Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Mapping metabolites from rough terrain: laser ablation electrospray ionization on non-flat samples†
Benjamin
Bartels
a,
Purva
Kulkarni
ab,
Norbert
Danz
c,
Sebastian
Böcker
b,
Hans Peter
Saluz
d and
Aleš
Svatoš
 *a
*a
aResearch Group Mass Spectrometry/Proteomics, Max Planck Institute for Chemical Ecology, Hans-Knöll-Strasse 8, D-07745, Jena, Germany. E-mail: svatos@ice.mpg.de
bChair for Bioinformatics, Friedrich Schiller University Jena, Ernst-Abbe-Platz 2, D-07743, Jena, Germany
cFraunhofer Institute for Applied Optics and Precision Engineering IOF, Albert-Einstein-Strasse 7, D-07745 Jena, Germany
dDepartment Cell and Molecular Biology, Leibniz Institute for Natural Product Research and Infection Biology, Adolf-Reichwein-Straße 23, D-07745, Jena, Germany
First published on 27th January 2017
Abstract
Established laser-based ionization experiments require the surface of a sample to be as flat as possible to guarantee optimal laser focus. A laser ablation electrospray ionization (LAESI) source was custom-built to accommodate the topography of non-flat sample surfaces. Employing a confocal distance sensor, a height profile of the surface in question was recorded prior to the actual ionization experiment. The robustness of the system was evaluated by the metabolic profiling of radish (Raphanus sativus) leaves, chosen due to their pronounced surface features and known content of specialized metabolites. After the ionization experiments, light microscopy imaging was performed to evaluate ablation crater size and position. Reproducible ablation mark diameters of 69 ± 7 μm in average have been achieved. Mass spectrometric imaging capability has been proven on R. sativus leaf samples as well.
Introduction
Laser ablation electrospray ionization (LAESI), developed by Nemes and Vertes,1 is a secondary ionization technique that ablates molecules from water-rich substrates with an infrared laser and ionizes the formed plume in an electrospray cloud (ESI). The ESI-like mechanism allows both small metabolites and peptides/proteins to be detected quantitatively,1 and the infrared laser provides desorption capabilities that do not depend on an externally applied matrix. Instead, the indigenous water of the sample is used as a makeshift matrix by exploiting the absorption maximum of water at 2940 nm wavelength.2 The versatility of LAESI has been proven on microbial,3 plant,1,4 and animal5,6 samples over the last decade.Because LAESI, in principle, allows spatially-resolved acquisition of mass spectra, it can be applied in mass spectrometry imaging (MSI) of biological samples. In these types of experiments,7 the acquired mass spectra can be linked to their position of origin within the imaged sample. With this spatial context, two-dimensional (2-D) ion intensity maps can be generated that provide additional information about the distribution and localization of chemical moieties within the sample. MSI is a busy field in both clinical research8,9 as well as other fields such as plant biology.10
Apart from experiments on thin tissue sections, MSI faces the challenge that the surface of the sample imaged has topography. While tissue sections cut by a microtome are flat, with almost no topography to speak of, other kinds of samples, e.g. whole leaves or microbial communities, usually feature uneven surfaces with a distinct height profile. Laser-based ionization techniques applied in MSI experiments are influenced by such a height profile, because the beam divergence needed to focus a laser beam makes the area of sample irradiated by the incident laser beam a function of the distance between sample surface and focusing lens. By choosing a focusing lens with a sufficiently long focal length, this effect may be reduced. However, a lens with longer focal length and equal diameter has a lower numerical aperture and thus focusses an incident laser beam less tightly than a lens with shorter focal length of otherwise comparable nature. For highly resolved MSI experiments, a tightly focused laser beam is mandatory as lateral resolution is determined by the laser spot size.
On the scale relevant for MSI experiments (μm), the surface relief can be measured with a confocal distance sensor (CDS). These sensors have been employed in the profilometry of semiconductors for over a decade.11 Commercially available chromatic CDSs separate white light into its component wavelengths by controlled chromatic aberration and subsequently analyze the reflected light through confocal optics to measure the distance to the reflective surface.12 By scanning the target surface prior to the laser ablation step, a height profile can be recorded to guide the sample surface on a sample stage in three dimensions to optimal laser focus.
