Open Access Article
Open Access ArticleMatrix-isolation and theoretical study of the HXeCCXeH⋯HCCH and HXeCC⋯HCCH complexes†
Luís Duarte and
Leonid Khriachtchev*
and
Leonid Khriachtchev*
Department of Chemistry, University of Helsinki, P.O. Box 55, FI-00014 Helsinki, Finland. E-mail: leonid.khriachtchev@helsinki.fi
First published on 3rd January 2017
Abstract
We report on the identification of the complexes of two noble-gas hydrides, HXeCCXeH and HXeCC, with acetylene. These complexes were prepared by photolysis (250 nm) and annealing (55–65 K) of HCCH/Xe matrices. The experimentally observed monomer-to-complex shifts of the H–Xe stretching modes of the HXeCCXeH⋯HCCH (about +17 cm−1) and HXeCC⋯HCCH complexes (from +20 to +46 cm−1) indicate the stabilization of the H–Xe bond relatively to the monomers. The CCSD/cc-pVTZ calculations predict two structures for each complex. The HXeCCXeH⋯HCCH complex has quasi T-shaped and linear structures, with the H–Xe stretching modes blue-shifted and red-shifted by about +27 and −9 cm−1, respectively. The HXeCC⋯HCCH complex has bent and T-shaped structures, with the H–Xe stretching modes blue-shifted by about +46 and +42 cm−1, respectively. Based on the calculations, the experimental bands of the HXeCCXeH⋯HCCH and HXeCC⋯HCCH complexes are assigned to the quasi T-shaped and bent structures, respectively. Complexes of an open-shell noble-gas hydride and of a molecule with two noble-gas atoms are reported for the first time.
1. Introduction
Noble-gas hydrides constitute a remarkable family of noble-gas compounds with the general formula HNgY where Ng is a noble-gas atom and Y is an electronegative atom or fragment. The first noble-gas hydrides were prepared in low-temperature matrices by Pettersson et al. in 1995 (ref. 1) and, by now, more than 30 compounds of this type have already been reported,2–8 including an argon molecule HArF.9 We can also mention the preparation and identification of organokrypton (HKrCCH and HKrC4H)10,11 and organoxenon (HXeCCH, HXeCC, HXeCCXeH and HXeC4H)11–13 hydrides.Matrix isolation is an ideal tool to study non-covalent interactions.14 Due to the relatively weak bonding and large dipole moments, the HNgY molecules can be strongly affected by the interaction with other species and the hosts.2,4,15–24 The H–Ng stretching frequency is sensitive to the complex formation and can provide important insights on intermolecular interactions. In principle, the interaction with other species can enhance the kinetic stability of the HNgY molecules and may be considered as a possible route to their preparation under normal conditions.25 These complexes are usually characterized by blue shifts of the H–Ng stretching mode. For example, the H–Xe stretching mode of the HXeCCH⋯HCCH complex in a xenon matrix exhibits a shift of +19 to +28 cm−1 in comparison with HXeCCH monomer.20 The HKrCCH⋯HCCH complex has been identified in a krypton matrix with a complexation-induced shift of the H–Kr stretching mode of +60 cm−1.21 Some other complexes (e.g., HXeOH⋯H2O, HKrCl⋯HCl, and HXeBr⋯HBr) are characterized by much larger experimental blue shifts of the H–Ng stretching mode (>100 cm−1).22–24
HXeCC and HXeCCXeH are unusual noble-gas hydrides because they represent an open-shell species and a molecule with two noble-gas atoms, respectively. Therefore, the interaction of these molecules with other species is of particular interest. Here we report on the identification of the HXeCCXeH⋯HCCH and HXeCC⋯HCCH complexes.
