In-plane molecular organization of hydrated single lipid bilayers: DPPC:cholesterol†
Berta
Gumí-Audenis
 abcd,
Luca
Costa
abcd,
Luca
Costa
 e,
Lorena
Redondo-Morata
f,
Pierre-Emmanuel
Milhiet
e,
Fausto
Sanz
bad,
Roberto
Felici
e,
Lorena
Redondo-Morata
f,
Pierre-Emmanuel
Milhiet
e,
Fausto
Sanz
bad,
Roberto
Felici
 g,
Marina I.
Giannotti
g,
Marina I.
Giannotti
 *abd and
Francesco
Carlà
*c
*abd and
Francesco
Carlà
*c
aInstitute for Bioengineering of Catalonia (IBEC), Barcelona, Spain. E-mail: migiannotti@ibecbarcelona.eu
bMaterials Science and Physical Chemistry Department, Universitat de Barcelona, Barcelona, Spain
cESRF, The European Synchrotron, Grenoble, France. E-mail: francesco.carla@esrf.fr
dCentro de Investigación Biomédica en Red (CIBER), Madrid, Spain
eCentre de Biochimie Structurale, CNRS UMR 5048 – INSERM UMR 1054, Montpellier, France
fUnité 1006, INSERM, Aix-Marseille Université, FR-13009 Marseille, France
gIstituto SPIN-CNR, Roma, Italy
First published on 24th November 2017
Abstract
Understanding the physical properties of cholesterol–phospholipid systems is essential to gain a better knowledge of the function of each membrane constituent. We present a novel, simple and user-friendly setup that allows for the straightforward grazing incidence X-ray diffraction characterization of hydrated individual supported lipid bilayers. This configuration minimizes the scattering from the liquid and allows the detection of the extremely weak diffracted signal of the membrane, enabling the differentiation of the coexisting domains in DPPC:cholesterol single bilayers.
Cell membranes are composed mainly of a mixture of lipids and proteins. Lateral segregation of membrane components into domains of lipids enriched in cholesterol (chol) and sphingolipids is involved in many membrane functions, for instance signaling, remodeling and trafficking.1,2 Chol is responsible for controlling the phase behavior as well as the lipid organization, regulating the fluidity and permeability of the membrane while increasing its mechanical resistance.3–8 In this context, it is of great interest to understand the physical properties of chol–phospholipid systems to gain a better knowledge on the role of chol in the membrane.
Membranes comprising phospholipids and chol have been extensively studied, including simplified models based on two components. In particular, temperature–composition phase diagrams of DPPC (1,2-dipalmitoyl-sn-glycero-3-phosphocholine)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chol have been defined using different techniques such as nuclear magnetic resonance (NMR), differential scanning calorimetry (DSC), and neutron and X-ray (XR) scattering.9–14 Yet, discrepancies on the determination of a complete phase diagram able to cover all the compositional space and temperature range still remain. Atomic force microscopy (AFM) and force spectroscopy (AFM-FS) have provided insights into the thermal transition of DPPC
chol have been defined using different techniques such as nuclear magnetic resonance (NMR), differential scanning calorimetry (DSC), and neutron and X-ray (XR) scattering.9–14 Yet, discrepancies on the determination of a complete phase diagram able to cover all the compositional space and temperature range still remain. Atomic force microscopy (AFM) and force spectroscopy (AFM-FS) have provided insights into the thermal transition of DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chol supported lipid bilayers (SLBs) at the nanometric scale. AFM has the ability to operate under environmentally controlled conditions,3,4 to characterize the topology and nanomechanics, as well as to define the coexistence of different domains, facilitating the linking between the chol content and the lateral organization of the membrane.3,4,15,16
chol supported lipid bilayers (SLBs) at the nanometric scale. AFM has the ability to operate under environmentally controlled conditions,3,4 to characterize the topology and nanomechanics, as well as to define the coexistence of different domains, facilitating the linking between the chol content and the lateral organization of the membrane.3,4,15,16
Similar information about phase segregation in lipid bilayers can also be in principle gathered by XR scattering techniques. XR scattering techniques are very powerful, noninvasive techniques that have been extensively used in lipid bilayer studies to probe length scales ranging from angstroms to microns. A large part of the XR-based experiments has been focused so far on determining the electronic vertical structure of lipid monolayers, bilayers and stacks of bilayers (or multi-bilayers), at the liquid–air and solid–liquid interfaces, respectively, by means of XR reflectivity (XRR), which is a well-established technique in this field.17–21
Information about the lateral in-plane structure of such systems can be instead obtained by grazing incidence XR diffraction (GIXD). Nevertheless, the requirement of wetting preservation to guarantee the stability of biological membranes at the solid–liquid interface makes the in-plane structural characterization of a single lipid bilayer extremely challenging. The presence of a wetting layer makes the use of high energy XR necessary to increase the transmission through the liquid, resulting in a weaker signal from the organic molecules.19 Additionally, the scattering generated by the liquid environment increases the background level complicating the detection of the signal scattered by the bilayer structure. For this reason, most of the reported structural information relative to lipid membranes has been extrapolated from experiments conducted on multi-bilayers22,23 or on monolayers at the liquid–air interface.24 However, lipid monolayers do not represent the lamellar nature of the membrane, and the physical and chemical properties of the multi-bilayers also differ from those of single bilayers. Because of their ease of formation and single lamellar arrangement, SLBs are among the most common models for native membranes,25 in addition to large and giant unilamellar vesicles (LUVs and GUVs, respectively). In this context, only a few successful GIXD studies on single hydrated bilayers have been reported, achieving their goals by using complex methods and controlling humidity conditions during the XR measurements.26–28
The aim of the present work is twofold: to propose a novel user-friendly setup based on a thin layer cell configuration that allows the successful acquisition of GIXD data on SLBs under aqueous conditions and to help further understanding the DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chol system that has been the subject of a large number of studies with structural data.9–12
chol system that has been the subject of a large number of studies with structural data.9–12
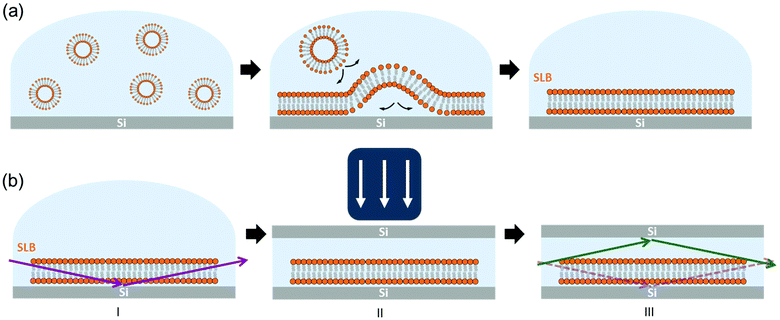
A schematic drawing of the setup used in this work is shown in Fig. 1. Hydrated SLBs were first prepared on Si wafer substrates with the (100) orientation by means of the vesicle rupture method (Fig. 1(a)),29,30 transferred onto the diffractometer stage – at the ID03 beamline of ESRF – and aligned with the XR beam (Fig. 1(b)-I). A second Si substrate was then placed on top of the first Si wafer and pressed against the sample with an external load (Fig. 1(b)-II). This Si–SLB–Si arrangement allows the confinement of a thin layer of liquid between the sample and the flat Si surface. The lateral dimensions of the upper Si substrate were defined small enough to prevent the XR diffracted beam passing through this second Si before the detector, therefore avoiding any reflection/refraction (Fig. 1(b)-III).
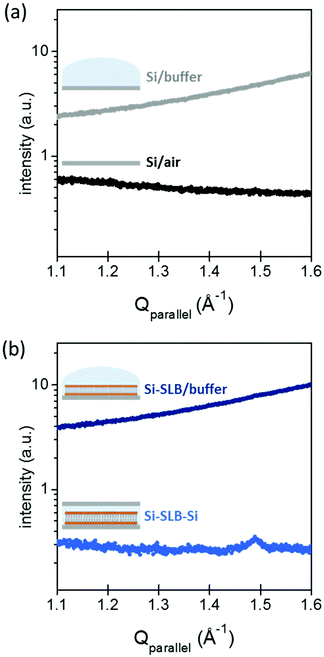
The gap between the two Si substrates was measured with the XR beam by performing a height scan in transmission configuration, and it resulted to be only a few microns thick (see ESI Fig. S1†). In this configuration, the SLB is confined in a controlled geometry (Fig. 1(b)-III) and the thickness of the liquid layer is reduced enough to minimize the background signal generated by the liquid environment, allowing the detection of the diffracted signal of the bilayer. The effect of the liquid (buffer solution) on the GIXD background level is shown in Fig. 2. The presence of the buffer on a bare Si surface produces an increase of the background of about one order of magnitude compared to the Si/air interface (Fig. 2(a)). In this experimental configuration, it is basically impossible to detect any diffracted signal from an SLB, as observed in Fig. 2(b) (dark blue). However, when the second Si substrate is placed on top and pressed against the bottom Si–SLB system, the GIXD signal from the bilayer becomes visible (Fig. 2(b), light blue). It is worth noticing that, although the setup allows one to access any Qparallel value, the access to high Qperpendicular values is limited by geometrical factors, such as the in-plane size of the two Si wafers and the distance between them (see ESI 1.3†).
Using this Si–SLB–Si configuration, we acquired the GIXD patterns of single SLBs composed of DPPC and chol at different molar ratios (DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chol: 100
chol: 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, 90
0, 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, 80
10, 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, 60
20, 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 and 50
40 and 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) in 20 mM HEPES, 150 mM NaCl and 20 mM MgCl2 buffer solution of pH 7.4, at room temperature. All the GIXD measurements performed on the different DPPC
50) in 20 mM HEPES, 150 mM NaCl and 20 mM MgCl2 buffer solution of pH 7.4, at room temperature. All the GIXD measurements performed on the different DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chol SLBs successfully revealed a 2D order as shown in Fig. 3 for the DPPC
chol SLBs successfully revealed a 2D order as shown in Fig. 3 for the DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chol 90
chol 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
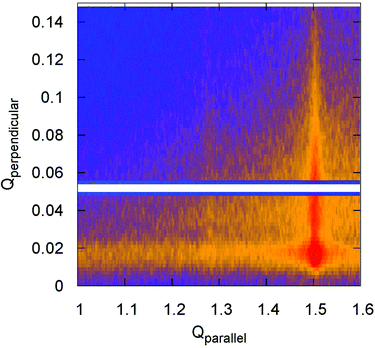
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 SLB. It is clearly visible that the diffracted intensity appears as two rods at the Qparallel values of 1.28 Å−1 and 1.50 Å−1 perpendicular to the substrate surface, indicating the 2D nature of the samples that was verified by XRR (see ESI 3†). For a pure DPPC SLB, the XRR curve (Fig. S2†) reveals a film thickness of 5.9 nm, comparable to a single gel-like state lipid bilayer,3,4 validating the absence of multi-bilayer structures.
10 SLB. It is clearly visible that the diffracted intensity appears as two rods at the Qparallel values of 1.28 Å−1 and 1.50 Å−1 perpendicular to the substrate surface, indicating the 2D nature of the samples that was verified by XRR (see ESI 3†). For a pure DPPC SLB, the XRR curve (Fig. S2†) reveals a film thickness of 5.9 nm, comparable to a single gel-like state lipid bilayer,3,4 validating the absence of multi-bilayer structures.
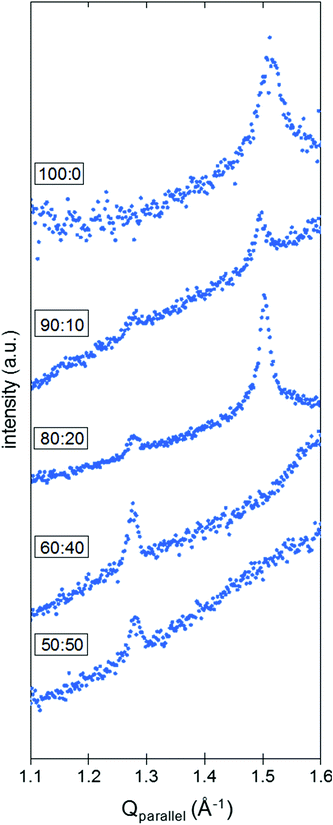
When integrated over Qperpendicular (see ESI 1.3† for experimental details), the diffraction pattern obtained for pure DPPC SLBs presents one peak at Qparallel = 1.50 Å−1 (Fig. 4). Although the characterization method seems pretty robust and allows the correct determination of the position of the peaks along the Qparallel direction, it is difficult to extract further information based on the intensity of the peaks. As a matter of fact, the surface of the samples may not be perfectly homogeneous at the millimeter scale and this lack of homogeneity, originating from areas not covered by the SLB, influences the quality of the data and the intensities measured (see ESI 4, Fig. S3† for further details). While the intensity of the peak can vary significantly with the sample orientation, the single peak positions in Qparallel remain constant, giving rise to the assumption of a hexagonal packing for the DPPC molecules on a Si–SLB under liquid conditions. In this context, a corresponding d-spacing of 0.48 nm was obtained. The molecular organization of DPPC has been reported by studying different DPPC configurations, i.e. monolayers at the liquid–air interface, or bilayers and stacks of bilayers at the solid–liquid interface. In general, both rectangular and hexagonal geometries have been assumed for DPPC packing. Some GIXD studies of pure DPPC monolayers at the liquid–air interface reported a rectangular geometry to define the packing, with values of 0.43 nm and 0.46 nm,24,27,31 or a distorted hexagonal lattice with geometrical parameters between 0.51 and 0.50 nm depending on the lateral pressure.32 Hexagonal packing for DPPC and DPPE SLBs in a liquid environment has been identified using GIXD, with a d-spacing of 0.50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 32 and 0.48
32 and 0.48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 28 nm, respectively, and using frequency modulation-AFM defining a lateral spacing of 0.49 nm.33 The value extracted from the GIXD experiments in this work is in good agreement with the literature.
28 nm, respectively, and using frequency modulation-AFM defining a lateral spacing of 0.49 nm.33 The value extracted from the GIXD experiments in this work is in good agreement with the literature.
The binary DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chol SLBs were also investigated for different compositions: 90
chol SLBs were also investigated for different compositions: 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, 80
10, 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, 60
20, 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 and 50
40 and 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 molar ratios. It is known that the chol content in gel-like state membranes, such as DPPC SLBs at room temperature, determines the phase behavior in homogeneous or phase-segregated bilayers. In the presence of chol, we identified a second peak at Qparallel = 1.28 Å−1 in the GIXD pattern for DPPC
50 molar ratios. It is known that the chol content in gel-like state membranes, such as DPPC SLBs at room temperature, determines the phase behavior in homogeneous or phase-segregated bilayers. In the presence of chol, we identified a second peak at Qparallel = 1.28 Å−1 in the GIXD pattern for DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chol SLBs on Si (Fig. 4). At low contents of chol (10 and 20 mol%), this peak coexists with the one previously observed for pure DPPC SLBs, with no variation in the Qparallel position within ±0.02 Å−1. Hence, considering two different hexagonal packings, we obtain d-spacings of 0.57 and 0.48 nm from the Qparallel values of 1.28 and 1.50 Å−1, respectively. On the other hand, when the chol content is above 30 mol% (60
chol SLBs on Si (Fig. 4). At low contents of chol (10 and 20 mol%), this peak coexists with the one previously observed for pure DPPC SLBs, with no variation in the Qparallel position within ±0.02 Å−1. Hence, considering two different hexagonal packings, we obtain d-spacings of 0.57 and 0.48 nm from the Qparallel values of 1.28 and 1.50 Å−1, respectively. On the other hand, when the chol content is above 30 mol% (60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 and 50
40 and 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 molar ratios), the GIXD patterns shown in Fig. 4 display only one peak. The peak corresponding to the DPPC-rich phase (high Qparallel) is no longer visible, whereas the one at 1.28 Å−1 (d-spacing of 0.57 nm) is still observable. These behaviors match with chol intercalating between the DPPC molecules and increasing the average distance between the DPPC moieties, in agreement with the GIXD reports on the DPPC
50 molar ratios), the GIXD patterns shown in Fig. 4 display only one peak. The peak corresponding to the DPPC-rich phase (high Qparallel) is no longer visible, whereas the one at 1.28 Å−1 (d-spacing of 0.57 nm) is still observable. These behaviors match with chol intercalating between the DPPC molecules and increasing the average distance between the DPPC moieties, in agreement with the GIXD reports on the DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chol monolayers at the liquid–air interface, that show a hexagonal packing displaying d-spacing values that depend on the amount of chol, for more than 25 mol% chol.24
chol monolayers at the liquid–air interface, that show a hexagonal packing displaying d-spacing values that depend on the amount of chol, for more than 25 mol% chol.24
Our observations are consistent with most phase diagrams for the binary mixture of DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chol,9–12 where low contents of chol of up to 30 mol% lead to separation into two different phases that coexist at room temperature and concentrations of chol of higher than 30 mol% appear to be a unique liquid ordered (lo) phase in any temperature range studied. This condition has also been observed by means of temperature-controlled AFM on SLBs on mica substrates, as well as with AFM-FS,4 demonstrating an enhanced nanomechanical resistance of both the chol-rich domains, for low chol content bilayers, and the homogeneous lo phase, for high chol content bilayers. Such an increase of nanomechanical resistance is associated with a strong lateral interaction mediated by chol molecules placed between the DPPC ones, due to a highly stable structure with most probably an equimolar DPPC
chol,9–12 where low contents of chol of up to 30 mol% lead to separation into two different phases that coexist at room temperature and concentrations of chol of higher than 30 mol% appear to be a unique liquid ordered (lo) phase in any temperature range studied. This condition has also been observed by means of temperature-controlled AFM on SLBs on mica substrates, as well as with AFM-FS,4 demonstrating an enhanced nanomechanical resistance of both the chol-rich domains, for low chol content bilayers, and the homogeneous lo phase, for high chol content bilayers. Such an increase of nanomechanical resistance is associated with a strong lateral interaction mediated by chol molecules placed between the DPPC ones, due to a highly stable structure with most probably an equimolar DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chol ratio. In addition, a fixed d-spacing, independent of the chol concentration, is also consistent with a well-defined interaction between DPPC and chol, as suggested in the condensed complex model.34
chol ratio. In addition, a fixed d-spacing, independent of the chol concentration, is also consistent with a well-defined interaction between DPPC and chol, as suggested in the condensed complex model.34
Although it is known that the underlying substrate may strongly affect the SLB order and the transition temperature range of the bilayer,9,35,36 AFM and fluorescence microscopy measurements proved that the DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chol SLBs on Si behave in the same way as those observed on mica or in vesicles (see ESI 5†). While the roughness of the Si substrates makes it difficult to detect the domains for the low chol content bilayers (90
chol SLBs on Si behave in the same way as those observed on mica or in vesicles (see ESI 5†). While the roughness of the Si substrates makes it difficult to detect the domains for the low chol content bilayers (90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 and 80
10 and 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 molar ratios) from the AFM topographical images (Fig. S4†), as they differ in height by ∼0.2–0.3 nm,4 fluorescence images evidenced domains on the DPPC
20 molar ratios) from the AFM topographical images (Fig. S4†), as they differ in height by ∼0.2–0.3 nm,4 fluorescence images evidenced domains on the DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chol 90
chol 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 SLBs (selected concentration as representative for bilayers with a low chol content, Fig. S5†). Conversely, both AFM and fluorescence measurements of DPPC
10 SLBs (selected concentration as representative for bilayers with a low chol content, Fig. S5†). Conversely, both AFM and fluorescence measurements of DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chol SLBs with high concentrations of chol (60
chol SLBs with high concentrations of chol (60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 and 50
40 and 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 molar ratios) on Si substrates show homogeneous membranes (Fig. S4 and S5†). Correlative AFM-fluorescence characterization of a DPPC
50 molar ratios) on Si substrates show homogeneous membranes (Fig. S4 and S5†). Correlative AFM-fluorescence characterization of a DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chol 90
chol 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 SLB on a glass substrate also allows the correlation of low fluorescence intensity regions with higher topographical domains (Fig. S6†), previously identified as enriched in chol,4 which also suggested a decrease in membrane fluidity for the chol-rich regions.
10 SLB on a glass substrate also allows the correlation of low fluorescence intensity regions with higher topographical domains (Fig. S6†), previously identified as enriched in chol,4 which also suggested a decrease in membrane fluidity for the chol-rich regions.
Conclusions
The structural characterization of a single lipid membrane at the solid–liquid interface by GIXD is extremely challenging, and only a few successful GIXD studies on single hydrated lipid bilayers have been reported,26–28 where complex setups and controlled humidity conditions are required during the measurements. Here, we present a novel, simple and user-friendly setup that allows for the straightforward GIXD characterization of hydrated individual SLBs, based on a Si–SLB–Si configuration. This allows the reduction of scattering from the liquid, revealing the extremely weak diffracted signal of the lipid bilayer, capable of detecting different coexisting domains in phase-segregated membranes. We recorded GIXD patterns on DPPC bilayers supported on Si substrates with various contents of chol. Two d-spacing values were assigned to the DPPC intermolecular distance of each phase in these phase-segregated bilayers (DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chol 90
chol 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 and 80
10 and 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 molar ratios), while a single d-spacing value was obtained for homogeneous bilayers (DPPC and DPPC
20 molar ratios), while a single d-spacing value was obtained for homogeneous bilayers (DPPC and DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chol 60
chol 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 and 50
40 and 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50), in accordance with the phase diagram of this binary system.4,24,33 The higher d-spacing corresponds to the chol-enriched phase, where DPPC and chol molecules may intercalate in a nearly stoichiometric ratio. This represents a reasonable scenario opening new avenues of research on the structure as well as the dynamic processes of cell membranes in a physiological environment.
50), in accordance with the phase diagram of this binary system.4,24,33 The higher d-spacing corresponds to the chol-enriched phase, where DPPC and chol molecules may intercalate in a nearly stoichiometric ratio. This represents a reasonable scenario opening new avenues of research on the structure as well as the dynamic processes of cell membranes in a physiological environment.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge financial support from the Catalan government (grant 2014SGR-1251) and the Spanish Ministry of Economy and Competitiveness (MINECO) and FEDER (CTQ2015-66194-R MINECO/FEDER) projects, and the Instituto de Salud Carlos III, through “Acciones CIBER”. The Networking Research Center on Bioengineering, Biomaterials and Nanomedicine (CIBER-BBN) is an initiative funded by the VI National R&D&I Plan 2008–2011, Iniciativa Ingenio 2010, Consolider Program, CIBER Actions and financed by the Instituto de Salud Carlos III with assistance from the European Regional Development Fund. LC acknowledges financial support from the Labex EpiGenMed program ANR-10-LABX-12-01. The CBS is a member of the France-BioImaging (FBI) and the French Infrastructure for Integrated Structural Biology (FRISBI), two national infrastructures supported by the French National Research Agency (ANR-10-INBS-04-01 and ANR-10-INBS-05, respectively). The GIXD experiments described here were conducted at the ID03 surface diffraction beamline of the ESRF, the European Synchrotron in Grenoble (France) (ESRF experiments SC-4144, IHSI-964). We are also grateful to Fabio Comin for his fruitful discussions and to Alain Panzarella (ESRF, Grenoble) for his technical assistance.Notes and references
- G. van Meer, D. R. Voelker and G. W. Feigenson, Nat. Rev. Mol. Cell Biol., 2008, 9, 112–124 CrossRef CAS PubMed.
- D. Lingwood and K. Simons, Science, 2010, 327, 46–50 CrossRef CAS PubMed.
- B. Gumi-Audenis, L. Costa, F. Carla, F. Comin, F. Sanz and M. I. Giannotti, Membranes, 2016, 6, 58.
- L. Redondo-Morata, M. I. Giannotti and F. Sanz, Langmuir, 2012, 28, 12851–12860 CrossRef CAS PubMed.
- T. Rog, M. Pasenkiewicz-Gierula, I. Vattulainen and M. Karttunen, Biochim. Biophys. Acta, Biomembr., 2009, 1788, 97–121 CrossRef CAS PubMed.
- W.-C. Hung, M.-T. Lee, F.-Y. Chen and H. W. Huang, Biophys. J., 2007, 92, 3960–3967 CrossRef CAS PubMed.
- J. J. Pan, S. Tristram-Nagle and J. F. Nagle, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2009, 80, 021931.
- P. E. Milhiet, M. C. Giocondi and C. Le Grimellec, J. Biol. Chem., 2002, 277, 875–878 CrossRef CAS PubMed.
- T. P. W. McMullen and R. N. McElhaney, Biochim. Biophys. Acta, Biomembr., 1995, 1234, 90–98 CrossRef.
- S. Karmakar, V. A. Raghunathan and S. Mayor, J. Phys.: Condens. Matter, 2005, 17, S1177–S1182 CrossRef CAS.
- Y.-W. Chiang, A. J. Costa and J. H. Freed, J. Phys. Chem. B, 2007, 111, 11260–11270 CrossRef CAS PubMed.
- D. Marsh, Biochim. Biophys. Acta, Biomembr., 2010, 1798, 688–699 CrossRef CAS PubMed.
- D. Marquardt, F. A. Heberle, J. D. Nickels, G. Pabst and J. Katsaras, Soft Matter, 2015, 11, 9055–9072 RSC.
- P. F. Almeida, Biophys. J., 2011, 100, 420–429 CrossRef CAS PubMed.
- S. Garcia-Manyes and F. Sanz, Biochim. Biophys. Acta, Biomembr., 2010, 1798, 741–749 CrossRef CAS PubMed.
- R. M. A. Sullan, J. K. Li, C. Hao, G. C. Walker and S. Zou, Biophys. J., 2010, 99, 507–516 CrossRef CAS PubMed.
- F. Evers, C. Jeworrek, K. Weise, M. Tolan and R. Winter, Soft Matter, 2012, 8, 2170–2175 RSC.
- B. Gumí-Audenis, F. Carlà, M. V. Vitorino, A. Panzarella, L. Porcar, M. Boilot, S. Guerber, P. Bernard, M. S. Rodrigues, F. Sanz, M. I. Giannotti and L. Costa, J. Synchrotron Radiat., 2015, 22, 1364–1371 Search PubMed.
- C. E. Miller, J. Majewski, T. Gog and T. L. Kuhl, Phys. Rev. Lett., 2005, 94, 238104.
- E. Novakova, K. Giewekemeyer and T. Salditt, Phys. Rev. E, 2006, 74, 051911.
- J. Daillant, E. Bellet-Amalric, A. Braslau, T. Charitat, G. Fragneto, F. Graner, S. Mora, F. Rieutord and B. Stidder, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 11639–11644 CrossRef CAS PubMed.
- T. Salditt and G. Brotons, Anal. Bioanal. Chem., 2004, 379, 960–973 CrossRef CAS PubMed.
- M. A. Barrett, S. Zheng, L. A. Toppozini, R. J. Alsop, H. Dies, A. Wang, N. Jago, M. Moore and M. C. Rheinstadter, Soft Matter, 2013, 9, 9342–9351 RSC.
- A. Ivankin, I. Kuzmenko and D. Gidalevitz, Phys. Rev. Lett., 2010, 104, 108101 CrossRef PubMed.
- E. T. Castellana and P. S. Cremer, Surf. Sci. Rep., 2006, 61, 429–444 CrossRef CAS.
- R. Ziblat, K. Kjaer, L. Leiserowitz and L. Addadi, Angew. Chem., Int. Ed., 2009, 48, 8958–8961 CrossRef CAS PubMed.
- R. Ziblat, L. Leiserowitz and L. Addadi, J. Am. Chem. Soc., 2010, 132, 9920–9927 CrossRef CAS PubMed.
- C. E. Miller, J. Majewski, E. B. Watkins, D. J. Mulder, T. Gog and T. L. Kuhl, Phys. Rev. Lett., 2008, 100, 058103 CrossRef CAS PubMed.
- L. Redondo-Morata, M. I. Giannotti and F. Sanz, Mol. Membr. Biol., 2014, 31, 17–28 CrossRef CAS PubMed.
- P. E. Milhiet, V. Vié, M.-C. Giocondi and C. Le Grimellec, Single Mol., 2001, 2, 109–112 CrossRef CAS.
- F. Neville, M. Cahuzac, O. Konovalov, Y. Ishitsuka, K. Y. C. Lee, I. Kuzmenko, G. M. Kale and D. Gidalevitz, Biophys. J., 2006, 90, 1275–1287 CrossRef CAS PubMed.
- E. B. Watkins, C. E. Miller, D. J. Mulder, T. L. Kuhl and J. Majewski, Phys. Rev. Lett., 2009, 102, 238101 CrossRef CAS PubMed.
- H. Asakawa and T. Fukuma, Nanotechnology, 2009, 20, 264008 CrossRef PubMed.
- H. M. McConnell and A. Radhakrishnan, Biochim. Biophys. Acta, 2003, 1610, 159–173 CrossRef CAS.
- Z. V. Leonenko, E. Finot, H. Ma, T. E. S. Dahms and D. T. Cramb, Biophys. J., 2004, 86, 3783–3793 CrossRef CAS PubMed.
- H. M. Seeger, A. Di Cerbo, A. Alessandrini and P. Facci, J. Phys. Chem. B, 2010, 114, 8926–8933 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, GIXD measurements, XRR measurements, AFM imaging and fluorescence measurements. See DOI: 10.1039/c7nr07510c |
| This journal is © The Royal Society of Chemistry 2018 |