In this work we demonstrate the feasibility of adapting the laser ablation step in LAESI experiments to non-flat surfaces of samples such as leaves by means of profilometry. In the future, by relying on the matrix-free nature of LAESI, we hope to conduct MSI experiments directly at ambient conditions without having to take extensive sample preparation steps; such steps might influence the chemical constitution of the sample.
Experimental
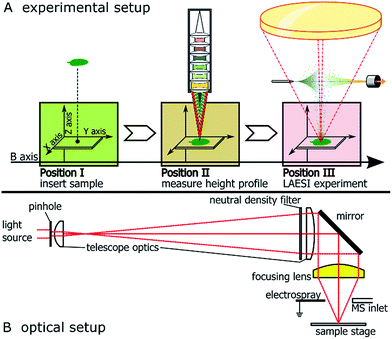
A custom-build LAESI source
The LAESI source was custom-built from commercially available optical elements (Thorlabs, Newton, NJ, USA and Layertec, Mellingen, Germany), motorized translation stages (Thorlabs), an OPO-pumped, infrared pulse laser operating at 2940 nm wavelength (Opotek, Carlsbad CA, USA), a syringe pump (Chemyx, Stafford, TX, USA) and electrospray emitter tips (New Objective, Woburn, MA, USA). The source was positioned in front of a LTQ Orbitrap XL mass spectrometer (ThermoScientific, La Jolla, CA, USA).Basic operating principle and beam path of the optical setup are shown in Fig. 1A and B, respectively. Please see the ESI† for a detailed description of assembly and used components.
Acquisition of height profiles
A confocal distance sensor (CDS) (μ-Epsilon, Ortenburg, Germany) was mounted in line with the optical axis of the focussing lens along the B-axis, offset by 108 mm. This way, the X, Y and Z-axes of the sample manipulation system were shared by the LAESI-source and the CDS. Alignment of the CDS′ measuring spot to the site of laser ablation was conducted by comparing the actual positions of ablation marks in white correction fluid (Stanger, Schwöllbogen, Germany) to the expected positions within the relative coordinate system of the CDS.Height profiles of sample surfaces were recorded by moving the sample below the CDS with the raster to be applied in the ablation experiment in the LAESI source. At every step, a Z-value was recorded and saved for the corresponding XY coordinates. For the purpose of visualization, false color representations of the recorded height profiles were generated by an in-house script written in R (version 3.2.3, 2015-12-10, R Foundation for Statistical Computing, http://www.r-project.org) and will here be henceforth referred to as topographic maps. These maps are represented using a standard ‘viridis’ color gradient scale13 where navy blue represents low intensity and the movement from green to yellow represents an increase in intensity for individual pixels on the imaged sample.
Missing values encountered while recording the height profiles of very rough sample surfaces were corrected using an in-house script written in R. The script performed missing value imputation using iterative mean substitution based on height profiles of the neighbouring pixels.