2. Materials and methods
2.1 Computational details
The quantum chemical calculations were performed at the M06-2X26,27 and CCSD28–30 levels of theory. The M06-2X functional has been shown to have a good performance for the systems, in which non-covalent interactions are important,26,27 and the CCSD level provides accurate geometries at a relatively affordable computationally cost.28–30 The cc-pVTZ and aug-cc-pVTZ basis sets were used for the light atoms.31 For Xe atoms, basis sets combined with effective core pseudopotentials (cc-pVTZ-PP and aug-cc-pVTZ-PP) were used.32 The basis sets and the pseudopotentials were taken from the EMSL Basis Set Library.33,34 The calculations were carried out using the Gaussian 09 (revision E.01) program35 and the MOLPRO (2015.1) program36,37 (for the HXeCCXeH⋯HCCH complex at the CCSD/cc-pVTZ-PP level). In order to probe the potential energy surface of the complexes, several initial geometries (with L-shaped, T-shaped, linear, and bent configurations) were analyzed.38 The geometry optimizations were followed by harmonic frequency calculations at the same level of theory, which also gave the zero-point vibrational energies (ZPVE) and verifies the nature of the obtained minima. The M06-2X calculations employed an ultrafine integration grid and very tight optimization convergence criteria. The interaction energies were calculated as the difference of the total energies of the complex and isolated species (with the geometries in the complex) and corrected for the ZPVE and the basis set superposition error (BSSE).39 The atomic charges were obtained using the natural population analysis (NPA)40 at the M06-2X level of theory.2.2 Experimental details
Acetylene (≥99.6%, AGA) and xenon (≥99.999%, AGA) were used without purification. The HCCH/Xe samples (mixture ratios of 1/250–1/2000) were deposited from the gas phase onto a CsI window at 30 K in a closed-cycle helium cryostat (DE-202A, APD). The matrix thickness was typically 100–150 μm. The FTIR spectra in the 4000–400 cm−1 range were measured at 9 K with a Nicolet 60 SX spectrometer by co-adding 500 scans at a spectral resolution of 1 cm−1. Photolysis of the matrices was performed at 9 K using an optical parametric oscillator (Continuum, OPO Sunlite with FX-1) at 250 nm with a pulse energy of ∼5 mJ and a repetition rate of 10 Hz. After photolysis, the matrices were annealed at different temperatures up to 65 K (typically, for 5 min) and then cooled down for spectral measurements. The products of photolysis and annealing were decomposed by a 488 nm argon-ion laser (Series 532, Model 35 LAS 450 230, Melles Griot) and a low-pressure mercury lamp (254 nm, HG-1, Ocean Optics).3. Results and discussion
3.1 Computational results
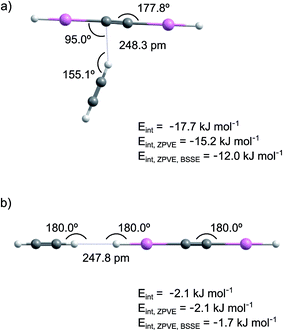
The calculated interaction energies are shown in Fig. 1 and Table S1 in ESI.† The interaction is much stronger in structure 1 than in structure 2. At the CCSD/cc-pVTZ-PP level of theory, the interaction energies of structures 1 and 2 are −12.0 and −1.7 kJ mol−1 after ZPVE and BSSE corrections. The M06-2X level leads to a similar interaction energy for structure 1 (−15.9 kJ mol−1 after ZPVE and BSSE corrections).
The NPA atomic charges and the bond lengths in HXeCCXeH and HCCH monomers and in the HXeCCXeH⋯HCCH complex are presented in Table 1 and S2 (ESI†), respectively. In structure 1, one of the positively charged H atoms of acetylene interacts with the negatively charged CC group of HXeCCXeH. Upon complexation, the charge-transfer character of HXeCCXeH is enhanced and the partial charges of the HXe and CC groups become more positive and more negative, respectively. The H–Xe and Xe–C bonds of HXeCCXeH in the complex are shorter by 0.45/0.68% and longer by 0.51/0.78% than those in the monomer (CCSD values here and below). The shortening of the H–Xe bond indicates its strengthening, which is connected with the increase of the positive charge of the HXe group. The change is bigger for the HXeC moiety located on the side, to which the acetylene molecule is tilted. The CC bond of HXeCCXeH slightly elongates (by 0.08%). For the acetylene molecule, the length of the CH bond interacting with HXeCCXeH increases by 0.75%, the CC bond elongates by 0.17%, and the other CH bond remains practically unchanged. In structure 2, the interaction occurs between H atoms of acetylene and HXeCCXeH. The H–Xe and Xe–C bonds, locating at the acetylene side, become longer and shorter by 0.56% and 0.69%, respectively, whereas the other H–Xe and Xe–C bonds become shorter and longer by 0.34% and 0.26%. The bond lengths of the acetylene molecule remain practically unchanged.
| Atom | HXeCCXeH⋯HCCH (structure 1) | |
|---|---|---|
| Charge | Change | |
| a T – atoms located on the side to which the acetylene molecule is tilted; Ac – atoms of acetylene; Ac-Int – atom of acetylene directly involved in the interaction. The change is calculated relatively to the values of the monomers. Structure 2 of the complex is not an energy minimum at this level. | ||
| H | −0.132 | +0.006 |
| Xe | +0.709 | +0.010 |
| C | −0.581 | −0.020 |
| CT | −0.579 | −0.018 |
| XeT | +0.714 | +0.014 |
| HT | −0.117 | +0.021 |
| HAc-Int | +0.253 | +0.022 |
| CAc | −0.234 | −0.003 |
| CAc | −0.260 | −0.029 |
| HAc | +0.226 | −0.004 |
The harmonic frequencies of HXeCCXeH and HCCH monomers and of the HXeCCXeH⋯HCCH complex are given in Table S3 (ESI†). The strongest mode of HXeCCXeH monomer is the H–Xe asymmetric stretch at 1559.8 cm−1 with a calculated intensity of ∼4800 km mol−1 (CCSD values). Other modes have much lower intensities or are outside the detection range and they are less important for the assignments. The calculated vibrational shifts of the H–Xe asymmetric stretching mode of the HXeCCXeH⋯HCCH complex are presented in Table 2. The CCSD calculations predict a blue shift of 26.7 cm−1 for structure 1 and a red shift of 9.0 cm−1 for structure 2. The increase of the frequency in structure 1 is explained by the shortening of the H–Xe bonds and by the increase of the positive charge of the HXe groups. In structure 2, the H–Xe bond involved in the interaction elongates while the other H–Xe bond slightly shortens, which probably leads to the calculated red shift of the H–Xe asymmetric stretching mode. The intensity of the H–Xe asymmetric stretching mode somewhat decreases in structure 1 and increases in structure 2. It should be noted that the calculated H–Xe stretching frequency of HXeCCXeH is overestimated with respect to the experimental value (∼1300 cm−1), which is typical for harmonic calculations of noble-gas hydrides.2
In addition, we calculated the HXeCCXeH⋯(HCCH)2 trimer at the M06-2X/aug-cc-pVTZ-PP level and found three energy minima with interaction energies of −32.7, −32.5, and −32.0 kJ mol−1 after the ZPVE and BSSE corrections (see Fig. S1 in ESI†). The calculated shifts of the H–Xe stretching mode of these structures relatively to HXeCCXeH monomer are +43, +38, and +31 cm−1, respectively (Table S3 in ESI†).
