Open Access Article
Open Access ArticleExperimental identification and computational characterization of a novel extracellular metalloproteinase produced by Clostridium sordellii†
Michael J. Aldape*ab,
Aoxiang Taoc,
Dustin D. Heeney a,
Eric R. McIndooa,
John M. Frencha and
Dong Xu
a,
Eric R. McIndooa,
John M. Frencha and
Dong Xu *c
*c
aVeterans Affairs Medical Center, Research and Development, Infectious Diseases Section, 500 West Fort St. (Bldg 117), Boise, ID 83702, USA. E-mail: mike.aldape@va.gov; Fax: +1-208-422-1425; Tel: +1-208-422-1000 ext. 7659
bNorthwest Nazarene University, Nampa, ID 83686, USA
cDepartment of Biomedical and Pharmaceutical Sciences, College of Pharmacy, Idaho State University, Meridian, ID 83642, USA. E-mail: xudong@isu.edu; Fax: +1-208-373-1834; Tel: +1-208-373-1832
First published on 2nd March 2017
Abstract
Clostridium sordellii is a lethal pathogen for both animals and humans. Severe capillary leakage, toxic shock syndrome, and an extreme leukemoid reaction (LR), are hallmark features of C. sordellii infections and contribute to its high mortality rate. Here we report the discovery of a previously unknown and uncharacterized metalloproteinase of C. sordellii (referred as Mcs1) that cleaves human vascular cell adhesion molecule (VCAM)-1 in vitro, an adhesion molecule critical to hematopoietic precursor retention and leukocyte diapedesis. We successfully identified the open reading frame encoding Mcs1 within the ATCC 9714 genome and developed an Δmcs1 mutant strain using the ClosTron mutagenesis technology. No VCAM-1 proteolysis was observed from exotoxins collected from mutant strain cultures. Using advanced protein structural modeling and molecular dynamics simulation techniques, the 3D molecular structure and conformational features of Mcs1 were also characterized. Our data demonstrates that Mcs1 proteolytic activity is controlled by the electrostatic interactions between Glu113 and Arg227 residues and the gating motions within its cleft region. This pilot interdisciplinary investigation provided crucial experimental evidence of the existence of Mcs1 in C. sordellii and molecular insights into its 3D structure and proteolytic activity. These findings have the potential to help advance new therapeutics and diagnostics against deadly C. sordellii infections. Follow-up in vitro and in vivo work is under way to further characterize Mcs1 enzymatic kinetics and its role in C. sordellii pathogenesis.
I. Introduction
Clostridium sordellii is a ubiquitous Gram positive anaerobic bacillus found in the soil or intestines of humans and animals (reviewed in ref. 1). In humans, C. sordellii causes rapidly progressive and lethal necrotizing soft tissue infections,1–3 including 100% mortality in women undergoing postpartum or gynecological procedures and medically-induced abortions. C. sordellii infections are characterized by a lack of fever, a weak innate immune inflammatory response and a marked leukemoid reaction (LR), where absolute circulating white blood cell counts commonly reach 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 per μL whole blood.4 LR, attributable to the expansion of the granulocytic population with a left shift to myelocytes, metamyeloctyes and band-type neutrophils, serve as the sole predictor of fatal outcome for C. sordellii infections.1 In addition, LR development also serves as a hallmark feature of Clostridium difficile and Clostridium novyi infections.5,6
000 per μL whole blood.4 LR, attributable to the expansion of the granulocytic population with a left shift to myelocytes, metamyeloctyes and band-type neutrophils, serve as the sole predictor of fatal outcome for C. sordellii infections.1 In addition, LR development also serves as a hallmark feature of Clostridium difficile and Clostridium novyi infections.5,6
C. sordellii infections are manifested by the lethal and hemorrhagic toxins (TcsL and TcsH, respectively). TcsL and TcsH are members of the large clostridial cytotoxins (LCC) family, which also consists of toxin A and toxin B from C. difficile and alpha-toxin by C. novyi. LCCs share high homology and possess glycosyltransferase activity, utilizing UDP-glucose or N-acetylglucosamine as their co-substrate to modify members of the Rho GTPase signaling superfamily. Once modified, Rho GTPases become inoperative, causing reorganization and degradation of the actin cytoskeleton, likely leading to the disruption of endothelial barrier function and the systemic capillary leakage characteristic of these clostridium infections.7–10
Two pairs of molecular interactions are critical to retaining polymorphonuclear leukocytes (PMNL) and PMNL precursors in the bone marrow during hematopoiesis: vascular cell adhesion molecule (VCAM)-1 to very late antigen (VLA)-4, and the chemokine/chemokine receptor pair of stromal cell derived factor (SDF)-1 (aka CXCL12) to its receptor CXCR4. VCAM-1 and SDF-1 are constitutively expressed by both bone marrow stromal cells and the sinusoidal venous endothelium of the hematopoietic compartment. Granulocytic precursors express high levels of CXCR4 and VLA-4, and during cell maturation these expression levels progressively decrease, thus facilitating an orderly release of mature granulocytes into the peripheral circulation. Several studies have demonstrated that targeted disruption of the VCAM-1/VLA-4 or CXCR4/SDF-1 axes results in a rapid, unregulated mobilization of hematopoietic precursors into the peripheral circulation.11–14
For unknown reasons, the C. sordellii LR drastically exceeds those observed during C. difficile and C. novyi infection, suggesting that exotoxins unique to C. sordellii contribute to this profound LR phenomenon. Our group recently demonstrated that the C. sordellii neuraminidase (NanS) both directly induces promyelocytic proliferation and modifies human VCAM-1 in vitro.15 However, animals infected with Δnans-mutant C. sordellii knockouts still developed LR, suggesting other virulence factors were responsible for augmenting the dramatic C. sordellii-associated LR (manuscript in progress).
In support of this hypothesis, we report here the identification of an uncharacterized metalloproteinase produced by C. sordellii, which we refer to as Mcs1 (metalloproteinase of C. sordellii – 1). We show that Mcs1 specifically cleaves human VCAM-1, an adhesion molecule critical to the retention of hematopoietic precursors to the bone marrow environment and leukocyte diapedesis. We successfully identified the Mcs1 open reading frame (ORF) in the C. sordellii ATCC 9714 strain genome. Inactivating this gene utilizing the ClosTron technology resulted in a loss of VCAM-1 cleaving ability. Using advanced computational modeling methods, we determined the three-dimensional atomic structure of Mcs1 and characterized the molecular features that are important to Mcs1 proteolytic activity.
II. Results
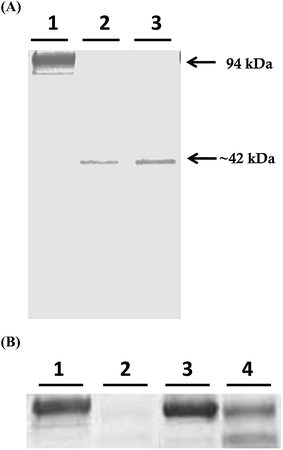
Mcs1 was originally identified following observations that crude, log phase C. sordellii exotoxin preparations grown in brain–heart infusion broth possessed proteolytic activity that cleaved human vascular cell adhesion molecule (VCAM)-1 in vitro (Fig. 1A, lane 2). Proteolytic cleavage of VCAM-1 by the unidentified protease generated a single, visibly truncated VCAM-1 species with a molecular mass of ∼42 kDa (Fig. 1A, lane 2), corresponding to about 47% of its complete form. Pre-treatment of the toxin preparation with protease inhibitors specific for serine, cysteine, aspartic, and aminopeptidase activities did not prevent VCAM-1 cleavage (Fig. 1A, lane 3). However, pre-treatment of the crude C. sordellii exotoxin preparation with EDTA completely prevented VCAM-1 cleavage (Fig. 2B; lane 3), suggesting the active moiety was a metalloproteinase. Heat inactivation of the toxin preparation partially abolished this effect (Fig. 2B; lane 4).Amino-terminal sequencing of the purified VCAM-1 cleavage identified the original N-terminus of the VCAM-1 molecule product (University of Oklahoma Health and Science Center: Proteomics Center), demonstrating that VCAM-1 was internally cleaved (approximately amino acid 381). Other cleavage products were not visible, suggesting that the remaining C-terminal portion of the VCAM-1 molecule was completely degraded or rendered unrecognizable by the detection antibody.
The Mcs1 open reading frame (ORF) was identified by searching the C. sordellii ATCC 9714 genome (Wasabi web-interfaced clostridium genome database; Victorian Bioinformatics Consortium, Department of Microbiology, Monash University, Australia) for genes annotated as putative or confirmed extracellular metalloproteinases. This search resulted in seven putative candidates, of which the ORF Cso_9714 00259 (ORF_00259; extracellular neutral metalloproteinase) was the single best potential Mcs1 candidate based on the predicted function and physical characteristics of the gene product (molecular weight of 56.9 kDa and isoelectric focusing point 4.79), and clear biochemical parallels to a partially purified form of the protease from crude exotoxin preparations (approximate molecular weight: 50.0 kDa, approximate isoelectric point: 4.2–5.3). The Mcs1 ORF was also identified in 5 out of 5 additional C. sordellii strains using PCR analysis and searching available annotated whole genome sequences (Wasabi web-interfaced clostridium genome database; see ESI Fig. S1†).
The ORF_00259 (now referred to as mcs1) contained 1539 base pairs (see ESI Fig. S2A†) that encoded for a 512-amino acid protein (see ESI Fig. S2B†). No identical sequence was found from GenBank for the full-length mcs1 sequence. Pfam and NCBI protein-BLAST analyses indicated that Mcs1 has 3 domains: a fungalysin/thermolysin propeptide (FTP) domain, a peptidase propeptide and YPEB (PEPSY) domain, and an M4 peptidase domain. No homologous structures of the FTP and PEPSY domains were found in the Protein Data Bank (PDB). Protein-BLAST searches against NCBI non-redundant protein PDB databases demonstrated that homology with Mcs1 was largely restricted to the catalytic M4 peptidase domain of the proteins. Bacillolysin (47% identity, 66% similarity) and elastase from Staphylococcus epidermidis (47% identity, 64% similarity), and aureolysin from Staphylococcus aureus (52% identify, 65% similarity) possessed the highest homology, suggesting that Mcs1 belongs to the M4 family of neutral peptidases (thermolysin-like metalloendopeptidases), a member of the zinc-dependent “GluZincin” superfamily. No Mcs1 homologues were identified in the C. difficile genome.
To determine if the translational product of the putative mcs1 gene (ORF_00259) was the sole metalloproteinase responsible for the observed VCAM-1-cleaving activity seen in the C. sordellii culture supernatant, an Mcs1 mutant strain was generated utilizing the ClosTron technique (see ESI Fig. S3A–C†). To confirm a single insertion of the group II intron within the mcs1 gene and genomic DNA of the mutant strain, a Southern blot hybridization was performed. As predicted, a single ∼3.9 kbp fragment was observed with the genomic DNA of the mcs1 mutant strain (see ESI Fig. S3D†).
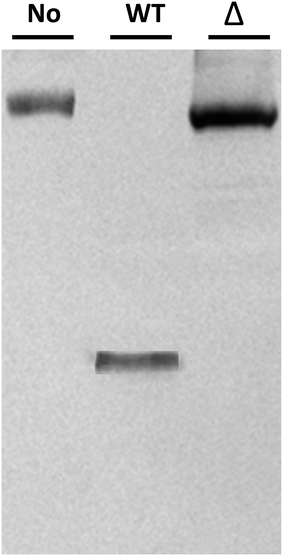
Potential Δmcs1 mutants were then screened for proteolytic activity using the VCAM-1 cleavage assay. Exotoxins from wild-type C. sordellii cultures cleaved VCAM-1, resulting in the 42 kDa truncated species (Fig. 2, lane 2). In contrast, no VCAM-1 proteolysis was identified from exotoxins collected from the Δmcs1 mutant strain culture (Fig. 2, lane 3). A slight 6.2 kDa shift in VCAM-1 was observed following exposure to the Δmcs1 mutant exotoxins (Fig. 2, lane 3), reflecting the enzymatic removal of sialic acid moieties from VCAM-1 via the C. sordellii neuraminidase.15 No differences in growth were observed between the wild-type and Δmcs1 mutant strains when cultured in BHI broth (data not shown).
When the Mcs1 protein sequence corresponding to the catalytic M4 peptidase domain was used, the NCBI protein-BLAST search against the PDB yielded the top ranked crystal structure (PDB ID: 1BQB)16 with 99% coverage, 4 × 10−92 E-values, and 52% sequence identity. This high-resolution structure (1.72 Å) of S. aureus aureolysin (referred as aureolysin) was selected as a template for homology modeling of the Mcs1 catalytic M4 peptidase domain. Amino acid sequence alignment between the catalytic domains of Mcs1 and aureolysin showed that, similar to aureolysin, the catalytic domain of Mcs1 is a single-chain enzyme consisting of 302 residues (Fig. 3). This high degree of similarity supported Mcs1's connection to the bacterial thermolysin family of metalloproteinases.
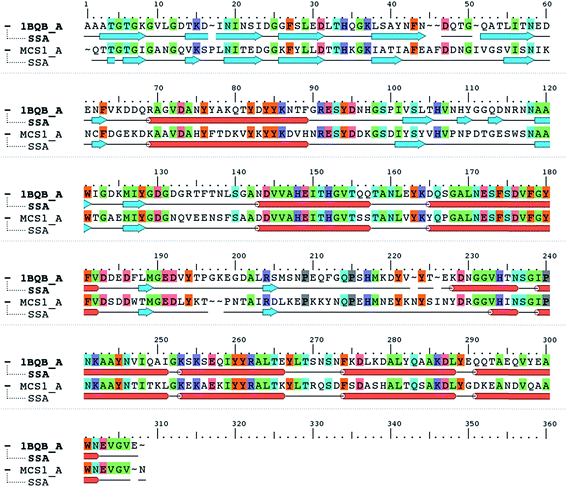 | ||
| Fig. 3 Protein sequence alignment of Staphylococcus aureus aureolysin (PDB: 1BQB) and C. sordellii Mcs1 catalytic domain. Conserved residues are highlighted in color. Cylinders represent α-helices and arrows represent β-strands. | ||
40 candidate structures were generated by Modeller and one candidate structure by PRIME. The energetic fitness of the 41 candidate structures, along with the template structure 1BQB as a reference, was evaluated using DOPE score. The PRIME generated structure was considered the best among all candidates and the Modeller structure #21 the second most suitable (Fig. 4A). DOPE per-residue based scores confirmed that the PRIME structure was energetically more favorable than Modeller structure #21 (Fig. 4B). Specifically, less per-residue deviation from template structure 1BQB was observed in the PRIME structure when compared to Modeller structure #21. The structural quality of the PRIME structure against the template structure was also evaluated using Ramachandran plots (Fig. 4C and D). Both the PRIME and template structure each had 100% of their residues within the allowed regions, with 81.4% and 87.9%, respectively, observed in the most favored region (Fig. 4C and D). Because of its comparable energetic and structural quality to the crystal structure template, the PRIME structure represented the best 3D atomic model for characterizing the functional peptidase domain of Mcs1.
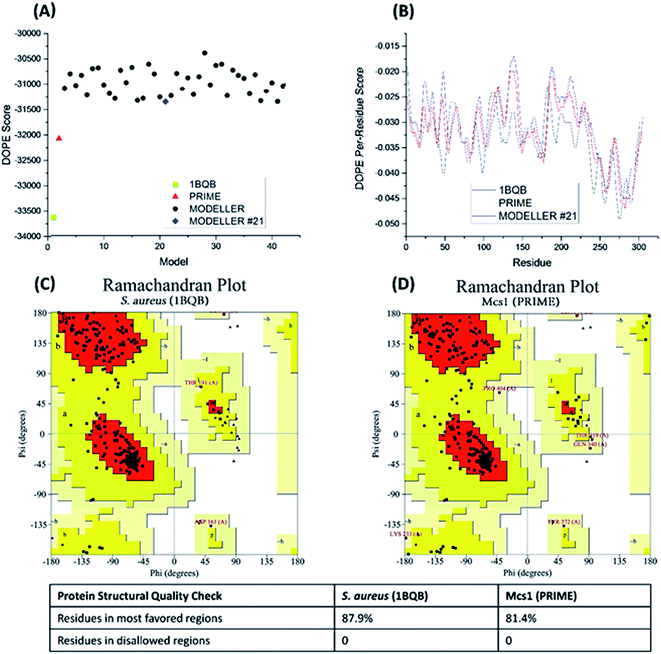 | ||
| Fig. 4 Energetic and structural quality of Mcs1 model structures and the S. aureus aureolysin crystal structure template (PDB: 1BQB) as a control. (A) Overall DOPE score (lower is better); (B) DOPE per-reside scores; (C) Ramachandran plot of 1BQB; and (D) Ramachandran plot of the best Mcs1 structure modelled using PRIME. | ||
Visual inspection of the Mcs1 model structure showed that the N-terminal subdomain (residues 1-156) consisted of mostly β-sheets and two α-helices. By comparison, the C-terminal subdomain (residues 157-302) contained primarily α-helices (Fig. 5A). The Mcs1 active site was located in a deep and narrow cleft between the N-terminal and the C-terminal subdomains. The entrance to this cleft was gated by loop structures found on both the N-terminal (loop-N) and C-terminal subdomains (loop-C) (Fig. 5A). The Mcs1 loop-C is longer than its counterpart in aureolysin due to insertions. The presence of one zinc and three calcium ions were located within the active site of Mcs1 and are essential to maintaining enzymatic activity. Additionally, the residues involved in coordinating the active site (i.e., zinc ion, substrate binding and catalysis) are conserved between Mcs1 and aureolysin (Fig. 3). The calcium ions located near the active site are a common feature shared by all members of thermolysin and neutral protease family (Fig. 5A).
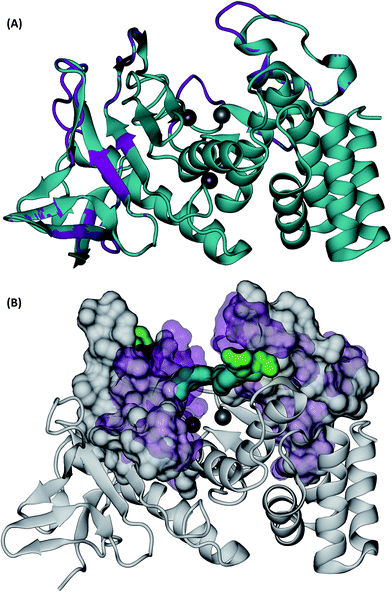 | ||
| Fig. 5 3D structures of C. sordellii Mcs1 (A) Mcs1 aligned with S. aureus aureolysin. C. sordellii Mcs1 (purple), Staphylococcus aureus aureolysin crystal structure 1BQB (cyan), zinc ion (silver), calcium ions (brown). Homology modeling was performed using PRIME. (B) Superimposition of the most open and the most closed conformations of Mcs1 extracted from MD simulations. Open cleft conformation (Mcs1: white solid surface, Glu113–Arg227: green solid surface) and closed cleft conformation (Mcs1: purple transparent surface; Glu113–Arg227: cyan solid surface). In the closed cleft conformation, Glu113–Arg227 interactions (cyan) completely block access to Mcs1 zinc cleavage site. | ||
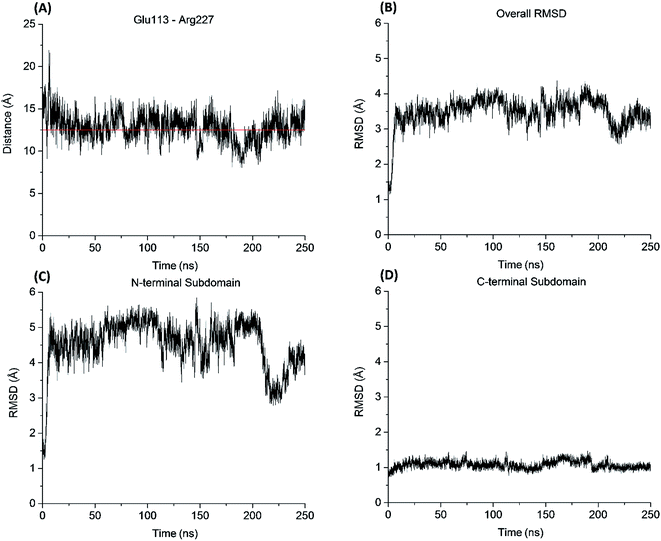
Similar to the aureolysin template structure, the Mcs1 active site cleft possesses a predominantly “closed” conformation. Cleft “opening” prompts Mcs1 enzymatic activity by allowing substrates to make contact with the zinc active site located at the bottom of the cleft. Our 250 nanosecond (ns) MD simulation demonstrated that cleft opening and closing is mostly dictated by the polar interactions between Glu113 and Arg227, located on the loop-N and loop-C subdomains, respectively, on opposing sides of the cleft. Maximal “open” and “closed” Mcs1 conformations illustrate how the accessibility of the zinc active site is “controlled” by the Glu113–Arg227 distance (Fig. 5B). MD simulation trajectory analysis also indicated that the Mcs1 active site is completed blocked when the Glu113–Arg227 distance is less than 12.5 Å. During the flexibility simulation, the Mcs1 active site was completely closed (Glu113–Arg227 distance < 12.5 Å) ∼50% of the time and only moderately accessible (Glu113–Arg227 distance = 12.5–15 Å) the other ∼50% (Fig. 6A). In very few instances was the Mcs1 cleft determined completely open (Glu113–Arg227 distance > 20 Å) (Fig. 6A).
The conformational flexibility of the subdomains and their contributions to the overall Mcs1 flexibility was also investigated (Fig. 6B). The C-terminal subdomain was far more rigid than that of the N-terminal subdomain (Fig. 6C and D). The overall root mean square deviation (RMSD) of the entire Mcs1 catalytic domain trended well with the RMSD of N-terminal domain, indicating the N-terminal subdomain is the driving force behind Mcs1's conformational changes (Fig. 6B and C).
III. Discussion
Bacterial metalloproteinases are important virulence factors for numerous human pathogens, including Vibrio cholerae (vibriolysin, PrtV and others), Bacillus anthracis (InhA – disrupts blood/brain barrier, causes lethal vascular hemorrhage; lethal toxin – a specific metalloproteinase that inactivates mitogen-activated protein kinase kinases in the host cells), Pseudomonas aeruginosa (LasB, cleaves PSGL-1 – the neutrophil ligand for endothelial P-selectin), Neisseria meningitidis (outer membrane protein NMBO135 is a zinc metalloproteinase and a target for vaccine development), Listeria monocytogenes (Mp1, a pH dependent metalloproteinase activated at low pH in the phagolysosome then activates broad range phospholipase C causing entrance of the pathogen into the cytosol of host cells), and the metalloproteinases of Clostridium botulinum type B and Clostridium tetani (reviewed in ref. 17). Aureolysin from S. aureus modulates biofilm formation and dispersal,18 contributes to the resistance of this pathogen to the innate immune system by cleaving human antimicrobial peptides19 and the complement protein, C3,20 and generates novel truncated versions of the staphylococcal immune modulator, phenol soluble modulin (PSM), having greater antimicrobial and immunostimulatory properties than the parent molecule.21Previous reports described C. sordellii as being a mildly proteolytic organism that digests gelatin, casein, coagulated egg albumen and meat in chopped meat medium.22 However, in this pilot work, we provided crucial experimental evidence of the existence of a novel extracellular metalloproteinase produced by the clinically important pathogen C. sordellii. To our knowledge, this is the first report of proteolytic activity attributable to a specific protein by C. sordellii, and the first description of a clostridial protease that acts specifically on host factors associated with leukocyte adherence.
Mcs1 displayed high homology to other proteases produced by significant bacterial pathogens, and we characterized the 3D atomic structure of Mcs1 catalytic domain using computational sequence analysis and homology modeling. The Mcs1 structure shares common features known to the thermolysin family. However, the cleft leading to the Mcs1 zinc active site is shown to be deeper and narrower than other thermolysin proteases. Our MD simulation demonstrated that the Glu113–Arg227 interactions control the accessibility to the Mcs1 zinc cleavage site, and the N-terminal subdomain dictates the overall conformational dynamics of the Mcs1 cleft region. A potential hinge motion between the N-terminal and the C-terminal subdomains could also contribute to the Mcs1 cleft opening and closing. The presence of numerous charged resides near and within the cleft suggests that Mcs1 substrate recognition is primarily driven by electrostatic interactions. The fact that Mcs1 rarely adopts a completely open conformation indicates that the Mcs1 cleft opening is likely induced by a series of coordinated molecular motions and interactions between Mcs1 residues and its substrates such as VCAM-1 (manuscript in preparation).
These current findings suggest Mcs1 may contribute to the development of the extreme LR observed during C. sordellii infection. The mechanisms driving the C. sordellii LR are not well understood. We previously reported that mice infected with C. sordellii displayed a robust increase in circulating granulocyte-colony stimulating factor (G-CSF). G-CSF induces the mobilization of leukocytes, both mature and immature, from the bone marrow into circulation in both mice and humans.11,12 The ability of Mcs1 to cleave human VCAM-1 could augment, at least in part, the premature egress already caused by G-CSF-induced elastase release. VCAM-1 also functions as a critical adhesion in the multistep cascade controlling PMNL recruitment to sites of infection (reviewed in ref. 23). We recently demonstrated that, in addition to VCAM-1, Mcs1 also cleaves other adhesions critical to extravasation, including ICAM-1, which is structurally similar to VCAM-1 (manuscript in progress). Therefore, it is plausible that Mcs1 may also play a role in the attenuated PMNL perfusion response commonly observed during fatal C. sordellii infection.1
Attempts at generating a recombinant form of Mcs1 have been challenging because basal expression of Mcs1 is cytotoxic to the E. coli expression host. However, small scale VCAM-1 cleavage assays using lysates collected from induced E. coli organisms cleave VCAM-1, but to a lesser degree than that caused by the C. sordellii culture supernatant alone. Partially purified native Mcs1 from C. sordellii culture supernatant also cleave VCAM-1. We are currently optimizing both the expression and purification conditions to improve the yields and specific activity for the native and recombinant forms of Mcs1 for future studies.
In conclusion, our findings suggest that the C. sordellii Mcs1 may be an important new member of the thermolysin-like family of metalloproteinases. The function and characterization of Mcs1 in LR development and pathogenesis of C. sordellii infection is now a current focus of our laboratory. Better understanding the molecular mechanisms, substrate specificity and in vivo role of Mcs1 will add to the growing body of knowledge of C. sordellii pathogenesis. Work is currently under way to investigate the molecular mechanisms of Mcs1 and VCAM-1 recognition. The experimental evidence and molecular insights gained from this pilot study are expected to open the door to new therapeutic interventions targeting Mcs1 proteolytic activity, which could potentially reduce the LR response associated with C. sordellii infection.
IV. Methods
Bacterial culture preparation
C. sordellii strain 9714 (ATCC, Manassas, VA) and the respective mcs1 mutant strain were grown anaerobically in Brain Heart Infusion (BHI; Becton Dickinson, San Jose, CA) media, and supplemented with thiamphenicol or erythromycin at 30 μg mL−1 and 2.5 μg mL−1, respectively when necessary. A Bactron II anaerobic chamber (Sheldon Manufacturing, Cornelius, OR) was utilized to maintain an anaerobic environment for all experiments involving C. sordellii growth and manipulation. Escherichia coli strains BL21(DE3), Top10 and CA434 were grown aerobically in LB media, supplemented with chloramphenicol or tetracycline at 25 μg mL−1 and 50 μg mL−1, respectively.VCAM-1 cleavage activity assay
Crude exotoxin culture supernatant samples were screened for presence of Mcs1 proteinase activity by cleavage of recombinant human VCAM-1 (R&D systems, Minneapolis, MN). Briefly, 20 μL of VCAM-1 (30 μg mL−1) was incubated with an equal volume C. sordellii exotoxins collected from wild type or Δmcs1 mutant strain overnight cultures. Samples were placed in a 37 °C water bath for 30 min. The reaction was terminated by addition of SDS-PAGE sample buffer containing no reducing agents, and samples were run on a 12% SDS-PAGE gel, transferred to a polyvinylidene difluoride membrane. Western blots were developed using a monoclonal mouse anti-human VCAM-1 antibody (R&D systems) followed by detection with the Pierce ECL2 Western Blotting Substrate (Thermo Fisher Scientific, Waltham, MA).Creation of the Mcs1 mutant strain
The Mcs1 mutant strain was generated in the C. sordellii ATCC 9714 background using the ClosTron group II intron gene knockout system developed by Heap et al.24 The target site and primer sequences for mcs1 gene insertional mutation was selected using the web based intron design tool based on the Perutka method located on the ClosTron website (http://clostron.com; Table 1). The selected target site was in the sense direction between the bases 780 and 781 from the start codon of the open reading frame (ORF) encoding mcs1. Primers 780-IBS, 780-EBS-1d, 780-EBS-2, and EBS-U were used for overlap extension PCR using the intron PCR template supplied in the Sigma Targetron kit and the manufacturer's recommended instructions [primers 780-IBS and 780-EBS1d at 0.2 μM, primers 780-EBS2 and 780-EBSU at 0.04 μM, 1 μL template, 1× GoTaq polymerase (Promega), 55 °C annealing, 30 cycles]. Both the ∼350 bp PCR product and ClosTron based pMTL007C-E5 knockout vector (supplied by Dr Nigel Minton, University of Nottingham, Nottingham, UK) were digested with HindIII and BsrGI, and the resulting digested products were ligated using standard molecular biological methods to produce the pCT-Mcs1 mutagenesis vector. The sequence and orientation of the resulting retargeted region of the group II intron was verified using DNA sequencing.| Primer | Sequence |
|---|---|
| MCS1F | AACTCTCAGTTTTCTAGCACCTT |
| MCS1R | ACTCCAACCTCATTCCATGC |
| 780-IBS | AAAAAAGCTTATAATTATCCTTAGGTATCGTAGGAGTGCGCCC AGATAGGGTG |
| 780-EBS-1d | CAGATTGTACAAATGTGGTGATAACAGATAAGTCGTAGGAAGT AACTTACCTTTCTTTGT |
| 780-EBS-2 | TGAACGCAAGTTTCTAATTTCGATTATACCTCGATAGAGGAAAG TGTCT |
| EBS-U | CGAAATTAGAAACTTGCGTTCAGTAAAC |
| PRBF | ATGTCAGACGCATGGCTTTC |
| PRBR | ATTACTGTGACTGGTTTGCACCACCCTCTTCG |
pCT-Mcs1 was transformed into conjugation competent E. coli strain CA434 and grown overnight in LB broth supplemented with 25 μg mL−1 chloramphenicol. 1 mL of overnight growth was pelleted, washed with PBS and re-suspended in 200 μL of an overnight C. sordellii ATCC 9714 culture. The mixed culture was spotted (∼10 μL per spot) onto a BHI agar plate and incubated anaerobically at 37 °C for 8 hours. Following, 2 mL of pre-reduced PBS was added to the plate, and the cell slurry was lifted from the agar using a cell scraper and plated onto BHI agar supplemented with 10 μg mL−1 thiamphenicol, 250 μg mL−1 cycloserine, and 4 μg mL−1 gentamycin. After 18 hours, thiamphenicol resistant colonies were streaked onto BHI agar supplemented with 2.5 μg mL−1 erythromycin, 250 μg mL−1 cycloserine, and 4 μg mL−1 gentamycin. Erythromycin resistant colonies appearing 3 days later were screened by PCR for insertion of the group II intron into mcs1.
Southern blot analysis
Briefly, 10 μg of genomic DNA from wild type C. sordellii 9714 or Δmcs1 was digested using PacI and separated on a 0.8% agarose gel. DNA was transferred to a Hybond-N+ nylon membrane (Amersham Biosciences) using the alkaline capillary method. Briefly, the agarose gel was treated twice with 0.25 N HCl for and twice with 0.4 M NaOH for 15 min each. The gel was placed on the capillary apparatus and overlaid with a positively charged nylon membrane (Roche, San Francisco, CA) pre-wetted with 0.4 N NaOH. After 6 h of transfer with 0.4 M NaOH, the nylon membrane was removed and washed for 5 min in 2× SSC and allowed to dry. A biotinylated DNA probe spanning the erm RAM and 3′ region of the group II intron was created using primers PRBF and PRBR supplied in the North2 South Complete Biotin Random Prime Labeling and Detection Kit (Thermo Fisher Scientific). The probe was used at a final concentration of 30 μg mL−1 and the membrane hybridized at 55 °C overnight, according to manufacturer's recommendations.Mcs1 protein sequence analysis
The Mcs1 ORF DNA sequence identified from C. sordellii ATCC 9714 genome was used to predict the Mcs1 protein sequence using the Translate Tool in the Expasy Proteomics Server.25 The translated protein sequence was initially used to putatively characterize the active site and possible protein characteristics. Pfam (Protein families database) was used to confirm the predicted motifs.26 Protein secondary structure prediction based on position-specific scoring matrices was performed on the PSI-PRED server.27 The protein domains and sequences were further characterized by protein-BLAST on National Center for Biotechnology Information (NCBI) server.28 We queried the Mcs1 amino acid sequence against the non-redundant database using default parameters and the results were manually inspected.Mcs1 homology modeling
The 3D structure for the Mcs1 protein was built by homology modeling. Initially, a protein-BLAST search against the Protein Data Bank (PDB) was performed on the NCBI server to identify candidate structural templates. Three important properties were considered to define a suitable template structure for Mcs1: query coverage, E-value score, and sequence identity. Two popular homology modeling software packages, Modeller v9.15 (ref. 29) and PRIME module of Schrodinger suite 2015-3,30 were used to perform the pairwise alignment of the Mcs1 and the template structure sequences using default parameters. The resulting alignments were inspected and the Mcs1 molecular models were subsequently constructed using Modeller and PRIME. As zinc and calcium ions are essential for Mcs1 proteolytic activity, they were monitored throughout homology modeling steps. A set of 40 candidate structures were generated by Modeller and one candidate structure was generated by PRIME's energy-based model building. These 41 Mcs1 candidate structures were evaluated based on energetic and structural quality measured in terms of Discrete Optimized Protein Energy (DOPE) score31 and Ramachandran plots,32 respectively. The best scored model was used as the starting structure for the MD simulation described below. Molecular visualization was performed with VMD33 and MAESTRO module of Schrodinger suite 2015-3.34Mcs1 molecular dynamics simulation
To further optimize the best scored structure and assess the structural characteristics of Mcs1, a 250 ns all-atom explicit solvent MD simulation was performed. Protonation states of the Mc1 protein residues were determined at pH 7 with PROPKA.35 The protonated Mcs1 structure was solvated in an orthorhombic TIP3P water box, leaving 10 Å between the solute surface and the box boundary. The Mcs1 protein and metal ions were parameterized with the OPLS 2005 force field.36 After the system was neutralized, an ionic concentration of 0.15 M NaCl was introduced to mimic experimental assay conditions. DESMOND 4.3 module of Schrodinger suite 2015-3 package37 was used to carry out the MD simulation. Before the production run, a 5-step simulation protocol was performed to relax the system: (1) 100 ps NVT ensemble with Brownian dynamics at 10 K with solute non-hydrogen atoms restrained; (2) 12 ps NVT ensemble using a 10 K Berendsen thermostat with a fast temperature relaxation constant, velocity resampling every 1 ps, and non-hydrogen solute atoms restrained; (3) 12 ps NPT ensemble using a 10 K Berendsen thermostat and a 1 atm Berendsen barostat with a fast temperature relaxation constant, a slow pressure relaxation constant, velocity resampling every 1 ps, and non-hydrogen solute atoms restrained; (4) 12 ps NPT ensemble using a 300 K Berendsen thermostat and a 1 atm Berendsen barostat with a fast temperature relaxation constant, a slow pressure relaxation constant, velocity resampling every 1 ps, and non-hydrogen solute atoms restrained; (5) 24 ps NPT ensemble using a 300 K Berendsen thermostat and a 1 atm Berendsen barostat with a fast temperature relaxation constant, a normal pressure relaxation constant. Finally, the 250 ns production run was performed in NPT ensemble without restraint. 300 K temperature and 1 atm pressure were maintained with Nosé–Hoover chain dynamics and Martyna–Tobias–Klein barostat.38 A 2 fs time step and periodic boundary conditions were applied in conjunction with particle-mesh Ewald39 to treat long-range electrostatics. A multiple-time-stepping algorithm was employed, in which bonded interactions short-range nonbonded interactions were evaluated at every time step and long-range electrostatic interactions were evaluated every three time steps at a cutoff of 9 Å.Author contributions
MA and DX wrote the main manuscript text, and MA helped prepare Fig. 1 and 2 and DX Fig. 3–6.AT conducted the experiments associated with the computer modeling of Msc1 and was involved in preparing Fig. 3–6.
DH, EM and JF were all involved in generating the Mcs1 mutant strain in C. sordellii and characterizing (via PCR, Southern blot and VCAM-1 cleavage assays) mutant strain candidates. Additionally, DH, EM and JF all helped prepare Fig. 1 and 2.
Additional information
(1) Competing financial interest statement: no authors involved with this manuscript have a commercial or other association that might pose a conflict of interest.(2) Financial support: this work was supported in part by the U. S. Department of Veterans Affairs, Office of Research and Development, Biomedical Laboratory Research Program (MJA, AEB, DLS), by the National Institutes of General Medical Sciences of the National Institutes of Health under Grant #P20GM103408 and #P20GM109007, by the Idaho Beef Council Research Grant (DX), and by Idaho State University College of Pharmacy.
(3) Presentation of information: portions of this work was presented at the Anaerobe Society of the Americas Bi-annual Conference in Chicago, IL in June of 2014 and at the 251st American Chemical Society National Meeting in San Diego, CA in March of 2016.
(4) Reprints request: a request for reprints should be made to the corresponding authors: Dr Michael J. Aldape, Veterans Affairs Medical Center, Infectious Diseases Section, 500 West Fort St. (Bldg 117; mail stop 151), Boise, ID 83702. Phone: +1-208-422-1000 ext. 7659; Fax: +1-208-422-1365; e-mail: E-mail: mike.aldape@va.gov.
Dr Dong Xu, Department of Biomedical and Pharmaceutical Sciences, College of Pharmacy, Idaho State University, Meridian, ID, 83642. Phone: +1-208-373-1832; Fax: +1-208-373-1834; e-mail: E-mail: xudong@isu.edu.
(5) Current affiliations: Dustin D. Heeney is now a graduate student at the University of California, Davis, One Shields Ave, Davis CA, 95616.
Acknowledgements
This material is based upon work supported in part by the U.S. Department of Veterans Affairs, Office of Research and Development Biomedical Laboratory Research Program, by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under Grant #P20GM103408 and #P20GM109007, by Idaho Beef Council Research Grant, and by Idaho State University College of Pharmacy. A special thanks to Drs Dena Lyras and Torsten Seemann (Victorian Bioinformatics Consortium, Department of Microbiology, Monash University, Australia) for providing access to the Wasabi clostridium genome database, and Dr Nigel Minton for providing the pMTL007-E5 knockout plasmid, E. coli strain CA434 and use of his ClosTron mutagenesis system.References
- M. J. Aldape, A. E. Bryant and D. L. Stevens, Clostridium sordellii infection: epidemiology, clinical findings, and current perspectives on diagnosis and treatment, Clin. Infect. Dis., 2006, 43, 1436–1446 CrossRef CAS PubMed.
- D. Sicard, Deaths from Clostridium sordellii after medical abortion, N. Engl. J. Med., 2006, 354, 1645–1647 CrossRef PubMed.
- C. Sinave, T. G. Le, D. Blouin, F. Leveille and E. Deland, Toxic shock syndrome due to Clostridium sordellii: a dramatic postpartum and postabortion disease, Clin. Infect. Dis., 2002, 35, 1441–1443 CrossRef PubMed.
- M. M. Wintrobe, The leukocytes, in Clinical Hematology, ed. M. M. Wintrobe, Lee & Febiger, Philadelphia, 6th edn, 1967 Search PubMed.
- M. A. Marinella, S. D. Burdette, R. Bedimo and R. J. Markert, Leukemoid reactions complicating colitis due to Clostridium difficile, South. Med. J., 2004, 97, 959–963 CrossRef PubMed.
- K. Murray-Lillibridge, J. Barry, S. Reagan, D. O'flanagan, G. Sayers, C. Bergin, E. Keenan, S. O'briain, P. Plunkett, G. McMahon, C. Keane, P. O'sullivan, D. Igoe, L. Mullen, M. Ward, A. Smith and M. Fischer, Epidemiological findings and medical, legal, and public health challenges of an investigation of severe soft tissue infections and deaths among injecting drug users – Ireland, 2000, Epidemiol. Infect., 2006, 134, 894–901 CrossRef CAS PubMed.
- R. H. Adamson, F. E. Curry, G. Adamson, B. Liu, Y. Jiang, K. Aktories, H. Barth, A. Daigeler, N. Golenhofen, W. Ness and D. Drenckhahn, Rho and rho kinase modulation of barrier properties: cultured endothelial cells and intact microvessels of rats and mice, J. Physiol., 2002, 539, 295–308 CrossRef CAS.
- J. Waschke, W. Baumgartner, R. H. Adamson, M. Zeng, K. Aktories, H. Barth, C. Wilde, F. E. Curry and D. Drenckhahn, Requirement of Rac activity for maintenance of capillary endothelial barrier properties, Am. J. Physiol.: Heart Circ. Physiol., 2004, 286, H394–H401 CrossRef CAS PubMed.
- S. Hippenstiel, S. Tannert-Otto, N. Vollrath, M. Krull, I. Just, K. Aktories, C. von Eichel-Streiber and N. Suttorp, Glucosylation of small GTP-binding Rho proteins disrupts endothelial barrier function, Am. J. Physiol., 1997, 272, L38–L43 CAS.
- B. Geny, H. Khun, C. Fitting, L. Zarantonelli, C. Mazuet, N. Cayet, M. Szatanik, M. C. Prevost, J. M. Cavaillon, M. Huerre and M. R. Popoff, Clostridium sordellii lethal toxin kills mice by inducing a major increase in lung vascular permeability, Am. J. Pathol., 2007, 170, 1003–1017 CrossRef CAS PubMed.
- S. Siddiq, D. Pamphilon, S. Brunskill, C. Doree, C. Hyde and S. Stanworth, Bone marrow harvest versus peripheral stem cell collection for haemopoietic stem cell donation in healthy donors, Cochrane Database Syst. Rev., 2009, CD006406 Search PubMed.
- C. Carlo-Stella, N. M. Di, M. Magni, P. Longoni, M. Milanesi, C. Stucchi, L. Cleris, F. Formelli and M. A. Gianni, Defibrotide in combination with granulocyte colony-stimulating factor significantly enhances the mobilization of primitive and committed peripheral blood progenitor cells in mice, Cancer Res., 2002, 62, 6152–6157 CAS.
- J. P. Levesque, J. Hendy, Y. Takamatsu, P. J. Simmons and L. J. Bendall, Disruption of the CXCR4/CXCL12 chemotactic interaction during hematopoietic stem cell mobilization induced by GCSF or cyclophosphamide, J. Clin. Invest., 2003, 111, 187–196 CrossRef CAS PubMed.
- I. Petit, M. Szyper-Kravitz, A. Nagler, M. Lahav, A. Peled, L. Habler, T. Ponomaryov, R. S. Taichman, F. Arenzana-Seisdedos, N. Fujii, J. Sandbank, D. Zipori and T. Lapidot, G-CSF induces stem cell mobilization by decreasing bone marrow SDF-1 and up-regulating CXCR4, Nat. Immunol., 2002, 3, 687–694 CrossRef CAS PubMed.
- M. J. Aldape, A. E. Bryant, Y. Ma and D. L. Stevens, The leukemoid reaction in Clostridium sordellii infection: neuraminidase induction of promyelocytic cell proliferation, J. Infect. Dis., 2007, 195, 1838–1845 CrossRef CAS PubMed.
- A. Banbula, J. Potempa, J. Travis, C. Fernandez-Catalan, K. Mann, R. Huber, W. Bode and F. Medrano, Amino-acid sequence and three-dimensional structure of the Staphylococcus aureus metalloproteinase at 1.72 A resolution, Structure, 1998, 6, 1185–1193 CrossRef CAS PubMed.
- S. Miyoshi and S. Shinoda, Microbial metalloproteases and pathogenesis, Microbes Infect., 2000, 2, 91–98 CrossRef CAS PubMed.
- K. J. Lauderdale, B. R. Boles, A. L. Cheung and A. R. Horswill, Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation, Infect. Immun., 2009, 77, 1623–1635 CrossRef CAS PubMed.
- M. Sieprawska-Lupa, P. Mydel, K. Krawczyk, K. Wojcik, M. Puklo, B. Lupa, P. Suder, J. Silberring, M. Reed, J. Pohl, W. Shafer, F. McAleese, T. Foster, J. Travis and J. Potempa, Degradation of human antimicrobial peptide LL-37 by Staphylococcus aureus-derived proteinases, Antimicrob. Agents Chemother., 2004, 48, 4673–4679 CrossRef CAS PubMed.
- A. J. Laarman, M. Ruyken, C. L. Malone, J. A. van Strijp, A. R. Horswill and S. H. Rooijakkers, Staphylococcus aureus metalloprotease aureolysin cleaves complement C3 to mediate immune evasion, J. Immunol., 2011, 186, 6445–6453 CrossRef CAS PubMed.
- D. J. Gonzalez, C. Y. Okumura, A. Hollands, R. Kersten, K. Akong-Moore, M. A. Pence, C. L. Malone, J. Derieux, B. S. Moore, A. R. Horswill, J. E. Dixon, P. C. Dorrestein and V. Nizet, Novel phenol-soluble modulin derivatives in community-associated methicillin-resistant Staphylococcus aureus identified through imaging mass spectrometry, J. Biol. Chem., 2012, 287, 13889–13898 CrossRef CAS PubMed.
- L. D. S. Smith, Clostridium sordellii, in The pathogenic anaerobic bacteria, ed. L. D. S. Smith, Springfield, IL, Charles C. Thomas Publishing, 1975, pp. 291–298 Search PubMed.
- W. A. Muller, Mechanisms of transendothelial migration of leukocytes, Circ. Res., 2009, 105, 223–230 CrossRef CAS PubMed.
- J. T. Heap, O. J. Pennington, S. T. Cartman, G. P. Carter and N. P. Minton, The ClosTron: a universal gene knock-out system for the genus Clostridium, J. Microbiol. Methods, 2007, 70, 452–464 CrossRef CAS PubMed.
- P. Artimo, M. Jonnalagedda, K. Arnold, D. Baratin, G. Csardi, C. E. de, S. Duvaud, V. Flegel, A. Fortier, E. Gasteiger, A. Grosdidier, C. Hernandez, V. Ioannidis, D. Kuznetsov, R. Liechti, S. Moretti, K. Mostaguir, N. Redaschi, G. Rossier, I. Xenarios and H. Stockinger, ExPASy: SIB bioinformatics resource portal, Nucleic Acids Res., 2012, 40, W597–W603 CrossRef CAS PubMed.
- R. D. Finn, P. Coggill, R. Y. Eberhardt, S. R. Eddy, J. Mistry, A. L. Mitchell, S. C. Potter, M. Punta, M. Qureshi, A. Sangrador-Vegas, G. A. Salazar, J. Tate and A. Bateman, The Pfam protein families database: towards a more sustainable future, Nucleic Acids Res., 2016, 44, D279–D285 CrossRef PubMed.
- D. W. Buchan, F. Minneci, T. C. Nugent, K. Bryson and D. T. Jones, Scalable web services for the PSIPRED Protein Analysis Workbench, Nucleic Acids Res., 2013, 41, W349–W357 CrossRef PubMed.
- M. Johnson, I. Zaretskaya, Y. Raytselis, Y. Merezhuk, S. McGinnis and T. L. N. C. B. I. Madden, BLAST: a better web interface, Nucleic Acids Res., 2008, 36, W5–W9 CrossRef CAS PubMed.
- B. Webb and A. Sali, Comparative Protein Structure Modeling Using MODELLER, Current Protocols in Bioinformatics, 2014, 47, 5–32 Search PubMed.
- Schrodinger Release 2015-3: Prime, version 4.1, New York, NY, Schrodinger, LCC, 2015 Search PubMed.
- M. Y. Shen and A. Sali, Statistical potential for assessment and prediction of protein structures, Protein Sci., 2006, 15, 2507–2524 CrossRef CAS PubMed.
- R. A. Laskowski, M. W. MacArthur, D. S. Moss and J. M. Thornton, PROCHECK: a program to check the stereochemical quality of protein structures, J. Appl. Crystallogr., 1993, 283–291 CrossRef CAS.
- W. Humphrey, A. Dalke and K. Schulten, VMD: visual molecular dynamics, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS PubMed.
- Schrodinger Release 2015-3: Maestro, version 10.3, New York, NY, Schrodinger, LCC, 2015 Search PubMed.
- H. Li, A. D. Robertson and J. H. Jensen, Very fast empirical prediction and rationalization of protein pKa values, Proteins: Struct., Funct., Genet., 2005, 61, 704–721 CrossRef CAS PubMed.
- J. L. Banks, H. S. Beard, Y. Cao, A. E. Cho, W. Damm, R. Farid, A. K. Felts, T. A. Halgren, D. T. Mainz, J. R. Maple, R. Murphy, D. M. Philipp, M. P. Repasky, L. Y. Zhang, B. J. Berne, R. A. Friesner, E. Gallicchio and R. M. Levy, Integrated Modeling Program, Applied Chemical Theory (IMPACT), J. Comput. Chem., 2005, 26, 1752–1780 CrossRef CAS PubMed.
- Schrodinger Release 2015-3: Desmond Molecular Dynamics System, version 4.3, New York, NY, D. E. Shaw Research, 2015 Search PubMed.
- G. J. Martyna, M. L. Klein and M. Tuckerman, Nose–Hoover chains: the canonical ensemble via continuous dynamics, J. Chem. Phys., 1992, 97, 2635–2643 CrossRef.
- U. Essmann, L. Perera, M. L. Berkowitz, T. Darden, H. Lee and L. G. Pedersen, A smooth particle mesh Ewald method, J. Chem. Phys., 1995, 103, 8577–8593 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra27654g |
| This journal is © The Royal Society of Chemistry 2017 |