Metabolic profiling experiments
Young leaves from Raphanus sativus (radish, piccolo cultivar) bought at the local grocery store (REWE, Germany) were put on microscopy slides without further preparation and without the use of adhesives. An area of 0.3 × 3.1 mm2 was analyzed with a lateral resolution of 100 μm. The LAESI experiments were conducted with an average laser power 550 ± 33 μW at 10 pulses and 20 Hz repetition rate. This corresponds roughly to a laser energy of 27 μJ per pulse, taking a pulse width of 7 ns into account. Power measurements were conducted after the laser light passed through all optical elements. Per-pulse energy stability was not assessable with the available equipment. A PicoTip emitter (New Objective) facilitated the electrospray with 33.3% propan-2-ol in deionized water with 0.4% ammonium acetate at 0.4 μl min−1 flow rate and a voltage of −2.1 kV. Mass spectra were continuously acquired in negative ion mode with a LTQ Orbitrap XL mass spectrometer (ThermoScientific). Optical images were captured after the experiments with an AxioZoom macro-stereoscope (Zeiss, Jena, Germany).Mass spectrometry imaging
The acquired mass spectra from the LAESI experiment were assembled in the form of a data cube and exported as a matrix to a *.csv file. For visualization, 2-D ion intensity maps for ions of interest were generated by an in-house script written in R using the Cardinal package.14 These 2-D ion intensity maps are represented using a standard gray-scale gradient where dark shades represent low intensities and lighter shades represent an increase in intensity for individual pixels on the imaged sample.Results
Confocal distance sensors and colored samples
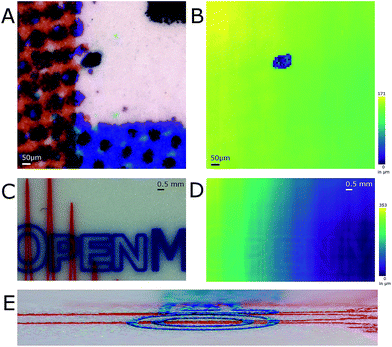
Optimizing the operational parameters of the CDS was crucial for consistent recording of height profiles on varying surface structures and colors. The sensor was operated at a dual acquisition mode with a primary exposure value of 100 μs for regular distance measurements and a secondary exposure value of 1000 μs as a fall-back option when the surface of the sample absorbed too much light and the intensity was too low for evaluation. For every measurement cycle, the median of 9 measured values was taken as height value. The observable measurement speed was thus 1.1 kHz on most surfaces. Height profile recording was artificially slowed down to evaluate the performance such as sample stage movement and possible influences from movement-induced vibrations.The influence of surface color on the distance measurement in the used CDS was evaluated by recording the height profile of a color graph printed on paper (Fig. 2A and B). A section of a bar chart featuring blue, red, black and white colors was profiled after burning a hole of 120 μm diameter into the paper. The hole was burned by IR-laser as a reference for both the optical image and the topographic map. The pattern printed by the color pigments on the piece measured was not discernible when the topographic map was viewed (Fig. 2B). On average, the height difference measured between the bottom of the hole and its surroundings was 113 μm as the stage is tilted. A USB storage device with colored markings was additionally profiled (Fig. 2C and D). In this case, the markings colored red and blue were discernible in the topographic map. Stereo-macroscopic imaging revealed the markings, which were elevated from the metal surface, formed surface features. In regions where the red and the blue markings crossed, elevation was found to be higher than in regions with only one colored marker (Fig. 2E).
Laser ablation step on flat samples
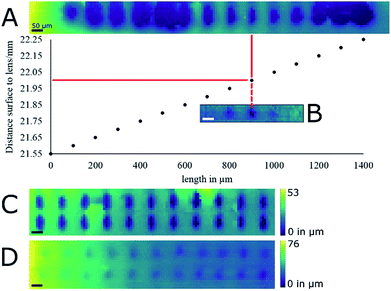
