incubation on the SiNW arrays, and the expression levels of integrin and focal adhesion kinase (FAK) were upregulated.44 These provided an explanation for the restricted cell spread due to the enhancement of cell adhesion on the Si nanowire array.
Among the four STNS tested in this study, the porous STNS were capable of promoting both cell size expansion and filopodia generation. This ability is closely related to the surface convexity as analysed by image processing. The numerical distribution of the grey-level values can classify the surface to be convex dominated or concave dominated. In addition, FFT can distinguish the surfaces further when it is difficult to tell their degree of convexity by morphology and evaluation of grey-level values. A convex surface with limited concave area is preferred for both cell spreading and filopodia growth. On the convex-dominated surface, less fluctuation on the surface leads to better cell growth and filopodia generation. However, if the surface needs to be concave-area dominated, more fluctuations are preferred because they bring in more convex area. Also, the filopodia preferred to grow on the consecutively raised surface, avoiding growth on the sinking area, which was consistent with the size extension preference.
Here, we brought forth the use of histogram and FFT to evaluate the ability of certain topological nanostructures to support cell extension and filopodia generation. This simple method allowed a more rational design and application of various nanostructures in biological fields. This kind of evaluation and prediction method had been rare, as compared to the numerous studies on the material characterization or biological response aspects. Buch-Månson et al. proposed a prediction model for cell settling on the nanowire array based on the density and single dimension of the nanowires.45 In future, more prediction methods are expected because they can greatly facilitate the application of nanostructures in biomedical fields.
4. Conclusion
A series of different topological nanostructures were prepared on silicon wafer using metal-assisted chemical etching. Rat bone mesenchymal stem cells were cultured on these structures, and the preferences for cell size and filopodia growth were studied. A surface with dominating convex area and limited concave area was best at promoting cell growth and filopodia generation and extension. The influence of the convexity of the topological structures on cell growth can be qualitatively predicted by simply doing image processing, including grey-level value histogram and FFT. For convex STNS, more fluctuation in the surface, as revealed by FFT, was less beneficial for the cells; but for concave STNS, more fluctuation in the surface was more beneficial for the cells. Our study provides a fast and simple evaluation standard to conduct topological nanostructure design for cell behaviour modulation.Acknowledgements
This work was supported by the national key R & D project from Minister of Science and Technology, China (2016YFA0202703), NSFC (31571006), NSFC (81601629), Beijing Talents Fund (2015000021223ZK21) and “Thousands Talents” program for pioneer researcher and his innovation team. Yang Zou and Hongqing Feng contributed equally to this work.Notes and references
- C. R. Jacobs, S. Temiyasathit and A. B. Castillo, Annu. Rev. Biomed. Eng., 2010, 12, 369–400 Search PubMed.
- A. R. Stern, M. M. Stern, M. E. van Dyke, K. Jähn, M. Prideaux and L. F. Bonewald, Biotechniques, 2012, 52, 361–373 CAS.
- S. L. Goodman, P. A. Sims and R. M. Albrecht, Biomaterials, 1996, 17, 2087–2095 CrossRef CAS PubMed.
- E. Pamuła, V. De Cupere, Y. F. Dufrêne and P. G. Rouxhet, J. Colloid Interface Sci., 2004, 271, 80–91 CrossRef.
- L. Bozec, G. van der Heijden and M. Horton, Biophys. J., 2007, 92, 70–75 CrossRef CAS PubMed.
- M. K. Gunnewiek, E. M. Benetti, A. Di Luca, C. A. Van Blitterswijk, L. Moroni and G. J. Vancso, Langmuir, 2013, 29, 13843–13852 CrossRef PubMed.
- M. J. Dalby, M. O. Riehle, H. Johnstone, S. Affrossman and A. S. G. Curtis, Biomaterials, 2002, 23, 2945–2954 CrossRef CAS PubMed.
- H. V Unadkat, M. Hulsman, K. Cornelissen, B. J. Papenburg, R. K. Truckenmüller, A. E. Carpenter, M. Wessling, G. F. Post, M. Uetz, M. J. T. Reinders, D. Stamatialis, C. A. van Blitterswijk and J. de Boer, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 16565–16570 CrossRef PubMed.
- M. J. P. Biggs, R. G. Richards, S. Mcfarlane and C. D. W. Wilkinson, J. R. Soc., Interface, 2008, 5, 1231–1242 CrossRef CAS PubMed.
- M. Unal, Y. Alapan, H. Jia, A. G. Varga, K. Angelino, M. Aslan, I. Sayin, C. Han, Y. Jiang, Z. Zhang and U. A. Gurkan, Nanobiomedicine, 2014, 1, 1–5 CrossRef.
- D. Qin, Y. Xia and G. M. Whitesides, Nat. Protoc., 2010, 5, 491–502 CrossRef CAS PubMed.
- C. Vieu, F. Carcenac, A. Pepin, Y. Chen, M. Mejias, A. Lebib, L. Manin-Ferlazzo, L. Couraud and H. Launois, Appl. Surf. Sci., 2000, 164, 111–117 CrossRef CAS.
- H. Becker and U. Heim, Sens. Actuators, A, 2000, 83, 130–135 CrossRef CAS.
- K. Y. Baik, S. Y. Park, S. Namgung, D. Kim, D. Cho and M. Lee, Nano Convergence, 2014, 1, 28 CrossRef.
- C. J. Bettinger, R. Langer and J. T. Borenstein, Angew. Chem., Int. Ed., 2009, 48, 5406–5415 CrossRef CAS PubMed.
- F. C. M. J. M. van Delft, F. C. van den Heuvel, W. A. Loesberg, J. te Riet, P. Schön, C. G. Figdor, S. Speller, J. J. W. A. van Loon, X. F. Walboomers and J. A. Jansen, Microelectron. Eng., 2008, 85, 1362–1366 CrossRef CAS.
- W. T. Su, Y. F. Liao and I. M. Chu, Micron, 2007, 38, 278–285 CrossRef CAS PubMed.
- S. Watari, K. Hayashi, J. A. Wood, P. Russell, P. F. Nealey, C. J. Murphy and D. C. Genetos, Biomaterials, 2012, 33, 128–136 CrossRef CAS PubMed.
- M. Devices, Sensors, 2012, 12, 15947–15982 CrossRef PubMed.
- A. Thapa, T. J. Webster and K. M. Haberstroh, J. Biomed. Mater. Res., Part A, 2003, 67, 1374–1383 CrossRef PubMed.
- A. Saez, M. Ghibaudo, A. Buguin and P. Silberzan, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 8281–8286 Search PubMed.
- S. H. Ku, J. Ryu, S. K. Hong, H. Lee and C. B. Park, Biomaterials, 2010, 31, 2535–2541 Search PubMed.
- P. Wang, J. Yu, J. Lin and W. Tsai, Acta Biomater., 2011, 7, 3285–3293 CrossRef CAS PubMed.
- P. Y. Wang, L. R. Clements, H. Thissen, A. Jane, W. B. Tsai and N. H. Voelcker, RSC Adv., 2012, 2, 12857–12865 RSC.
- A. Gao, R. Hang, X. Huang, L. Zhao, X. Zhang, L. Wang, B. Tang, S. Ma and P. K. Chu, Biomaterials, 2014, 35, 4223–4235 CrossRef CAS PubMed.
- S. Wu, Z. Weng, X. Liu, K. W. K. Yeung and P. K. Chu, Adv. Funct. Mater., 2014, 24, 5464–5481 CrossRef CAS.
- P. Wang, W. Li, J. Yu and W. Tsai, J. Mater. Sci.: Mater. Med., 2012, 23, 3015–3028 CrossRef CAS PubMed.
- P. Y. Wang, L. R. Clements, H. Thissen, A. Jane, W. B. Tsai and N. H. Voelcker, Adv. Funct. Mater., 2012, 22, 3414–3423 CrossRef CAS.
- J. H. Kim, H. W. Kim, K. J. Cha, J. Han, Y. J. Jang, D. S. Kim and J. Kim, ACS Nano, 2016, 10, 3342–3355 CrossRef CAS PubMed.
- C. Y. Tay, H. Gu, W. S. Leong, H. Yu, H. Q. Li, B. C. Heng, H. Tantang, S. C. J. Loo, L. J. Li and L. P. Tan, Carbon, 2010, 48, 1095–1104 CrossRef CAS.
- Z. Huang, N. Geyer, P. Werner, J. De Boor and U. Gösele, Adv. Mater., 2011, 23, 285–308 CrossRef CAS PubMed.
- B. Z. Huang, H. Fang and J. Zhu, Adv. Mater., 2007, 19, 744–748 CrossRef.
- H. Park, H. Jeong, Y. H. Jung and D. Seo, Science, 2015, 12356 Search PubMed.
- H. Assender, V. Bliznyuk and K. Porfyrakis, Science, 2002, 297, 973–976 CrossRef CAS PubMed.
- C. S. Chen, M. Mrksich, S. Huang, G. M. Whitesides and D. E. Ingber, Science, 1997, 276, 1425–1428 CrossRef CAS PubMed.
- L. Gao, R. McBeath and C. S. Chen, Stem Cells, 2010, 28, 564–572 CAS.
- C. K. Miranti and J. S. Brugge, Nat. Cell Biol., 2002, 4, E83–E90 CrossRef CAS PubMed.
- P. K. Mattila and P. Lappalainen, Nat. Rev. Mol. Cell Biol., 2008, 9, 446–454 CrossRef CAS PubMed.
- S. Oh, K. S. Brammer, Y. S. Li, D. Teng, A. J. Engler, S. Chien and S. Jin, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 2130–2135 CrossRef CAS PubMed.
- M. Maletic-Savatic, R. Malinow and K. Svoboda, Science, 1999, 283, 1923–1927 CrossRef CAS PubMed.
- H. Kress, E. H. K. Stelzer, D. Holzer, F. Buss, G. Griffiths and A. Rohrbach, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 11633–11638 CrossRef CAS PubMed.
- W. Luo and M. N. Yousaf, Chem. Commun., 2009, 1237–1239 RSC.
- A. Hart, Æ. N. Gadegaard, Æ. C. D. W. Wilkinson, R. O. C. Oreffo and Æ. M. J. Dalby, J. Mater. Sci.: Mater. Med., 2007, 18, 1211–1218 CrossRef CAS PubMed.
- Y. M. Qi Suijian, Y. Changqing, J. Shenglin and F. Chi-Chun, ACS Appl. Mater. Interfaces, 2009, 1, 30–34 Search PubMed.
- N. Buch-Månson, S. Bonde, J. Bolinsson, T. Berthing, J. Nygård and K. L. Martinez, Adv. Funct. Mater., 2015, 25, 3246–3255 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra00542c |
| ‡ Yang Zou and Hongqing Feng contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |