Open Access Article
Open Access ArticleAn oxime-based glycocluster microarray†
Antoine
Hoang
a,
Eugénie
Laigre
 b,
David
Goyard
b,
David
Goyard
 b,
Eric
Defrancq
b,
Eric
Defrancq
 b,
Françoise
Vinet
a,
Pascal
Dumy
c and
Olivier
Renaudet
b,
Françoise
Vinet
a,
Pascal
Dumy
c and
Olivier
Renaudet
 *bd
*bd
aLETI-CEA-Grenoble, 17 rue des Martyrs, 38054 Grenoble Cedex 9, France
bUniv. Grenoble Alpes, CNRS, DCM UMR 5250, F-38000 Grenoble, France
cEcole Nationale Supérieure de Chimie de Montpellier, 8 Rue de l'Ecole Normale, 34000 Montpellier, France
dInstitut Universitaire de France, 103 boulevard Saint-Michel, 75005 Paris, France. E-mail: olivier.renaudet@univ-grenoble-alpes.fr
First published on 5th June 2017
Abstract
Carbohydrate microarrays represent powerful tools to study and detect carbohydrate-binding proteins, pathogens or cells. In this paper, we report two original oxime-based methods to prepare surfaces displaying well-defined structures and valency in a given microspot with improved recognition potency with lectins. In a first “direct” approach, fully synthetic aminooxylated glycoclusters have been coated onto aldehyde-activated SiO2 (silicium substrate doped with 50 nm thermal oxide layer). To improve the preparation of the microarray in terms of rapidity and simplicity and to provide addressable surfaces on which sugars can be linked chemoselectively as clusters at defined plots, a second “indirect” strategy has been developed using successive oxime ligation steps. In both cases, binding assays with labelled lectins have revealed more potent and selective interaction due to the clustered presentation of sugars. The observed differences of interaction have been confirmed in solution by ITC.
Introduction
Carbohydrate microarrays have attracted intensive interest during the last decade.1 Their utility has been widely demonstrated to probe carbohydrate-binding proteins such as lectins, antibodies or enzymes and even pathogens or cells.2–5 Typically, carbohydrate moieties are immobilized on a glass slide at their anomeric position through a linker to both keep the sugar away from the surface and allow its presentation in a multivalent fashion to ensure high affinity with the biological target by means of the ‘glycoside cluster effect’.6 However, despite the large variety of linkers and chemical methodologies used in this area, the resulting two-dimensional multivalent organization and density of sugars on the surface is difficult to control and can thus strongly alter the avidity and selectivity of the interaction process.7,8 To overcome these limitations and improve the performance of these systems, recent publications have reported the utilization of glycocluster-based microarrays. This approach is indeed interesting since it allows the controlled presentation of sugars in a well-defined three-dimensional arrangement instead of randomized spacing between glycans. For example, it has been demonstrated that the spatial arrangement of dendrimeric structures offers better interactions with biological targets than monovalent ligands when immobilized onto surfaces.8 So far, a few glycocluster-arrays have been explored successfully with model lectins and multivalent glycoconjugates, sometimes with debatable molecular definition.9–12In this paper, we report on the preparation of glycocluster arrays following two oxime-based methods to obtain surfaces displaying well-defined structures and valency in a given microspot. In the first approach (Fig. 1A), tetravalent cyclopeptide-based glycoclusters presenting an aminooxy group have been directly immobilized onto aldehyde-activated SiO2 slides through oxime ligation. To provide readily addressable arrays on which sugars can be linked chemoselectively as clusters at defined plots, a second strategy (Fig. 1B) has been developed. An unglycosylated cyclopeptide scaffold was first immobilized onto the surface, and then successively treated to generate aldehyde functions and to covalently immobilize aminooxylated carbohydrates as clusters. In both cases, binding interactions have been evaluated with labelled lectins, thus confirming the recognition properties of the resulting glyco-surfaces.
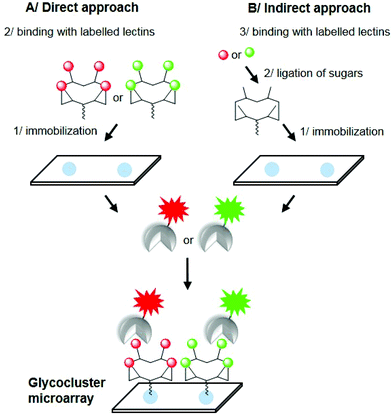 | ||
| Fig. 1 Direct (A) and indirect (B) approach for the preparation of glycocluster microarrays using oxime ligation. | ||
Results and discussion
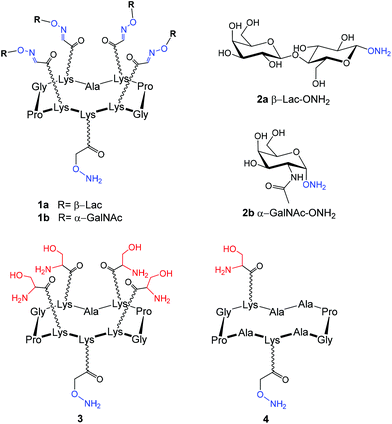
Modified glycans can be easily immobilized onto a large panel of commercially available supports including silica, aluminium oxide13 and gold surfaces14 displaying functional groups such as activated ester,15,16 epoxide,17,18 aldehyde,19 azide,7,20 alkyne21 or thiol.22,23 In addition, it was also demonstrated that oligonucleotide-,24 streptavidin-25,26 and polymer-functionalized27 surfaces can also serve in non-covalent immobilization but may cause unspecific interactions with the biological target. In the present study, we have decided to design our experiments on SiO2 slides modified using a previously reported protocol that was found suitable to immobilize oligonucleotides by oxime ligation.28,29 Briefly, the support was treated by silanization using 5,6-epoxyhexyltriethoxysilane at 80 °C to anchor epoxide functions onto the surfaces. Subsequent hydrolysis of the epoxide moieties and oxidation of the so-formed vicinal diols with sodium periodate generated aldehyde groups as confirmed by multireflexion IR experiments (i.e. presence of the carbonyl band at 1720 cm−1). Atomic Force Microscopy (AFM) was allowed to measure the roughness of the silane's layer on the surface at about 5–6 nm.With these aldehyde surfaces in hand, four aminooxy-glycosylated structures have been synthesized to be immobilized. On the basis of recent binding studies performed with lectin and cyclopeptide-based glycoclusters in solution30–37 and on solid-support,38–41 we have selected similar structures (Fig. 2) for this study that are: (i) tetravalent cyclopeptides displaying β-lactose 1a or α-N-acetylgalactosamine 1b and (ii) monomeric glycans 2a–b![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 which are selective for lectins PNA from Arachis hypogaea (peanut) and HPA (Helix pomatia agglutinin) respectively.
40 which are selective for lectins PNA from Arachis hypogaea (peanut) and HPA (Helix pomatia agglutinin) respectively.
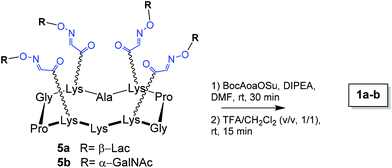
Cyclopeptide scaffolds 1a–b have been prepared from 5a–b by acylation of the free lysine with N-hydroxysuccinimidyl ester of Boc-aminooxy (BocAoaOSu)40 in DMF as shown in Scheme 1. After Boc deprotection in a mixture of 50% of TFA in CH2Cl2 and reverse-phase HPLC purification, the aminooxylated structures 1a–b were obtained in ∼40% yield.
Several parameters were evaluated next to immobilize and study these structures on aldehyde coated surfaces, such as the coating buffer, the coating concentration on the plate and the protein concentration. These preliminary experiments have indicated that the ideal coating concentration for the aminooxylated ligands is 50 μM and the minimal detectable concentration of the protein is approximately 1/10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 dilution of a solution at 1 mg mL−1. Following these conditions, compounds 1a–b and 2a–b have been manually coated horizontally (Fig. 3, lines 1–4) at the same concentration (0.2 μL at a concentration of 50 μM in water) and in triplicate on the modified surface with a spot diameter in the range of millimeters. For comparison we have also immobilized compound 2b at a higher concentration (i.e. 250 μM, Fig. 3, line 5). After 12 hours at room temperature, the slide was washed with phosphate buffer (pH 7) containing 1% tween and the surface was saturated successively with methoxylamine and bovine serum albumin (BSA) to prevent unspecific interactions of the resulting surfaces with the lectins. Finally, the full slide was incubated with the lectin HPA labeled with FITC in phosphate buffer (pH 7) for 1 hour at 37 °C and the presence of the lectin on the spot was visualized using a fluorescence microarray scanner.
000 dilution of a solution at 1 mg mL−1. Following these conditions, compounds 1a–b and 2a–b have been manually coated horizontally (Fig. 3, lines 1–4) at the same concentration (0.2 μL at a concentration of 50 μM in water) and in triplicate on the modified surface with a spot diameter in the range of millimeters. For comparison we have also immobilized compound 2b at a higher concentration (i.e. 250 μM, Fig. 3, line 5). After 12 hours at room temperature, the slide was washed with phosphate buffer (pH 7) containing 1% tween and the surface was saturated successively with methoxylamine and bovine serum albumin (BSA) to prevent unspecific interactions of the resulting surfaces with the lectins. Finally, the full slide was incubated with the lectin HPA labeled with FITC in phosphate buffer (pH 7) for 1 hour at 37 °C and the presence of the lectin on the spot was visualized using a fluorescence microarray scanner.
As shown in line 3, HPA binds strongly to the tetravalent cluster 1b displaying GalNAc. This observation first suggests that the immobilization of the glycocluster does not affect its recognition properties for HPA, which is indeed in excellent agreement with previous binding assays on resin beads.41 In addition, when similar compounds displaying Lac instead of GalNAc were spotted, no trace of lectin was observed on the corresponding plots (lines 1 and 2, Fig. 3), thus confirming the specificity of the interaction. More interestingly, only a little interaction with HPA can be detected when monomeric GalNAc 2b alone is immobilized even at a high concentration (lines 4 and 5, i.e. five times more concentrated than 1b). These results indicate that the presentation of GalNAc as a cluster strongly favors the interaction with HPA in comparison with the monosaccharide. To explain this effect, we hypothesize that the interaction is not only due to the local concentration of GalNAc on the array but also to a more appropriate spatial arrangement of the sugar at the surface of the cyclopeptide scaffold, which may promote multivalent interaction with HPA. It should be finally mentioned that different plot diameters are observed when 1b and 2b are spotted, presumably due to the difference in viscosity of both solutions leading to different drop spreading on the surface.
Because oxime ligation is a highly reproducible and quantitative reaction that can be performed under mild aqueous conditions, we next aimed at developing another strategy to prepare readily addressable arrays on which sugars can be linked chemoselectively as clusters at defined positions. Such an “indirect” approach would indeed present the advantage to avoid time-consuming full synthesis of the glycoclusters in solution since the glycocluster is assembled on the surface, and to prevent the manipulation (i.e. purification, solution storage, etc.) of such compounds that may be unstable.36 Instead, only simple aminooxylated building blocks are required in this case. The feasibility of this approach has been validated previously on resin beads.41 For this purpose, the unglycosylated cyclopeptide scaffold 3 which presents four serine residues (Fig. 1) was first immobilized through oxime ligation on the full aldehyde surface by dipping the glass slide in water. After washing and blocking with methoxylamine and BSA, the slide was treated with sodium periodate in water to convert serine residues into aldehydes and thus obtain surfaces fully covered with cyclopeptide presenting four aldehyde functions. We next manually added in triplicate aminooxylated Lac 2a (Fig. 4A, line 1) and GalNAc 2b (Fig. 4A, line 2) to generate glycoclusters 1a and 1b on the spotting area through oxime ligation with the aldehyde groups. To evaluate the efficiency of this process, we have compared the binding result, after incubation with the FITC-labelled PNA lectin, with a control experiment realized following the direct immobilization method described above, i.e. compounds 1a (Fig. 4A, line 1) and 1b (Fig. 4B, line 2) anchored to aldehyde slides.
As shown in Fig. 4, the presence of PNA is clearly visualized on expected line 1 (Lac) and not on line 2 (GalNAc) which again confirms the specificity of the interaction. More remarkably, the fluorescence intensity is similar as shown in Fig. 4A and B which undoubtedly indicates that the full synthesis of glycocluster 1a on the surface was efficient as no difference in interaction is observed when the direct approach is used (Fig. 4B). To our knowledge, this result represents the first example of glycocluster-microarray synthesis on slides.
We finally used the undirected approach to compare the recognition potency between the tetravalent scaffold and a monovalent analog and thus confirm that the difference in interaction observed in Fig. 3 is not due to the fact that the monovalent sugar is far closer to the surface than the tetravalent system (Fig. 4C). To do this, the cyclopeptide 4 functionalized with an oxyamine and bearing only one serine residue was immobilized on the slide and treated as described above with GalNAc 2b. As observed in Fig. 3, the binding with FITC-HPA has revealed a stronger interaction on the plot functionalized with tetravalent structures (Fig. 4C, two spots above). This result demonstrates again that the clustered presentation is more favorable for the interaction than monovalent conjugates (Fig. 4C, two spots below). All these data are in excellent agreement with previous studies performed on different surfaces and using different sugar densities and lectins.38,39,41,42 However, to confirm the difference in interaction, the binding affinities of the tetravalent and monovalent cyclopeptide-based structures (6 and 7, Fig. S9†) as well as of the corresponding monosaccharide (GalNAc) have been measured by isothermal titration calorimetry (ITC) with HPA (Fig. S10–12†). As expected, this experiment has indicated a Kd of approximately 548 nM for the tetravalent compound 6, which corresponds to a 757-fold binding improvement compared to the monovalent compound 7 (Kd of 415 μM). It is also interesting to note that the binding constant measured for GalNAc is in good agreement with the literature (205 μM)43 and is rather comparable to the monovalent structure 7, thus suggesting that the scaffold itself has only a limited influence in the binding. Altogether, ITC experiments clearly validate the reliability of the microarray methodology.
Conclusion
Herein we present two approaches based on oxime ligation to prepare glycocluster-based microarrays and the binding studies with FITC-labelled lectins. On one side, fully synthetic aminooxylated glycoclusters displaying four copies of Lac or GalNAc have been coated on aldehyde SiO2 slides under mild aqueous conditions. Binding assays have revealed both potent and selective interaction with HPA due to the multivalent effect as observed previously on different surfaces.38,39,41,42 Binding constants have been measured by ITC and have confirmed the reliability of the microarray experiments. On the other side, we have developed an indirect method to generate glycoclusters on surfaces using successive oxime ligation steps. Comparative interaction studies between both direct and indirect approaches with PNA have confirmed the formation of the glycocluster with a similar binding efficiency. This indirect approach represents a unique and significant improvement for the preparation of glycocluster microarrays in terms of rapidity and simplicity without affecting recognition properties. In our opinion, it is indeed advantageous in the fact that it only requires simple building blocks and highly efficient ligation reactions to generate universal surfaces on which sugars can be linked chemoselectively as clusters at defined plots. Further studies are currently under development in our laboratory with the aim to increase valency and diversity of the immobilized structures.Acknowledgements
This work was supported by CNRS, Université Grenoble Alpes, the “Communauté d'agglomération Grenoble-Alpes Métropole” (Nanobio program) and Labex ARCANE (ANR-11-LABX-0003-01). O. R. acknowledges the french Agence Nationale de la Recherche (ANR-12-JS07-0001-01 “VacSyn”) and the European Research Council Consolidator Grant “LEGO” (647938) for D. G. and E. L.References
- S. Park, J. C. Gildersleeve, O. Blixt and I. Shin, Chem. Soc. Rev., 2013, 42, 4310 RSC.
- J. Hirabayashi, M. Yamada, A. Kuno and H. Tateno, Chem. Soc. Rev., 2013, 42, 4443 RSC.
- K. T. Pilobello, L. Krishnamoorthy, D. Slawek and L. K. Mahal, ChemBioChem, 2005, 6, 985 CrossRef CAS PubMed.
- L. D. Huang, A. K. Adak, C. C. Yu, W. C. Hsiao, H. J. Lin, M. L. Chen and C. C. Lin, Chem. – Eur. J., 2015, 21, 3956 CrossRef CAS PubMed.
- N. Laurent, J. Voglmeir and S. L. Flitsch, Chem. Commun., 2008, 4400 RSC.
- J. J. Lundquist and E. J. Toone, Chem. Rev., 2002, 102, 555 CrossRef CAS PubMed.
- K. Godula, D. Rabuka, K. T. Nam and C. R. Bertozzi, Angew. Chem., Int. Ed., 2009, 48, 4973 CrossRef CAS PubMed.
- S. N. Narla, H. Nie, Y. Li and X. L. Sun, Glycoconjugate J., 2015, 32, 483 CrossRef CAS PubMed.
- H. M. Branderhorst, R. Ruijtenbeek, R. M. J. Liskamp and R. J. Pieters, ChemBioChem, 2008, 9, 1836 CrossRef CAS PubMed.
- A. L. Martin, B. Li and E. R. Gillies, J. Am. Chem. Soc., 2009, 131, 734 CrossRef CAS PubMed.
- K. Godula and C. R. Bertozzi, J. Am. Chem. Soc., 2012, 134, 15732 CrossRef CAS PubMed.
- X. Zhou, C. Turchi and D. Wang, J. Proteome Res., 2009, 8, 5031 CrossRef CAS PubMed.
- S. H. Chang, J. L. Han, S. Y. Tseng, H. Y. Lee, C. W. Lin, Y. C. Lin, W. Y. Jeng, A. H. J. Wang, C. Y. Wu and C. H. Wong, J. Am. Chem. Soc., 2010, 132, 13371 CrossRef CAS PubMed.
- D. H. Min, J. Su and M. Mrksich, Angew. Chem., Int. Ed., 2004, 116, 6099 CrossRef.
- O. Blixt, S. Head, T. Mondala, C. Scanlan, M. E. Huflejt, R. Alvarez, M. C. Bryan, F. Fazio, D. Calarese and J. Stevens, et al. , Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 17033 CrossRef CAS PubMed.
- C. Y. Huang, D. A. Thayer, A. Y. Chang, M. D. Best, J. Hoffmann, S. Head and C. H. Wong, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 15 CrossRef CAS PubMed.
- O. Oyelaran, Q. Li, D. Farnsworth and J. C. Gildersleeve, J. Proteome Res., 2009, 8, 3529 CrossRef CAS PubMed.
- Y. Zhang, C. Campbell, Q. Li and J. C. Gildersleeve, Mol. BioSyst., 2010, 6, 1583 RSC.
- S. E. Tully, M. Rawat and L. C. Hsieh-Wilson, J. Am. Chem. Soc., 2006, 128, 7740 CrossRef CAS PubMed.
- O. Michel and B. J. Ravoo, Langmuir, 2008, 24, 12116 CrossRef CAS PubMed.
- O. J. Barrett, A. Pushechnikov, M. Wu and M. D. Disney, Carbohydr. Res., 2008, 343, 2924 CrossRef CAS PubMed.
- S. Park, M. Lee, S. J. Pyo and I. Shin, J. Am. Chem. Soc., 2004, 126, 4812 CrossRef CAS PubMed.
- L. G. Harris, W. C. E. Schofield, K. J. Doores, B. G. Davis and J. P. S. Badyal, J. Am. Chem. Soc., 2009, 131, 7755 CrossRef CAS PubMed.
- Y. Chevolot, C. Bouillon, S. Vidal, F. Morvan, A. Meyer, J. P. Cloarec, A. Jochum, J. P. Praly, J. J. Vasseur and E. Souteyrand, Angew. Chem., Int. Ed., 2007, 46, 2398 CrossRef CAS PubMed.
- K. Godula and C. R. Bertozzi, J. Am. Chem. Soc., 2010, 132, 9963 CrossRef CAS PubMed.
- O. E. Galanina, M. Mecklenburg, N. E. Nifantiev, G. V. Pazynina and N. V. Bovin, Lab Chip, 2003, 3, 260 RSC.
- O. Norberg, I. H. Lee, T. Aastrup, M. Yan and O. Ramström, Biosens. Bioelectron., 2012, 34, 51 CrossRef CAS PubMed.
- E. Defrancq, A. Hoang, F. Vinet and P. Dumy, Bioorg. Med. Chem. Lett., 2003, 13, 2683 CrossRef CAS PubMed.
- N. Dendane, A. Hoang, L. Guillard, E. Defrancq, F. Vinet and P. Dumy, Bioconjugate Chem., 2007, 18, 671 CrossRef CAS PubMed.
- M. Monestier, P. Charbonnier, C. Gateau, M. Cuillel, F. Robert, C. Lebrun, E. Mintz, O. Renaudet and P. Delangle, ChemBioChem, 2016, 17, 590 CrossRef CAS PubMed.
- B. Thomas, C. Pifferi, G. C. Daskhan, M. Fiore, N. Berthet and O. Renaudet, Org. Biomol. Chem., 2015, 13, 11529 CAS.
- N. Berthet, B. Thomas, I. Bossu, E. Dufour, E. Gillon, J. Garcia, N. Spinelli, A. Imberty, P. Dumy and O. Renaudet, Bioconjugate Chem., 2013, 24, 1598 CrossRef CAS PubMed.
- M. Fiore, N. Berthet, A. Marra, E. Gillon, P. Dumy, A. Dondoni, A. Imberty and O. Renaudet, Org. Biomol. Chem., 2013, 11, 7113 CAS.
- I. Bossu, N. Berthet, P. Dumy and O. Renaudet, J. Carbohydr. Chem., 2011, 30, 458 CrossRef CAS.
- S. André, O. Renaudet, I. Bossu, P. Dumy and H. J. Gabius, J. Pept. Sci., 2011, 17, 427 CrossRef PubMed.
- I. Bossu, M. Šulc, K. Křenek, E. Dufour, J. Garcia, N. Berthet, P. Dumy, V. Křen and O. Renaudet, Org. Biomol. Chem., 2011, 9, 1948 CAS.
- O. Renaudet and P. Dumy, Org. Lett., 2003, 5, 243 CrossRef CAS PubMed.
- N. Dendane, A. Hoang, O. Renaudet, F. Vinet, P. Dumy and E. Defrancq, Lab Chip, 2008, 8, 2161 RSC.
- M. Wilczewski, A. Van der Heyden, O. Renaudet, P. Dumy, L. Coche-Guérente and P. Labbé, Org. Biomol. Chem., 2008, 6, 1114 CAS.
- H. Ide, K. Akamatsu, Y. Kimura, K. Michiue, K. Makino, A. Asaeda, Y. Takamori and K. Kubo, Biochemistry, 1993, 32, 8276 CrossRef CAS PubMed.
- O. Renaudet and P. Dumy, Org. Biomol. Chem., 2006, 4, 2628 CAS.
- M. P. Dubois, C. Gondran, O. Renaudet, P. Dumy, H. Driguez, S. Fort and S. Cosnier, Chem. Commun., 2005, 4318 RSC.
- J. F. Sanchez, J. Lescar, V. Chazalet, A. Audfray, J. Gagnon, R. Alvarez, C. Breton, A. Imberty and E. P. Mitchell, J. Biol. Chem., 2006, 281, 20171 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ob00889a |
| This journal is © The Royal Society of Chemistry 2017 |