White correction liquid (Stanger) painted on microscopic slides was the first target for evaluating laser ablation performance on a reasonably flat surface. The paint formed a permanent uniform layer that could be ablated by IR laser, allowing height profiling for extended periods of time. The reproducibility of the height profiling was assessed by measuring the same ablation marks multiple times and by comparing them to stereo-macroscopic images. Laser energy, repetition rate and focus were not chosen with any particular rationale in mind apart from their ability to produce an ablation mark. The biggest contributor to uncertainty in the recording of height profiles was found to be the lead-screw backlash of the stage. The backlash distance is advertised by the manufacturer (Thorlabs) to be smaller than 6 μm. Positioning errors as well as uncertainties in the measurement of height profiles of ca. 5 μm were observed when repeatedly recording and measuring the height profile of the same area. By adopting a line-by-line scanning raster and thus approaching the region of interest (ROI) always from the same side, the positioning errors within the XY-axes were reduced so as to be unobservable by means of profilometry with our system. The uncertainty in the recorded height profiles depended on both the positioning of the sample with the Z-axis and the uncertainty of the chromatic confocal measurement principle itself. The former was of the same magnitude as the X- and Y-axes, and the latter was found to be in the range of a micrometer. Positional errors in the Z-axis during height profile recording were mostly negligible because displacement along the Z-axis was only needed when the target surface left the working distance of the CDS. Positional errors of roughly 5 μm that have to be assumed when the Z-axis displaces the sample up and down, did not lead to any observable differences in any of the laser ablation experiments.With the height profiling thus made reasonably reliable, the measurement spot of the CDS was aligned with the focal point of the laser beam by recording the height profile of ablation marks where the distance between paint surface and laser focusing lens was increased by a 50 μm increment between two subsequent ablation marks at a laser energy of 93 μJ per pulse (Fig. 3A) using 10 pulses at 20 Hz repetition rate. In the used correction fluid these laser settings resulted in well-defined ablation marks. To more precisely determine focal point position relative to the CDS, the experiment was repeated with 10 μm increments between subsequent ablation marks. In a second experiment, the height profile of an ablation mark array was measured at optimal laser focus with laser energy of 93 μJ per pulse with 10 pulses at 20 Hz repetition rate (Fig. 3C). The average ablation mark was found to be elliptical in shape with a major axis of 85 ± 5 μm and a minor axis of 40 ± 5 μm. Both experiments were later repeated at a lower energy of 27 μJ per pulse (Fig. 3B and D) that turned out to be more relevant for the laser ablation of biological samples (data not shown).
LAESI(-MSI) on non-flat surfaces of radish leaves
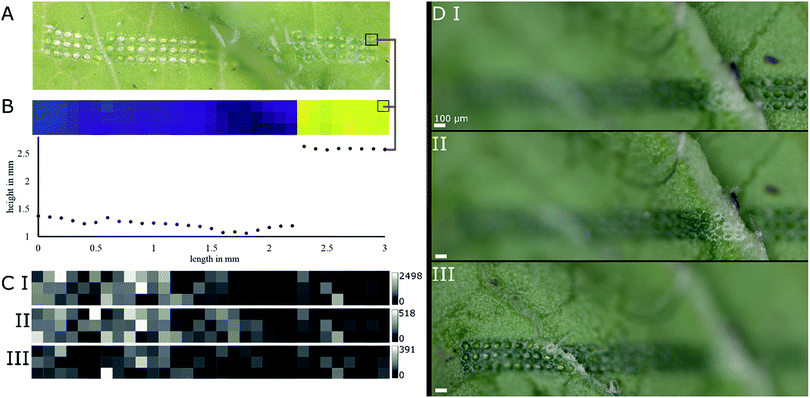
Young radish (R. sativus) leaves were subjected to LAESI experiments to test the height-profile-guided sample positioning on a biological sample with demanding surface topography and previously characterized specialized metabolites for validation.Mass spectra acquired in negative ion mode featured several m/z values corresponding well with sum formulas of glucosinolates typically found in radish.15 The sum formulas were putatively assigned by accurate mass and isotope patterns and are shown in Table 1. The mass features m/z 418.051, 434.024, and 447.054, corresponding well to sum formulas of glucoraphanin, glucoraphenin and glucobrassicin, respectively, are of biological interest. These glucosinolates are typically found in R. sativus cultivars15 and thought to play a role in herbivore deterrence.16 In a ROI of a representative sample (Fig. 4A and B) average-sized diameters of ablation marks were 69 ± 7 μm on top of a ledge and 69 ± 6 μm below the feature. Fig. 4B displays both a topographic map of the ROI and the height profile of the first row prior to the LAESI experiment. The ROI was deliberately chosen around a ledge-like surface feature, to include flat surfaces and sharp features to demonstrate the system's ability to adapt automatically to both situations. The maximum Z-distance between two particular ablation marks was 1.43 mm. Based on the results shown in Fig. 3B, this distance represents a height difference that would increase the diameter of the ablation mark roughly seven times, if the sample position were not to be adapted along the Z-axis.
| m/z | Ion | Sum formula | Compound |
|---|---|---|---|
| 146.046 | [M − H]− | C5H9NO4 | Glutamic acid |
| 175.025 | [M − H]− | C6H8O6 | Ascorbic acid |
| 179.056 | [M − H]− | C6H12O6 | Hexose |
| 418.051 | [M − H]− | C12H23NO10S3 | Glucoraphanin |
| 434.024 | [M − H]− | C12H21NO10S3 | Glucoraphenin |
| 447.054 | [M − H]− | C16H20N2O9S2 | Glucobrassicin |
| 477.064 | [M − H]− | C17H22N2O10S2 | Neoglucobrassicin |
As surface sampling happened in discrete steps, the experimental setup offered the possibility to conduct MSI experiments. Basic profiling of the sample and conducting a MSI experiment differed only in processing of the data after acquisition. In Fig. 4C, ion intensity maps generated from the data of the previously mentioned ROI are shown for previously mentioned mass features m/z 418.051, 434.024, and 447.054, namely glucoraphanin, glucoraphenin and glucobrassicin, respectively. A discrepancy can be spotted when comparing the optical image and the topographic map in Fig. 4A and B. Investigating other replicates of the same experiment confirmed the sample positioning was working correctly (Fig. 4D), even when sharp edges were involved.
Discussion
MSI on biological samples is usually connected to a distinct surface topography that hampers analysis with laser-based ionization methods. The typical approach is to section the sample into pieces that are then subjected more easily to laser ablation. This approach becomes increasingly complicated when the aim of analysis is the study of molecular interactions rather than the distribution of one specific compound of interest, especially if the interaction takes place between two different organisms. Even on relatively uniform surfaces, the correct focussing of the laser within ca. 100 μm range is critical to get a quantitative picture when studying single cells, as a high numerical aperture lens is critical in proper focussing.In this study, sample preparation was deliberately kept to a minimum, as this is the way to minimize interference prior to analysis. The only preparation step was cutting pieces from the sample that fit into the ion source. Although inconsistencies in the data were proven to originate from sample displacement while being removed from the LAESI source for optical inspection, a case can be made for sample fixation. Finding adhesives that do not create profound background signals in an ESI source and that comply with the no-chemical interference policy the source is developed under, is challenging. A possible solution may be the use of carbon based adhesives that are applied in electron microscopy, for example. Another possibility may be including instrumentation for optical observation into the experimental setup itself, removing the need to move the sample at all once it has been put into the source.
While testing the described experimental setup directly on a real world sample might seem hasty at first, it has to be considered that the basic mechanisms of LAESI have already been explored and published in greater detail.1,2,17–19 The methodology to be validated was in fact not the LAESI concept but focusing a laser onto an undisturbed surface of a biological sample. By demonstrating consistent laser focus on a real world sample while detecting the mass features expected to be found in said sample validated the feasibility of the experimental setup.
With our work, the feasibility of implementing profilometry in MS experiments to overcome the hurdle of a pronounced surface topography in laser-based ionization techniques is demonstrated, using LAESI imaging as an example. Although experimental details might differ in the other ionization techniques, such as uniform matrix deposition for AP-MALDI, the basic principle remains the same. Adapting a sample position based on a previously measured height profile not only helps in maintaining a consistent ablation mark diameter but also keeps the travel distance of created ions between sample surface and mass spectrometer inlet constant, as the mass spectrometer inlet is usually in a fixed position relative to the focusing lens. As the ablated material travels against gravity and is slowed down by ambient air, the ablated material is separated according to shape and mass. Again, keeping the travelling distance constant for all ablated spots is beneficial to data quality. In MALDI-based MSI of 3D samples on axial ion acceleration instruments, dramatic blue and red mass shifts occur and the obtained data must be corrected by software.20 Deviation in the travel distance of ions from different sample heights results in reduced mass accuracy that can be eliminated when the Z-axis compensates for surface topography.
