Open Access Article
Open Access ArticleCarbonylation as a novel method for the assembly of pyrazine based oligoamide alpha-helix mimetics†
Seger
Van Mileghem‡
a,
Brecht
Egle‡
a,
Philippe
Gilles
 a,
Cedrick
Veryser
a,
Cedrick
Veryser
 a,
Luc
Van Meervelt
a,
Luc
Van Meervelt
 b and
Wim M.
De Borggraeve
b and
Wim M.
De Borggraeve
 *a
*a
aMolecular Design and Synthesis, Department of Chemistry, KU Leuven, Celestijnenlaan 200F, box 2404, B-3001 Leuven, Belgium
bBiochemistry, Molecular and Structural Biology, Department of Chemistry, KU Leuven, Celestijnenlaan 200F, box 2404, B-3001 Leuven, Belgium. E-mail: wim.deborggraeve@kuleuven.be
First published on 29th November 2016
Abstract
The design and synthesis of oligoamide α-helix peptidomimetics is reported. The oligoamide type systems are prepared in a modular fashion by coupling the monomers using palladium-catalyzed carbonylation chemistry. This enabled us to use substrates with a low nucleophilicity, leading to previously unreported pyrazine based oligoamide α-helix mimetics. The proof of principle is given by synthesizing a small set of compounds. Various end-capping groups were introduced and also a mixed multimer was successfully prepared.
Introduction
Protein–protein interactions (PPI), because of their importance in biological processes, are interesting though difficult targets for potential therapeutic applications. With good reason PPI were once considered undruggable, mostly because of their large (1500–3000 Å2) and shallow but dynamic interaction surfaces.1 Fortunately, in most cases, the interaction is highly dependent on just a small number of key amino acid residues. These are located on the interaction surface, forming a so called hot spot.2 Currently, the number of small molecule protein–protein interaction inhibitors is increasing.1,3–5 Analysis of multiprotein complexes in the Protein Data Bank by the Arora group has shown that in more than 60% of the cases, α-helices are present in the interaction surfaces.6 Hence, mimetics of this secondary structure element show significant medicinal potential. Due to the nature of PPIs, small molecules targeting these interactions can lie beyond Lipinski's rule of five.7 Several approaches exist where the helical backbone is replaced by a small molecule core decorated with amino acid side chains in the same spatial positions as in an α-helix at relative positions i, i + 3/i + 4, i + 7 of the α-helix. A multitude of such minimalist peptidomimetics8–18 have been reported in the last two decades. A remarkable class of molecules are the oligoamide systems which have shown potency in the modulation of protein–protein interactions.19Central in the design of these oligoamide systems is the modularity in synthesis. This elegant concept provides an unambiguous strategy for the assembly of monomeric units to multimers. Among the oligoamide type systems described in the literature (some examples in ref. 20–23 and Scheme 1A), amide bond formation is not always straightforward.24 The amide synthesis is typically performed via acyl chlorides or via the corresponding carboxylic acid with specialty coupling reagents (e.g. Ph3PCl2,25 Ghosez's reagent26 and Mukaiyama reagent27) since the amine coupling partners are not always nucleophilic. Due to the elaborate synthetic work, significant efforts have been made towards solid phase synthesis and recently, late stage introduction of the amino acid residues.26,28
 | ||
| Scheme 1 (A) An α-helix next to known oligoamide type helix mimetics. (B) Our proposed strategy towards pyrazine based peptidomimetics. | ||
Here, we report on palladium-catalyzed carbonylation as an alternative strategy for the assembly of monomers (Scheme 1B). The use of catalytic carbonylation chemistry as a coupling method is advantageous since (1) no stoichiometric amounts of coupling reagents are used, (2) it offers a broad functional group tolerance and (3) it is able to handle substrates which are very low in nucleophilicity.29
Previous experience with peptidomimetics30 and carbonylation31 in our group motivated us to combine this knowledge to pursue the synthesis of unreported pyrazine based oligoamide α-helix peptidomimetics via palladium-catalyzed carbonylation. This scaffold has the interesting feature of increasing hydrophilicity and solubility by adding hydrogen bond acceptors that can contribute to a so called wet edge.32,33 The modularity in design by using carbonylation chemistry enabled us to synthesize both pyrazine based multimers as well as a hybrid dimer.
Results and discussion
Retrosynthesis
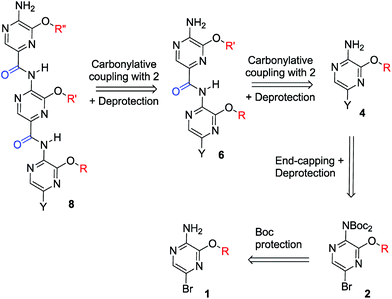
It was proposed that the pyrazine based multimeric structure (Scheme 2) mimics the amino acid residues at the same face of the helix as its phenyl and pyridine counterparts,34 namely at the i, i + 4 and i + 7 positions. Building block 1 (or its protected version 2) consists of a pyrazine core which enabled the development of a modular route (Scheme 2). The idea is inspired by peptide synthesis where a sequence of activation, coupling and deprotection is repeated a number of times to synthesize (oligo)peptides. Similarly, in our strategy, building blocks 2 serve as a substrate for the growing oligoamide multimers via a palladium-catalyzed carbonylation with CO gas,35 which are then deprotected to prepare them for the next coupling. The main reason for using aminocarbonylation as the coupling methodology is due to its ability to couple substrates which are low in nucleophilicity, such as aminopyrazine derivatives.36 Moreover, the reaction conditions of carbonylation chemistry are mild and therefore compatible with sensitive amino acid side chains present on the peptidomimetics.35 We recently reported a carbon monoxide precursor system based on the elegant two-chamber setup of the Skrydstrup group.31,37 In order to avoid the use of a CO lecture bottle and CO filled balloons, this setup was successfully used in a later stage for carbonylation couplings for the sake of safety (see the ESI†).In order to avoid homocoupling and polymerization, the first building block needs to have its halide group replaced with a moiety that cannot interfere in the following palladium-catalyzed coupling. This is a so called end-capping group, which in the peptide synthesis analogy corresponds to blocking the C-terminal amino acid by formation of e.g. an ester or attachment to a solid support. The end-capping can be done via carbonylation chemistry or other cross-coupling chemistry, such as Suzuki coupling38 forming biaryl type compounds (Y = Ar). Deprotection of an end-capped scaffold yields 4 that is ready to be coupled with another Boc-protected building block (2) via carbonylation. Finally, another iteration of the deprotection/coupling followed by a final deprotection yields the pyrazine based α-helix peptidomimetic trimer (8). To demonstrate this proof of principle, a range of monomeric building blocks was synthesized which were assembled into a set of multimers.
The Boc-protected aminopyrazines (2, Scheme 2) allow easy preparation of mixed multimers, containing both pyrazine and other types of monomers (e.g. phenyl scaffolds). This not only increases the potential structural diversity but also allows tuning the conformational rigidity of the multimers, which has been demonstrated to be advantageous for some PPI targets.39
Synthesis
In a first stage the synthesis of pyrazine building blocks is performed (Table 1). Aminopyrazine is dibrominated using NBS in DMSO.40 Following bromination, the desired amino acid mimicking side chain41 was attached using a fully regioselective SNAr in THF.40,42Table 1 summarizes the scope of the synthesized pyrazine building blocks. Monomers 1a–d were prepared in excellent yields. Following the synthesis of the monomers, we embarked upon the assembly of multimers. Some examples are depicted in Scheme 3. After Boc-protection of 1, an alkoxycarbonylation was performed to synthesize methyl ester end-capped building blocks 3 in good yields. Subsequently, the Boc groups were removed under acidic conditions. In a following step dimers 5a and 5b were synthesized and subsequently deprotected. The last two steps were repeated to obtain trimers 8a and 8b.X-ray analysis of 6b confirms the presence of intramolecular hydrogen bonds, in analogy to the oligoamide pyridine counterparts.34 An overlay image of our dimer 6b with a benzamide43 and pyridine analog44 is shown in Fig. 1 (see the ESI† for the refined structure with the according thermal ellipsoids). It was observed that the distance between the O-alkylated side chains is slightly smaller than is observed for the benzamide system (Fig. 1). This is a result of the additional hydrogen bond between the amide proton and pyrazine nitrogen, as was also the case for the pyridine dimer.44 The presence of this hydrogen bond is also responsible for the stronger downfield shift of the amide proton in comparison with the one in our hybrid system (10.09 ppm for 10vs. 8.45 ppm for 14) and the benzamide and pyridinyl amide systems described in the literature.34,44 Furthermore, a smaller angle of inclination is observed (155.9° versus 159.6° for our pyrazine system and the reported benzamide system, respectively). This clearly shows the structural resemblance between our and known systems. However, a more exact evaluation of which residues in a helix can be mimicked by these scaffolds would require a more detailed analysis (EKOS).45
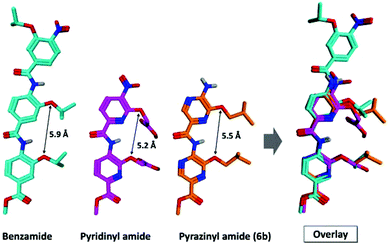 | ||
| Fig. 1 Overlay analysis of Wilson's benzamide crystal (CCDC 870274), Hamilton's pyridine crystal (CCDC 697087) and our pyrazine crystal (CCDC 1512413). | ||
In the case of dimer formation, an amount up to 30% of Buchwald–Hartwig amination46 side product was observed. This was confirmed by X-ray crystallography (see the ESI†). For the synthesis of 5b, the amination product (5b′, not shown) was isolated in 29% yield. These findings correlate with a report of Buchwald et al.,47 describing that in a palladium/xantphos system the preferred reaction conditions are electron deficient aryl halides combined with electron deficient nucleophiles, which is the case for our pyrazine substrates. On the other hand, the low trimer formation yields might be due to the poor solubility of the dimers in toluene, since significant amounts of the starting materials were recovered. Other solvents commonly used in carbonylation chemistry (such as dioxane, THF and DMF) did not improve the yield of these reactions.
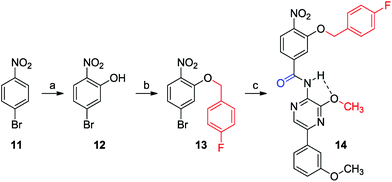
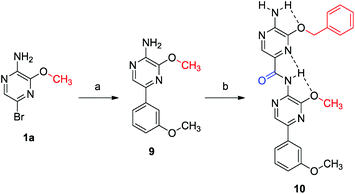
In a next example, Suzuki cross coupling was used as end-capping chemistry. This is demonstrated in Scheme 4. Building block 1a was end-capped to obtain compound 9. Boc-protected building block 2c was attached via carbonylation, which after deprotection led to dimer 10. Alternatively, other aryl building blocks can also be introduced as the second monomeric unit via this modular synthesis. This is demonstrated in Scheme 5. Phenyl monomer 13 was synthesized starting from p-nitrobromobenzene (11) via a vicarious nucleophilic substitution which was carried out in a mixture of THF and liquid ammonia.48 The resulting phenol 12 was treated with p-fluorobenzyl alcohol in a Mitsunobu reaction to yield 13.49 Finally, this monomer was coupled with 9, which gave rise to hybrid dimer 14.
Similar to the reported oligoamide systems, the water solubility of our multimers remains problematic. However, the introduction of a water solubilizing end capping group is possible via the carbonylation strategy. When end-capping 2d with N-methylpiperazine (not shown), only traces of the desired product were obtained. As it appears, secondary amines react with anilinic carbamate groups, forming unsymmetrical ureas at elevated temperatures and deprotecting the pyrazine.50 Fortunately, when performing this carbonylation with unprotected monomer 1d, the end-capped product 15a was obtained (Scheme 6). A similar result was achieved when using monomer 1a. Compound 15a was then coupled with 2b. After deprotection this gave 16, which mimics an α-helix with valine and leucine residues. The water solubility problem was however not solved by introducing the piperazine moiety.
Conclusion
In conclusion, a modular synthesis protocol has been developed towards previously unreported oligoamide pyrazine based α-helix peptidomimetics, as well as a hybrid type derivative. A coupling–deprotection sequence was used, utilizing a palladium-catalyzed aminocarbonylation to form the amide bonds between poorly nucleophilic aminopyrazine type building blocks. This methodology enables the flexible synthesis of a diverse set of (hetero)aryl oligoamide α-helix peptidomimetics. The synthesis also features the introduction of various end capping groups via (carbonylative) cross coupling reactions. Attempts to improve trimer formation yields are currently in progress.Acknowledgements
We are grateful to K. Duerinckx for the NMR measurements and to prof. Dr Jef Rozenski for HR-MS measurements. CV and BE thank FWO for PhD fellowships received. WDB thanks KU Leuven for financial support via project OT/14/067. LVM thanks the Hercules Foundation for supporting the purchase of the diffractometer through project AKUL/09/0035.Notes and references
- A. Voet, E. F. Banwell, K. K. Sahu, J. G. Heddle and K. Y. J. Zhang, Curr. Top. Med. Chem., 2013, 13, 989–1001 CrossRef CAS PubMed.
- T. Clackson and J. A. Wells, Science, 1995, 267, 383–386 CrossRef CAS PubMed.
- V. Azzarito, K. Long, N. S. Murphy and A. J. Wilson, Nat. Chem., 2013, 5, 161–173 CrossRef CAS PubMed.
- L. Y. Jin, W. R. Wang and G. W. Fang, Annu. Rev. Pharmacol. Toxicol., 2014, 54, 435–456 CrossRef CAS PubMed.
- M. R. Arkin, Y. Y. Tang and J. A. Wells, Chem. Biol., 2014, 21, 1102–1114 CrossRef CAS PubMed.
- B. N. Bullock, A. L. Jochim and P. S. Arora, J. Am. Chem. Soc., 2011, 133, 14220–14223 CrossRef CAS PubMed.
- B. C. Doak, J. Zheng, D. Dobritzsch and J. Kihlberg, J. Med. Chem., 2016, 59, 2312–2327 CrossRef CAS PubMed.
- Z. C. Zhang, X. Q. Li, T. Song, Y. Zhao and Y. G. Feng, J. Med. Chem., 2012, 55, 10735–10741 CrossRef CAS PubMed.
- W. Antuch, S. Menon, Q. Z. Chen, Y. C. Lu, S. Sakamuri, B. Beck, V. Schauer-Vukasinovic, S. Agarwal, S. Hess and A. Dömling, Bioorg. Med. Chem. Lett., 2006, 16, 1740–1743 CrossRef CAS PubMed.
- J. Becerril and A. D. Hamilton, Angew. Chem., Int. Ed., 2007, 46, 4471–4473 CrossRef CAS PubMed.
- M. D. Cummings, C. Schubert, D. J. Parks, R. R. Calvo, L. V. LaFrance, J. Lattanze, K. L. Milkiewicz and T. B. Lu, Chem. Biol. Drug Des., 2006, 67, 201–205 CAS.
- H. Yin, G. I. Lee, H. S. Park, G. A. Payne, J. M. Rodriguez, S. M. Sebti and A. D. Hamilton, Angew. Chem., Int. Ed., 2005, 44, 2704–2707 CrossRef CAS PubMed.
- H. Yin, G. I. Lee, K. A. Sedey, O. Kutzki, H. S. Park, B. P. Omer, J. T. Ernst, H. G. Wang, S. M. Sebti and A. D. Hamilton, J. Am. Chem. Soc., 2005, 127, 10191–10196 CrossRef CAS PubMed.
- H. Yin, B. P. Orner, H. S. Park and A. D. Hamilton, Abstr. Pap., Am. Chem. Soc., 2002, 224, U133–U133 Search PubMed.
- H. Moon and H.-S. Lim, Curr. Opin. Chem. Biol., 2015, 24, 38–47 CrossRef CAS PubMed.
- V. Azzarito, J. A. Miles, J. Fisher, T. A. Edwards, S. L. Warriner and A. J. Wilson, Chem. Sci., 2015, 6, 2434–2443 RSC.
- S. Rodriguez-Marin, N. S. Murphy, H. J. Shepherd and A. J. Wilson, RSC Adv., 2015, 5, 104187–104192 RSC.
- M. K. P. Jayatunga, S. Thompson and A. D. Hamilton, Bioorg. Med. Chem. Lett., 2014, 24, 717–724 CrossRef CAS PubMed.
- P. Ravindranathan, T. K. Lee, L. Yang, M. M. Centenera, L. Butler, W. D. Tilley, J. T. Hsieh, J. M. Ahn and G. V. Raj, Nat. Commun., 2013, 4, 1923 CrossRef PubMed.
- O. V. Kulikov, S. Kumar, M. Magzoub, P. C. Knipe, I. Saraogi, S. Thompson, A. D. Miranker and A. D. Hamilton, Tetrahedron Lett., 2015, 56, 3670–3673 CrossRef CAS.
- O. V. Kulikov, S. Thompson, H. Xu, C. D. Incarvito, R. T. W. Scott, I. Saraogi, L. Nevola and A. D. Hamilton, Eur. J. Org. Chem., 2013, 3433–3445 CrossRef CAS.
- S. Marimganti, M. N. Cheemala and J. M. Ahn, Org. Lett., 2009, 11, 4418–4421 CrossRef CAS PubMed.
- N. S. Murphy, P. Prabhakaran, V. Azzarito, J. P. Plante, M. J. Hardie, C. A. Kilner, S. L. Warriner and A. J. Wilson, Chem. – Eur. J., 2013, 19, 5546–5550 CrossRef CAS PubMed.
- G. M. Burslem and A. J. Wilson, Synlett, 2014, 324–335 CAS.
- J. Plante, F. Campbell, B. Malkova, C. Kilner, S. L. Warriner and A. J. Wilson, Org. Biomol. Chem., 2008, 6, 138–146 CAS.
- F. Campbell, J. P. Plante, T. A. Edwards, S. L. Warriner and A. J. Wilson, Org. Biomol. Chem., 2010, 8, 2344–2351 CAS.
- O. V. Kulikov and A. D. Hamilton, RSC Adv., 2012, 2, 2454–2461 RSC.
- G. M. Burslem, H. F. Kyle, P. Prabhakaran, A. L. Breeze, T. A. Edwards, S. L. Warriner, A. Nelson and A. J. Wilson, Org. Biomol. Chem., 2016, 14, 3782–3786 CAS.
- J. R. Martinelli, D. A. Watson, D. M. M. Freckmann, T. E. Barder and S. L. Buchwald, J. Org. Chem., 2008, 73, 7102–7107 CrossRef CAS PubMed.
- W. M. De Borggraeve, B. M. P. Verbist, F. J. R. Rombouts, V. G. Pawar, W. J. Smets, L. Kamoune, J. Alen, E. V. Van der Eycken, F. Compernolle and G. J. Hoornaert, Tetrahedron, 2004, 60, 11597–11612 CrossRef CAS.