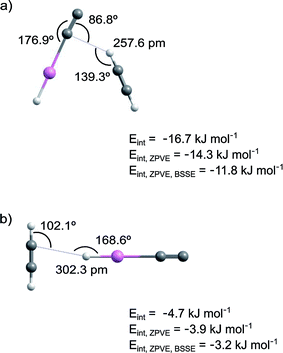
The calculated interaction energies are shown in Fig. 2 and Table S4 in ESI.† The interaction is much stronger in structure 1 than in structure 2. At the CCSD/cc-pVTZ-PP level of theory, the interaction energies of structures 1 and 2 are −11.8 and −3.2 kJ mol−1 after ZPVE and BSSE corrections. The M06-2X level leads to a similar interaction energy for structure 1 (−15.3 kJ mol−1 after ZPVE and BSSE corrections).
The NPA atomic charges and the bond lengths of the HCCH and HXeCC monomers and the HXeCC⋯HCCH complex are presented in Table 3 and S5 (ESI†), respectively. In structure 1, one of the positively charged H atoms of acetylene interacts with the negatively charged C atom of HXeCC. Upon the complex formation, the positive charge of the HXe group increases and the charge of the CC group of HXeCC becomes more negative. The H–Xe and Xe–C bond lengths decrease and increase by 0.69% and 1.18% (CCSD values), and the CC bond of HXeCC increases by 0.16%. For the acetylene molecule, the HC bond directly interacting with HXeCC increases by 0.47%, the CC bond increases by 0.17%, and the other CH bond remains practically unchanged. In structure 2, the interaction occurs between the H atom of HXeCC and the CC group of acetylene. Upon the complex formation, the H–Xe bond length decreases by 0.46% and the Xe–C bond elongates by 1.02%. The structural changes on the acetylene molecule are minor and the bond lengths change by ≤0.08%. As in the previous complex, the shortening of the H–Xe bond indicates its strengthening, which is connected with the increase of the positive charge of the HXe group.
| Atom | HXeCC⋯HCCH (structure 1) | |
|---|---|---|
| Charge | Change | |
| a Ac – atoms of acetylene; Ac-Int – atom of acetylene directly involved in the interaction. The change is calculated relatively to the values of the monomers. Structure 2 of the complex is not an energy minimum at this level. | ||
| H | −0.044 | +0.025 |
| Xe | +0.723 | +0.008 |
| C | −0.725 | −0.005 |
| C | +0.057 | −0.017 |
| HAc-Int | +0.252 | +0.022 |
| CAc | −0.234 | −0.004 |
| CAc | −0.257 | −0.026 |
| HAc | +0.228 | −0.003 |
The calculated frequencies of HXeCC and HCCH monomers and of the HXeCC⋯HCCH complex are presented in Table S6 in ESI.† The strongest vibrational mode of HXeCC corresponds to the H–Xe stretch at 1911.9 cm−1 with a calculated intensity of 771 km mol−1 (CCSD values). The calculated vibrational shifts for the H–Xe stretching mode are given in Table 4. The calculations predict blue shifts of 46.4 and 41.7 cm−1 for structures 1 and 2, respectively. The H–Xe stretching intensity decreases in both structures, which is connected with the blue shift of this mode, the shortening of the H–Xe bond, and the increase of the positive charge of the HXe group.
In addition, we studied the HXeCC⋯(HCCH)2 trimer at the M06-2X/aug-cc-pVTZ-PP level and found two energy minima with interaction energies of −31.3 and −28.1 kJ mol−1 after ZPVE and BSSE corrections (Fig. S2 in ESI†). The calculated shifts of the H–Xe stretching mode of these complexes compared to HXeCC monomer are +123 cm−1 and +77 cm−1, respectively (Table S6 in ESI†).
3.2 Experimental results and assignments
The HCCH/Xe (1/1000 and 1/2000) matrices deposited at 30 K contain essentially monomeric acetylene. The strongest absorptions of the acetylene monomer in a xenon matrix are at 3280.4 and 3266.8 cm−1 and at 727.6 cm−1.12,13,41 For higher acetylene concentrations (1/250 and 1/500), bands at 3251.9, 3248.8, 3229.3, 754.5, 751.6, 744.4, and 736.3 cm−1 belonging to acetylene dimers and multimers increase in intensity.41–48The HCCH/Xe matrices were irradiated at 250 nm typically for 15 minutes, which decomposed ∼25–30% of acetylene. Photolysis of acetylene leads to the appearance of CCH radicals (a broad band at ∼1852 cm−1),12,13,41 XeHXe+ (953.2, 842.6, and 730.5 cm−1),49 and Xe–CC (1767.4 cm−1).41 The CCH radicals are the primary products of photolysis of acetylene and their subsequent photodecomposition produces CC molecules that form the Xe–CC species.41 250 nm photolysis is known to produce higher amounts of Xe–CC relatively to 193 nm photolysis.12,50 The formation of C4 (1536.2 cm−1),51 C4H2 (3304.9, 1234.2, and 624.0 cm−1),11 and C8 (2057.0 cm−1)51 are also observed, especially in matrices with high acetylene concentrations. Decomposition of HCCH and CCH produces H atoms stabilized in the matrix.
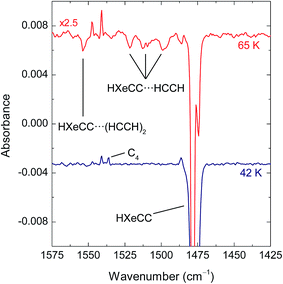
Annealing of the photolyzed matrices at ∼40 K mobilizes H atoms52,53 and leads to the formation of a number of known noble-gas hydrides (Fig. 3): HXeCCH (3273.5, 1486.3, 1480.7, and 626 cm−1), HXeCC (1747.7, 1478.3, and 1474.7 cm−1), HXeCCXeH (1305.8, 1300.9, and 1294.3 cm−1), and HXeH (1181.2 and 1166.4 cm−1).12,13,54 Using 250 nm photolysis, we can produce HXeCC and HXeCCXeH in relatively good amounts,12,50 which presumably increases the proportion of the complexes of HXeCC and HXeCCXeH with respect to the complexes of HXeCCH studied previously using 193 nm photolysis.20 To remind, the strongest bands of noble-gas hydrides originate from the H–Xe stretching modes and only these absorptions are expected to be observed for the studied complexes. Other products of annealing are C2H3 (1348.8, 891.0 cm−1),55 C4H (2050.4 cm−1),11,56 and C4H2.11
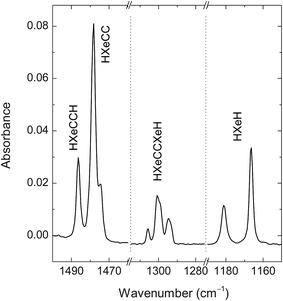 | ||
| Fig. 3 FTIR spectrum of HXeCCH, HXeCC, HXeCCXeH, and HXeH. The HCCH/Xe (1/1000) matrix was deposited at 30 K, photolyzed at 250 nm, and then annealed at 42 K. | ||
Annealing at higher temperatures (55–65 K) activates mobility of acetylene, which is evidenced by an increase of the bands of HCCH multimers. In particular, it leads to the appearance of bands at 1514.1, 1509.4, and 1505.4 cm−1 of the HXeCCH⋯HCCH complex reported previously (not shown here).20 The formation of the HXeCCH⋯HCCH complexes occurs via the attachment of mobile acetylene molecules to HXeCCH monomers. One can expect the attachment of acetylene also to HXeCC and HXeCCXeH. It should be mentioned that the high-temperature annealing changes the band structure of HXeCCXeH; in fact, the 1294 cm−1 band becomes the strongest component.
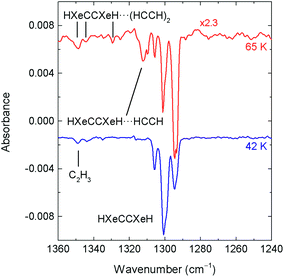
In this work, we present the difference decomposition spectra of noble-gas hydrides in the complexes under study. The same approach was used in our previous study of the HKrCCH⋯HCCH complex.21 Noble-gas hydrides are known to be very photolabile, which helps their identification. In fact, HXeCC can be decomposed using an argon-ion laser operating at 488 nm, whereas the HXeCCH and HXeCCXeH are practically stable under this irradiation. On the other hand, HXeCCH and HXeCCXeH can be decomposed by 254 nm light of a low-pressure mercury lamp.12,50 This selective photolysis of HXeCC at 488 nm is very important for the present work. Indeed, the bands of the HXeCC⋯HCCH complex can overlap with those of the HXeCCH⋯HCCH complex. In this situation, they cannot be clearly distinguished after annealing and by UV photolysis. Furthermore, the detected spectral features are rather weak and are not well visible in the annealing spectra (even for the HXeCCXeH⋯HCCH complex).
Fig. 4 shows the effect of irradiation at 488 nm on matrices annealed at 42 and 65 K in the range of HXeCC absorption. For the matrix annealed at 42 K, only the bands of HXeCC monomer are bleached. For the matrix annealed at 65 K, this irradiation consumes, in addition to the bands of HXeCC monomer, also the bands at 1521.6, 1512.8, and 1498.9 cm−1. These three bands are blue-shifted from the strongest H–Xe stretching band of HXeCC monomer by +44, +35, and +21 cm−1. These experimental shifts are in good agreement with the values calculated for the HXeCC⋯HCCH complex (+46.4 and +41.7 cm−1 for structures 1 and 2 at the CCSD/cc-pVTZ level). The small difference between the shifts calculated for structures 1 and 2 (∼4 cm−1) does not allow structural assignment based on the spectroscopic evidence. However, taking into account that structure 1 has a four times stronger interaction than structure 2, it is reasonable to expect that it is the predominant structure in the experiment. Nevertheless, we cannot completely exclude the formation of structure 2. A band at 1553.8 cm−1 produced by annealing at high temperature is also decomposed at 488 nm. This absorption is enhanced at higher temperatures and in samples with higher amounts of acetylene (HCCH/Xe matrices with 1/500 ratios). The shift of this band from the band of HXeCC monomer is +76 cm−1. This band is tentatively assigned to the HXeCC⋯(HCCH)2 trimer having computational shifts of +123 or +77 cm−1 (Fig. S2 in ESI†).
Fig. 5 shows the effect of irradiation at 254 nm on matrices annealed at 42 and 65 K in the range of the HXeCCXeH absorption (after 488 nm irradiation). For the matrix annealed at 42 K, only the bands of HXeCCXeH monomer are bleached in this spectral region. For the matrix annealed at 65 K, this irradiation consumes, in addition to the bands of HXeCCXeH monomer, also the bands at 1312.3/1309.9 cm−1. These bands are assigned to structure 1 of the HXeCCXeH⋯HCCH complex. The experimental shift observed for these bands, relatively to HXeCCXeH monomer (the strongest band at 1294 cm−1 after annealing at 65 K) is about +17 cm−1, which is in agreement with the shift of +26.7 cm−1 calculated for structure 1. For structure 2, the predicted shift is −9.0 cm−1 and no bands suitable for this structure are found in the experimental spectra. In agreement, the interaction in structure 2 is much weaker than in structure 1. Additional bands in this region, at 1348.5/1344.1 cm−1 and 1329.7 cm−1 produced by annealing at high temperature, are also decomposed at 254 nm. These absorptions are enhanced at higher temperatures and in samples with higher amounts of acetylene. The shift of these bands from the strongest band of HXeCCXeH monomer band is +55 and +36 cm−1. These bands are suitable for the HXeCCXeH⋯(HCCH)2 trimer structures with the computational shifts of +43 cm−1, +38 cm−1 and +31 cm−1 (tentative assignment; Fig. S1 in ESI†).
In reference experiments, HCCH/Xe (1/1000) matrices were directly annealed at 65 K (i.e. without photolysis at 250 nm) and then irradiated at 488 and 254 nm. In this case, no bands assigned above to the new complexes were observed. This observation supports that the new bands are indeed due to HNgY complexes but not the result of acetylene aggregation. In addition, experiments involving longer irradiation times were performed. In this case, HCCH/Xe (1/2000) matrices were irradiated until approximately 90% of acetylene was decomposed and annealed at 65 K. In this case, the relative intensity of the bands assigned above to the new complexes significantly decrease, which is reasonably explained by the smaller amount of acetylene in the matrix.
4. Concluding remarks
In this work, we have identified complexes of HXeCCXeH and HXeCC with acetylene. These complexes were prepared by annealing-induced diffusion of acetylene molecules (55–65 K) to HXeCCXeH and HXeCC monomers formed at lower temperatures (∼40 K). The H–Xe asymmetric stretching absorption of the HXeCCXeH⋯HCCH complex has bands at 1312.3/1309.9 cm−1 that are blue-shifted by about 17 cm−1 from the strongest band of HXeCCXeH monomer. For the H–Xe stretching absorption of the HXeCC⋯HCCH complex, the bands at 1521.6, 1512.8, and 1498.9 cm−1, shifted by up to +46 cm−1, were observed. The observed blue shifts indicate the stabilization of the H–Xe bond upon complexation, which is characteristic of noble-gas hydrides.2,4,15,20–24 Complexes of an open-shell noble-gas hydride and of a molecule with two noble-gas atoms are reported for the first time. No bands suitable for the HXeH complexes with acetylene were observed in this study.The CCSD/cc-pVTZ calculations predict two structures of the HXeCCXeH⋯HCCH complex (see Fig. 1). The H–Xe asymmetric stretching mode of structure 1 is blue-shifted by 26.7 cm−1, whereas in structure 2, it is red-shifted by 9.0 cm−1. Structure 1 has a much bigger interaction energy (−12.0 kJ mol−1 after ZPVE and BSSE corrections) than the second structure (−1.7 kJ mol−1). For the HXeCC⋯HCCH complex, two structures were also found (see Fig. 2). The H–Xe stretching mode is blue-shifted by 46.4 and 41.7 cm−1 for structures 1 and 2, respectively. Structure 1 has a bigger interaction energy (−11.8 kJ mol−1) than the second structure (−3.2 kJ mol−1). Based on these calculations, the experimental bands at 1312.3/1309.9 cm−1 are assigned to the HXeCCXeH⋯HCCH complex with structure 1. The bands at 1521.6, 1512.8 and 1498.9 cm−1 are assigned to the HXeCC⋯HCCH complex and most probably belong to structure 1. The band splitting observed for both complexes most likely results from the matrix site effect caused by specific interactions of the complexes with matrix atoms in certain local morphologies.
We have also considered the HXeCCXeH⋯(HCCH)2 and HXeCC⋯(HCCH)2 trimers (see Fig. S1 and S2 in ESI† for the optimized structures). In this case, the complexation effect is stronger than in the corresponding 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexes. In fact, the calculated shifts are +43, +38 and +31 cm−1 for HXeCCXeH⋯(HCCH)2 and +123 and +77 cm−1 for HXeCC⋯(HCCH)2. In the experimental spectra, bands at 1348.5/1344.1 and 1329.7 cm−1 (shifts of +55 and +36 cm−1) and at 1553.8 cm−1 (shift of +76 cm−1) are tentatively assigned to these trimers, respectively.
1 complexes. In fact, the calculated shifts are +43, +38 and +31 cm−1 for HXeCCXeH⋯(HCCH)2 and +123 and +77 cm−1 for HXeCC⋯(HCCH)2. In the experimental spectra, bands at 1348.5/1344.1 and 1329.7 cm−1 (shifts of +55 and +36 cm−1) and at 1553.8 cm−1 (shift of +76 cm−1) are tentatively assigned to these trimers, respectively.
It can be seen that the experimental shifts tend to be smaller than the calculated ones. For the HXeCC⋯HCCH complex, the experimental shifts are from +21 to +44 cm−1, whereas the calculated shift is +46 cm−1. For the HXeCC⋯(HCCH)2 trimer, the experimental shift is +76 cm−1 compared to the calculated values of +123 and +77 cm−1. A qualitatively similar situation is observed for the HXeCCXeH complexes. However, this level of agreement is very reasonable taking into account various matrix effects. Indeed, the calculations are made for species in vacuum whereas the experiments are performed in a polarizable medium. In addition, limitation of the computational methods can be important, for example, the use of the harmonic approximation. Similarly to the present case, the calculated shift for the HXeCCH⋯HCCH complex (+34 cm−1) was found to be larger than the experimental value (+19 and +28 cm−1). For the HXeCCH⋯(HCCH)2 trimer, the calculated shift (+81 cm−1) was also larger than the experimental one (+51 cm−1).20
It is worth noting that both calculated and experimental shifts of the HXeCCH⋯HCCH complex are smaller than those of the HXeCC⋯HCCH complex. It is a remarkable fact because the H–Xe stretching frequencies of the monomers are practically the same, showing a similar strength of this bond. This difference in the effect of complexation may be connected with a stronger interaction in the HXeCC⋯HCCH complex, which has an interaction energy of −11.8 kJ mol−1 compared to −10.3 kJ mol−1 in HXeCCH⋯HCCH (CCSD/cc-pVTZ; after ZPVE and BSSE corrections).57 To remind, the correlation between the complexation-induced shift and the interaction energy were found for the HXeI⋯HX complexes (X = I, Br, and Cl).16,58
Acknowledgements
This work was supported by Project KUMURA of the Academy of Finland (No. 1277993). The CSC-IT Center for Science is thanked for computational resources.References
- M. Pettersson, J. Lundell and M. Räsänen, J. Chem. Phys., 1995, 102, 6423–6431 CrossRef CAS.
- L. Khriachtchev, M. Räsänen and R. B. Gerber, Acc. Chem. Res., 2009, 42, 183–191 CrossRef CAS PubMed.
- L. Khriachtchev, A. Domanskaya, J. Lundell, A. Akimov, M. Räsänen and E. Misochko, J. Phys. Chem. A, 2010, 114, 4181–4187 CrossRef CAS PubMed.
- W. Grochala, L. Khriachtchev and M. Räsänen, in Physics and Chemistry at Low Temperatures, ed. L. Khriachtchev, Pan Stanford Publishing, 2011, pp. 419–446 Search PubMed.
- L. Khriachtchev, S. Tapio, A. V. Domanskaya, M. Räsänen, K. Isokoski and J. Lundell, J. Chem. Phys., 2011, 134, 124307 CrossRef PubMed.
- M. Turowski, M. Gronowski, J.-C. Guillemin and R. Kołos, J. Mol. Struct., 2012, 1025, 140–146 CrossRef CAS.
- M. Gronowski, M. Turowski and R. Kołos, J. Phys. Chem. A, 2015, 119, 2672–2682 CrossRef CAS PubMed.
- C. Zhu, M. Räsänen and L. Khriachtchev, J. Chem. Phys., 2015, 143, 244319 CrossRef PubMed.
- L. Khriachtchev, M. Pettersson, N. Runeberg, J. Lundell and M. Räsänen, Nature, 2000, 406, 874–876 CrossRef CAS PubMed.
- L. Khriachtchev, H. Tanskanen, A. Cohen, R. B. Gerber, J. Lundell, M. Pettersson, H. Kiljunen and M. Räsänen, J. Am. Chem. Soc., 2003, 125, 6876–6877 CrossRef CAS PubMed.
- H. Tanskanen, L. Khriachtchev, J. Lundell, H. Kiljunen and M. Räsänen, J. Am. Chem. Soc., 2003, 125, 16361–16366 CrossRef CAS PubMed.
- L. Khriachtchev, H. Tanskanen, J. Lundell, M. Pettersson, H. Kiljunen and M. Räsänen, J. Am. Chem. Soc., 2003, 125, 4696–4697 CrossRef CAS PubMed.
- V. I. Feldman, F. F. Sukhov, A. Y. Orlov and I. V. Tyulpina, J. Am. Chem. Soc., 2003, 125, 4698–4699 CrossRef CAS PubMed.
- L. Khriachtchev, J. Phys. Chem. A, 2015, 119, 2735–2746 CrossRef CAS PubMed.
- A. Lignell and L. Khriachtchev, J. Mol. Struct., 2008, 889, 1–11 CrossRef CAS.
- M. Tsuge, S. Berski, M. Räsänen, Z. Latajka and L. Khriachtchev, J. Chem. Phys., 2013, 138, 104314 CrossRef PubMed.
- A. Cohen, M. Tsuge, L. Khriachtchev, M. Räsänen and R. B. Gerber, Chem. Phys. Lett., 2014, 594, 18–22 CrossRef CAS.
- B. Biswas and P. C. Singh, Phys. Chem. Chem. Phys., 2015, 17, 30632–30641 RSC.
- S. Mondal and P. C. Singh, RSC Adv., 2014, 4, 20752–20760 RSC.
- A. Domanskaya, A. V. Kobzarenko, E. Tsivion, L. Khriachtchev, V. I. Feldman, R. B. Gerber and M. Räsänen, Chem. Phys. Lett., 2009, 481, 83–87 CrossRef CAS.
- K. Willmann, T. Vent-Schmidt, M. Räsänen, S. Riedel and L. Khriachtchev, RSC Adv., 2015, 5, 35783–35791 RSC.
- A. V. Nemukhin, B. L. Grigorenko, L. Khriachtchev, H. Tanskanen, M. Pettersson and M. Räsänen, J. Am. Chem. Soc., 2002, 124, 10706–10711 CrossRef CAS PubMed.
- A. Corani, A. Domanskaya, L. Khriachtchev, M. Räsänen and A. Lignell, J. Phys. Chem. A, 2009, 113, 10687–10692 CrossRef CAS PubMed.
- A. Lignell, J. Lundell, L. Khriachtchev and M. Räsänen, J. Phys. Chem. A, 2008, 112, 5486–5494 CrossRef CAS PubMed.
- R. B. Gerber, E. Tsivion, L. Khriachtchev and M. Räsänen, Chem. Phys. Lett., 2012, 545, 1–8 CrossRef CAS.
- Y. Zhao and D. G. Truhlar, Theor. Chem. Acc., 2008, 120, 215–241 CrossRef CAS.
- Y. Zhao and D. G. Truhlar, Acc. Chem. Res., 2008, 41, 157–167 CrossRef CAS PubMed.
- G. D. Purvis and R. J. Bartlett, J. Chem. Phys., 1982, 76, 1910–1918 CrossRef CAS.
- G. E. Scuseria, C. L. Janssen and H. F. Schaefer, J. Chem. Phys., 1988, 89, 7382–7387 CrossRef CAS.
- J. Čížek, J. Chem. Phys., 1966, 45, 4256–4266 CrossRef.
- T. H. Dunning, J. Chem. Phys., 1989, 90, 1007–1023 CrossRef CAS.
- K. A. Peterson, D. Figgen, E. Goll, H. Stoll and M. Dolg, J. Chem. Phys., 2003, 119, 11113–11123 CrossRef CAS.
- K. L. Schuchardt, B. T. Didier, T. Elsethagen, L. Sun, V. Gurumoorthi, J. Chase, J. Li and T. L. Windus, J. Chem. Inf. Model., 2007, 47, 1045–1052 CrossRef CAS PubMed.
- D. Feller, J. Comput. Chem., 1996, 17, 1571–1586 CrossRef CAS.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery Jr, J. E. Peralta, F. Ogliaro, M. J. Bearpark, J. Heyd, E. N. Brothers, K. N. Kudin, V. N. Staroverov, R. Kobayashi, J. Normand, K. Raghavachari, A. P. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, N. J. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, Ö. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski and D. J. Fox, Gaussian 09, Revision E.01, Gaussian, Inc., Wallingford CT, 2013 Search PubMed.
- H. J. Werner, P. J. Knowles, G. Knizia, F. R. Manby and M. Schütz, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2012, 2, 242–253 CrossRef CAS.
- H. J. Werner, P. J. Knowles, G. Knizia, F. R. Manby, M. Schütz, P. Celani, W. Györffy, D. Kats, T. Korona, R. Lindh, A. Mitrushenkov, G. Rauhut, K. R. Shamasundar, T. B. Adler, R. D. Amos, A. Bernhardsson, A. Berning, D. L. Cooper, M. J. O. Deegan, A. J. Dobbyn, F. Eckert, E. Goll, C. Hampel, A. Hesselmann, G. Hetzer, T. Hrenar, G. Jansen, C. Köppl, Y. Liu, A. W. Lloyd, R. A. Mata, A. J. May, S. J. McNicholas, W. Meyer, M. E. Mura, A. Nicklass, D. P. O'Neill, P. Palmieri, D. Peng, K. Pflüger, R. Pitzer, M. Reiher, T. Shiozaki, H. Stoll, A. J. Stone, R. Tarroni, T. Thorsteinsson and M. Wang, MOLPRO, version, 2015, 1, a package of ab initio programs Search PubMed.
- The HXeCCXeH⋯HCCH initial geometries were optimized at the M06-2X/aug-cc-pVTZ and MP2/aug-pVDZ (not presented) levels. The HXeCC⋯HCCH initial geometries were optimized at the M06-2X/aug-cc-pVTZ and CCSD/cc-pVDZ levels. The energy minima found were then optimized at the CCSD/cc-pVTZ level.
- S. F. Boys and F. Bernardi, Mol. Phys., 1970, 19, 553–566 CrossRef CAS.
- E. D. Glendening, A. E. Reed, J. E. Carpenter and F. Weinhold, NBO Version 3.1 Search PubMed.
- G. Maier and C. Lautz, Eur. J. Org. Chem., 1998, 1998, 769–776 CrossRef.
- V. I. Feldman, F. F. Sukhov, A. Y. Orlov, I. V. Tyul'pina, E. A. Logacheva and D. A. Tyurin, Russ. Chem. Bull., 2005, 54, 1458–1466 CrossRef CAS.
- A. V. Golovkin, D. I. Davlyatshin, A. L. Serebrennikova and L. V. Serebrennikov, J. Mol. Struct., 2013, 1049, 392–399 CrossRef CAS.
- D. G. Prichard, R. N. Nandi and J. S. Muenter, J. Chem. Phys., 1988, 89, 115–123 CrossRef CAS.
- G. T. Fraser, R. D. Suenram, F. J. Lovas, A. S. Pine, J. T. Hougen, W. J. Lafferty and J. S. Muenter, J. Chem. Phys., 1988, 89, 6028–6045 CrossRef CAS.
- G. W. Bryant, D. F. Eggers and R. O. Watts, Chem. Phys. Lett., 1988, 151, 309–314 CrossRef CAS.
- D. Prichard, J. S. Muenter and B. J. Howard, Chem. Phys. Lett., 1987, 135, 9–15 CrossRef CAS.
- Y. Ohshima, Y. Matsumoto, M. Takami and K. Kuchitsu, Chem. Phys. Lett., 1988, 147, 1–6 CrossRef CAS.
- H. Kunttu, J. Seetula, M. Räsänen and V. A. Apkarian, J. Chem. Phys., 1992, 96, 5630–5635 CrossRef CAS.
- H. Tanskanen, L. Khriachtchev, J. Lundell and M. Räsänen, J. Chem. Phys., 2004, 121, 8291–8298 CrossRef CAS PubMed.
- J. Szczepanski, S. Ekern, C. Chapo and M. Vala, Chem. Phys., 1996, 211, 359–366 CrossRef CAS.
- L. Khriachtchev, H. Tanskanen, M. Pettersson, M. Räsänen, V. Feldman, F. Sukhov, A. Orlov and A. F. Shestakov, J. Chem. Phys., 2002, 116, 5708–5716 CrossRef CAS.
- J. Eberlein and M. Creuzburg, J. Chem. Phys., 1997, 106, 2188–2194 CrossRef CAS.
- M. Pettersson, J. Lundell and M. Räsänen, J. Chem. Phys., 1995, 103, 205–210 CrossRef CAS.
- H. Tanskanen, L. Khriachtchev, M. Räsänen, V. I. Feldman, F. F. Sukhov, A. Y. Orlov and D. A. Tyurin, J. Chem. Phys., 2005, 123, 064318 CrossRef PubMed.
- K. I. Dismuke, W. R. M. Graham and W. Weltner, J. Mol. Spectrosc., 1975, 57, 127–137 CrossRef CAS.
- The HXeCCH⋯HCCH energies mentioned in the text were obtained by optimizing the structure, reported in ref. 20, at the CCSD/cc-pVTZ level with the Molpro 2015.1 software. The calculated shift upon complexation is +38 cm−1.
- C. Zhu, M. Tsuge, M. Räsänen and L. Khriachtchev, J. Chem. Phys., 2015, 142, 144306 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Calculated interaction energies, bond lengths, vibrational frequencies, and infrared intensities of the complexes and monomers. Optimized structures of the trimers. See DOI: 10.1039/c6ra25266d |
| This journal is © The Royal Society of Chemistry 2017 |