The outlined experimental setup takes a two-step approach to performing profilometry and the corresponding LAESI experiment. Although conducting both steps in a single location might appear to be the more efficient and elegant solution, and also a way to keep down the physical dimensions of the ion source, we consider the two-step approach in physically separated locations to be more advantageous. There is no reduction in time needed to conduct the whole experiment, because in both cases a height value needs to be measured prior to the ionization for every coordinate position on the sample to be imaged. The time needed to move the XYZ-axes to the LAESI setup after profilometry of the ROI is negligible in relation to the overall duration of the whole experiment. By offsetting the profilometry instrumentation exactly one pixel ahead of the ionization experiment, the experimental time could be reduced considerably. Within individual rows of pixels, profilometry and ionization experiments could be conducted in parallel. Only at the beginning and the end of each row of pixels would the experimental procedure need to revert back to sequential one. The cost, however, would most likely be a reduced dynamic range for height value measurements along the Z-axis, as sample positioning along the Z-axis would be fixed to the ionization experiment to guarantee optimal laser focus and could not be used to expand the innate measuring range to which every kind of distance sensor is limited. The more accurate such a distance sensor, the smaller the measuring range tends to be. The instrumental realization of the distance sensor used is also a factor that needs to be considered. In a one-step setup, either the distance sensor or the focussing lens of the laser optics would need to sit at an angle smaller than 90° to the surface. Although not impossible from a technical point of view, such a setup is more susceptible to surface features either blocking the line of sight of the distance sensor21 – in this case, most likely a triangulation device – or blocking the beam path of the laser, further decreasing the dynamic range of height differences that are accessible with the setup.
By decoupling the profilometry step from the ionization experiment, height differences larger than the measuring range of the used distance sensor can be measured; the Z-axis is used to put the sample surface imaged back into the measuring range of the CDS. Positioning the CDS orthogonally to the surface also makes the setup less prone to artefacts caused by sharp surface features blocking the line of sight of the CDS. Free space along the axis used for moving the sample from the initial experimental step to the next could also be used for additional instrumentation for multi-modal imaging experiments. By using the same XYZ-manipulators and thus the same coordinate system in every experimental step along such an “assembly line”, the co-registration of the individual data sets would be greatly improved. The simplest addition would be an optical microscope or macro-stereoscope for capturing optical images prior to and after an MSI experiment.
The optical system here described will need some additional modification to get close to the target of a lateral resolution of 20 μm, but current results suggest this will soon be reachable. However, with a tighter focus, less material will be ablated and sensitivity will become a critical point in our experiments. Improving ion yield during ionization will require reducing the number of debris and metabolite clusters that are lost for ESI-assisted re-ionization. Heating the transfer capillary interface or adding ion funnels should improve ion transmission.22,23
Conclusions
We have successfully implemented profilometry in the LAESI setup, enabling accurate laser focusing during the profiling/imaging of metabolites from rough samples such as radish leaves. Under the current system configuration, well-defined spots are ablated close in size to plant cell diameter (69 ± 7 μm) with further room to improve. The experiments are performed in automatic regime under LabView control. We have developed a script for profilometry data rectification and for creating MSI images. This approach could be easily adapted to other desorption/ionization methods.Acknowledgements
We would like to acknowledge the contributions of Daniel Veit and Frank Müller in advising the design of all custom-made components and ultimately their manufacturing, Bernd Höfer (IOF Jena) for advising in the design of the laser optics, Josef Cvačka (IOCHB Prague) for suggesting μ-Epsilon sensors for laser triangulation, and Emily Wheeler for text editing. BB thanks the IMPRS for PhD fellowship and the Max-Planck-Gesellschaft is acknowledged for generous financial support.Notes and references
- P. Nemes and A. Vertes, Anal. Chem., 2007, 79, 8098–8106 CrossRef CAS PubMed.
- A. Vogel and V. Venugopalan, Chem. Rev., 2003, 103, 577–644 CrossRef CAS PubMed.
- S. A. Stopka, T. R. Mansour, B. Shrestha, E. Marechal, D. Falconet and A. Vertes, Anal. Chim. Acta, 2016, 902, 1–7 CrossRef CAS PubMed.
- R. Shroff, K. Schramm, V. Jeschke, P. Nemes, A. Vertes, J. Gershenzon and A. Svatos, Plant J., 2015, 81, 961–972 CrossRef CAS PubMed.
- P. Sripadi, J. Nazarian, Y. Hathout, E. P. Hoffman and A. Vertes, Metabolomics, 2009, 5, 263–276 CrossRef CAS.
- R. S. Jacobson, R. L. Thurston, B. Shrestha and A. Vertes, Anal. Chem., 2015, 87, 12130–12136 CrossRef CAS PubMed.
- B. Bartels and A. Svatos, Frontiers in Plant Science, 2015, 6, 7 CrossRef PubMed.
- J. L. Norris and R. M. Caprioli, Chem. Rev., 2013, 113, 2309–2342 CrossRef CAS PubMed.
- P. Neubert and A. Walch, Expert Rev. Proteomics, 2013, 10, 259–273 CrossRef CAS PubMed.
- B. A. Boughton, D. Thinagaran, D. Sarabia, A. Bacic and U. Roessner, Phytochem. Rev., 2016, 15, 445–488 CrossRef CAS PubMed.
- D. Vaughnn, C. Watkins and D. Anderson, Proc. SPIE, 2001, 4449, 169–177 CrossRef.
- A. K. Ruprecht, C. Pruss, H. J. Tiziani, W. Osten, P. Lucke, A. Last, E. Mohr and P. Lehmann, in Optical Measurement Systems for Industrial Inspection IV, Pts 1 and 2, ed. W. Osten, C. Gorecki and E. Novak, 2005, vol. 5856, pp. 128–135 Search PubMed.
- S. Garnier, Viridis: default color maps from ‘matplotlib’.R package version 0.3.4, 2016, https://CRAN.R-project.org/package=viridis Search PubMed.
- K. D. Bemis, A. Harry, L. S. Eberlin, C. Ferreira, S. M. van de Ven, P. Mallick, M. Stolowitz and O. Vitek, Bioinformatics, 2015, 31, 2418–2420 CrossRef CAS PubMed.
- E. Ciska, B. Martyniak-Przybyszewska and H. Kozlowska, J. Agric. Food Chem., 2000, 48, 2862–2867 CrossRef CAS PubMed.
- S. Textor and J. Gershenzon, Phytochem. Rev., 2009, 8, 149–170 CrossRef CAS.
- P. Nemes, A. A. Barton, Y. Li and A. Vertes, Anal. Chem., 2008, 80, 4575–4582 CrossRef CAS PubMed.
- A. Vogel and V. Venugopalan, in Optical Thermal Response of Laser-Irradiated Tissue, ed. A. J. Welch and M. J. C. VanGemert, Springer-Verlag Berlin, Berlin, 2nd edn, 2011, DOI:10.1007/978-90-481-8831-4_14, pp. 551–615.
- Z. Y. Chen, A. Bogaerts and A. Vertes, Appl. Phys. Lett., 2006, 89, 3 Search PubMed.
- P. Kulkarni, F. Kaftan, P. Kynast, A. Svatos and S. Bocker, Anal. Bioanal. Chem., 2015, 407, 7603–7613 CrossRef CAS PubMed.
- A. Schick and U. Breitmeier, in Optical Metrology in Production Engineering, ed. W. Osten and M. Takeda, 2004, vol. 5457, pp. 115–125 Search PubMed.
- H. Chen, N. N. Talaty, Z. Takáts and R. G. Cooks, Anal. Chem., 2005, 77, 6915–6927 CrossRef CAS PubMed.
- I. Marginean, J. S. Page, A. V. Tolmachev, K. Q. Tang and R. D. Smith, Anal. Chem., 2010, 82, 9344–9349 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra26854d |
| This journal is © The Royal Society of Chemistry 2017 |