- C. Veryser, S. Van Mileghem, B. Egle, P. Gilles and W. M. De Borggraeve, React. Chem. Eng., 2016, 1, 142–146 CAS.
- S. M. Biros, L. Moisan, E. Mann, A. Carella, D. Zhai, J. C. Reed and J. Rebek, Bioorg. Med. Chem. Lett., 2007, 17, 4641–4645 CrossRef CAS PubMed.
- P. Prabhakaran, A. Barnard, N. S. Murphy, C. A. Kilner, T. A. Edwards and A. J. Wilson, Eur. J. Org. Chem., 2013, 3504–3512 CrossRef CAS.
- J. T. Ernst, J. Becerril, H. S. Park, H. Yin and A. D. Hamilton, Angew. Chem., Int. Ed., 2003, 42, 535–539 CrossRef CAS PubMed.
- A. Brennfuhrer, H. Neumann and M. Beller, Angew. Chem., Int. Ed., 2009, 48, 4114–4133 CrossRef PubMed.
- V. Theodorou, M. Gogou, A. Giannoussi and K. Skobridis, ARKIVOC, 2014, 11–23 Search PubMed.
- P. Hermange, A. T. Lindhardt, R. H. Taaning, K. Bjerglund, D. Lupp and T. Skrydstrup, J. Am. Chem. Soc., 2011, 133, 6061–6071 CrossRef CAS PubMed.
- A. Suzuki, J. Organomet. Chem., 1999, 576, 147–168 CrossRef CAS.
- J. L. Yap, X. B. Cao, K. Vanommeslaeghe, K. Y. Jung, C. Peddaboina, P. T. Wilder, A. Nan, A. D. MacKerell, W. R. Smythe and S. Fletcher, Org. Biomol. Chem., 2012, 10, 2928–2933 CAS.
- B. Jiang, C. G. Yang, W. N. Xiong and J. Wang, Bioorg. Med. Chem., 2001, 9, 1149–1154 CrossRef CAS PubMed.
- G. A. Patani and E. J. LaVoie, Chem. Rev., 1996, 96, 3147–3176 CrossRef CAS PubMed.
- S. Scales, S. Johnson, Q. Y. Hu, Q. Q. Do, P. Richardson, F. Wang, J. Braganza, S. J. Ren, Y. D. Wan, B. J. Zheng, D. Faizi and I. McAlpine, Org. Lett., 2013, 15, 2156–2159 CrossRef CAS PubMed.
- V. Azzarito, P. Prabhakaran, A. I. Bartlett, N. S. Murphy, M. J. Hardie, C. A. Kilner, T. A. Edwards, S. L. Warriner and A. J. Wilson, Org. Biomol. Chem., 2012, 10, 6469–6472 CAS.
- I. Saraogi, C. D. Incarvito and A. D. Hamilton, Angew. Chem., Int. Ed., 2008, 47, 9691–9694 CrossRef CAS PubMed.
- D. Y. Xin, E. Ko, L. M. Perez, T. R. Ioerger and K. Burgess, Org. Biomol. Chem., 2013, 11, 7789–7801 CAS.
- A. S. Guram, R. A. Rennels and S. L. Buchwald, Angew. Chem., Int. Ed. Engl., 1995, 34, 1348–1350 CrossRef CAS.
- M. C. Harris, O. Geis and S. L. Buchwald, J. Org. Chem., 1999, 64, 6019–6022 CrossRef CAS.
- M. Makosza and K. Sienkiewicz, J. Org. Chem., 1998, 63, 4199–4208 CrossRef CAS.
- B. H. Lipshutz, D. W. Chung, B. Rich and R. Corral, Org. Lett., 2006, 8, 5069–5072 CrossRef CAS PubMed.
- M. Lamothe, M. Perez, V. Colovray-Gotteland and S. Halazy, Synlett, 1996, 507–508 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. CCDC 1512412–1512414. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c6ob02358d |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2017 |







![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
: