Open Access Article
Open Access ArticleDendrimer-encapsulated nanoparticle-core micelles as a modular strategy for particle-in-a-box-in-a-box nanostructures†
J. B.
ten Hove‡
 ab,
J.
Wang‡
a,
F. W. B.
van Leeuwen
ab and
A. H.
Velders
ab,
J.
Wang‡
a,
F. W. B.
van Leeuwen
ab and
A. H.
Velders
 *abc
*abc
aLaboratory of BioNanoTechnology, Wageningen University & Research, Axis, 6708 WG Wageningen, The Netherlands. E-mail: aldrik.velders@wur.nl
bInterventional Molecular Imaging Laboratory, Department of Radiology, Leiden University Medical Centre, The Netherlands
cIstituto Regional de Investigacion Cientifica Aplicada, Universidad de Castille-La Mancha, Ciudad Real, Spain
First published on 28th November 2017
Abstract
The hierarchically controlled synthesis and characterization of self-assembling macromolecules and particles are key to explore and exploit new nanomaterials. Here we present a versatile strategy for constructing particle-in-a-box-in-a-box systems by assembling dendrimer-encapsulated gold nanoparticles (DENs) into dendrimicelles. This is realized by combining positively charged PAMAM dendrimers with a negative-neutral block copolymer. The number of particles per dendrimicelle can be controlled by mixing DENs with empty PAMAM dendrimers. The dendrimicelles are stable in solution for months and provide improved resistance for the nanoparticles against degradation. The dendrimicelle strategy provides a flexible platform with a plethora of options for variation in the type of nanoparticles, dendrimers and block copolymers used, and hence is tunable for applications ranging from nanomedicine to catalysis.
Introduction
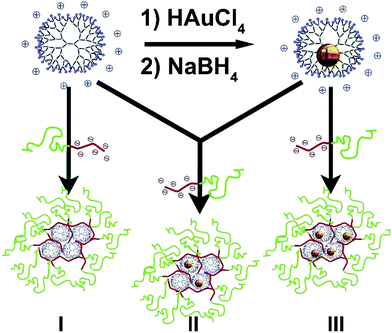
The hierarchical organization of molecules, macromolecules, nanoparticles and self-assembled systems is at the heart of supramolecular and nanoscale chemistry, with nanocontainers being among the most appealing ones for applications ranging from drug delivery to catalysis.1–4 Within the vast interest in obtaining well-defined nanoparticles in confined systems, micelles and dendrimers have proved to be excellent building blocks. Dendrimers are hyperbranched molecules and are archetypical soft nanoparticles in the 1–10 nanometer range with the potential to encapsulate molecules,5 or nanoparticles.6–8 Amis and Crooks and collaborators extensively studied the encapsulation of bare nanoparticles with polyamidoamino (PAMAM) dendrimers,8–10 and via various NMR techniques we unambiguously proved that (palladium) nanoparticles are formed and reside inside these dendrimers, thus forming so-called dendrimer-encapsulated nanoparticles (DENs).11,12 Micelles provide a method to self-assemble multiple, e.g., polymeric, molecular building blocks into well-defined structures typically in the 10–100 nanometer range.13,14 In our investigations of coacervate core micellar systems for e.g. medical applications,15,16 we recently showed the excellent control over the assembly of negatively charged, carboxylic acid-terminated, PAMAM dendrimers with a positive-neutral poly(vinylpyridine)-poly(ethylene oxide) diblock copolymer into well-defined micelles. These micelles contain 1–100 dendrimers per micelle, depending on the dendrimer generation used.17 We here present a methodology in which gold nanoparticles in amine-terminated dendrimers, AuDENs, can form micelles by assembling with negative-neutral block copolymers to form a nanoparticle-in-a-box-in-a-box system (Scheme 1).Results and discussion
PAMAM dendrimers exist with a variety of terminating groups.18,19 For the incorporation of AuNPs, the use of (amine-terminated) PAMAMs is preferred, as PAMAM dendrimers with carboxylic acid end groups tend to form dendrimer stabilized nanoparticles rather than DENS,20,21 and hydroxyl-terminated dendrimers lack the required surface charge for self-assembly into coacervate core micelles. We synthesized Au256DENs following the original work of Amis for the formation of well-defined single particles, focusing on amine-terminated 7th generation PAMAM.10 To prove that the AuNPs reside inside the PAMAM dendrimers, e.g. AuDENs as opposed to dendrimer-stabilized nanoparticles (Au-DSNPs), we used 1H-NMR spectroscopy and DOSY (Fig. S1†) to verify the complexation of metal ions in the interior of PAMAM and the consecutive formation of dendrimer-encapsulated nanoparticles.11,12 In addition, Transmission Electron Microscopy (TEM) showed that the AuDENs are ∼1.7 nm in diameter, in agreement with the expected size of Au256 NPs (Fig. S2†).Next, we assembled dendrimicelles by mixing the Au256DENs at pH 7 with an anionic–neutral block copolymer having a 64-subunit polymethacrylic acid block and a 885-subunit polyethyleneoxide block, pMAA64pEO885. pH 7 was chosen since both the pMAApEO block copolymer as well as the PAMAM dendrimers are charged at this pH.22,23 This allows electrostatic interactions of the positively charged amine-terminal groups of PAMAM to associate with the negatively charged methacrylic acid block of the pMAApEO, forming a coacervate core. The neutral PEO block forms the corona of the micelle and stabilizes the resulting nanostructure. Dynamic light scattering, DLS, reveals this self-assembly as an increase in the scattered light intensity and, concurrently, as an increase in size (Fig. 1, S3†).
For the optimal building block ratios, i.e. the preferred micelle composition (PMC), we measured DLS at different mixing ratios of block copolymer to PAMAM, keeping the amount of positive charge (PAMAM) constant, and varying the amount of negative pMAA64pEO885 (Fig. S4†). From this experiment the PMC was determined to be at a mixing ratio of 0.92, corresponding to a slight excess of PAMAM to block copolymer. This deviation from the theoretical molar charge neutrality might be attributed to the known presence of defects in higher generation PAMAM dendrimers, resulting in fewer charged end-groups and hence observed charge-stoichiometry at lower charge fraction.17,24,25
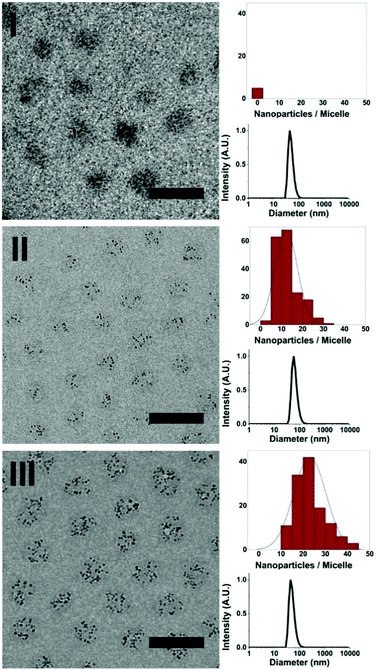
To investigate whether or not the presence of a nanoparticle inside a dendrimer affects the formation of dendrimicelles, we mixed Au256DENs with block copolymer at the previously determined PMC. DLS analysis showed that dendrimicelles formed and exhibited a similar size in solution as empty dendrimicelles. Additionally, we also prepared dendrimicelles by mixing a block copolymer with a dendrimer solution containing 50% Au256DENs and 50% empty PAMAM dendrimers at the PMC. Here, similarly sized dendrimicelles were again formed. DLS only indicates the formation of the dendrimicelles revealing the hydrodynamic diameter; however, it cannot discriminate whether dendrimicelles with only PAMAM and dendrimicelles with DENs were formed. Therefore, we characterized the obtained nanostructures in more detail and determined their stability under various conditions using both cryoTEM and DLS. CryoTEM allows for visualization of the obtained nanostructures in near-native state from which micelle properties such as shape, micelle core and corona size, and aggregation number can be derived. Representative CryoTEM micrographs and DLS data of the dendrimicelles discussed above are shown in Fig. 1 and S5–7,† revealing the dendrimicelle structures for the PAMAM and DEN systems, and that mixing PAMAM and DENs result in mixed-core dendrimicelles.
The dendrimicelle sample with empty PAMAM in the core can be visualized by cryoTEM (I, Fig. 1) since the dense coacervate core provides a higher electron-density, and hence contrast, in the cryoTEM images.26 However, the presence of electron dense AuNPs greatly improves the contrast and facilitates image analysis. In fact, Fig. 1 shows dendrimicelles made using 50% empty PAMAM mixed with 50% Au256DENs (II), and shows the dendrimicelles made of 100% Au256DENs (III). CryoTEM corroborates that the ∼50 nm-sized structures observed with DLS are indeed well-defined nano-assemblies with a core–shell structure.
Cryo-TEM micrographs of the micelles made from 100% Au256DENs (III, Fig. 1, S8†) show that the average core diameter is 27 ± 5 nm. DLS data show the hydrodynamic diameter of these micelles to be 52 nm. The expected size for a PEO885 corona is ∼14 nm,27 hence showing the commensurability for the core size between the DLS (hydrodynamic radius minus corona) and cryoTEM (visible core) results. On the other hand, the micelle size (core + corona) obtained from the micelle core–core distance in the cryoTEM images is 39 ± 5 nm (Fig. S9†), which indicates a corona thickness of ∼6 nm. This difference is attributed to the compression of the PEG shell as a result of cryoTEM sample preparation and packing.28
The formation of dendrimicelles straightforwardly allows for incorporation and tuning of the number of NPs per micelle by mixing DENs with empty, amine-terminated PAMAM. Moreover, the presence of only one AuNP per PAMAM allows for the direct determination of the aggregation number of the dendrimicelles by simply counting the number of nanoparticles per micelle. The number of Au256DENs per micelle is 23 ± 7 for the dendrimicelle sample made using 100% Au256DENs and 12 ± 5 for the sample made using 50% empty PAMAM and 50% Au256DENs. Assuming a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 association of positive and negative charges in the dendrimicelle core, this corresponds to an average micelle molecular weight of ∼12 MDa, roughly three times as heavy as the biggest proteins (Titins) found in nature.29
1 association of positive and negative charges in the dendrimicelle core, this corresponds to an average micelle molecular weight of ∼12 MDa, roughly three times as heavy as the biggest proteins (Titins) found in nature.29
Using DLS, we further studied the stability of dendrimicelles formed at the PMC against different environmental conditions. Fig. S10† shows the observed scattering intensity and size of dendrimicelles versus pH. The dendrimicelles are stable between pH 6 and 8, in correspondence with the pKa values of the charged building blocks (∼6 for methacrylic acid,23 and ∼9 for amine-terminated PAMAM22), supporting the hypothesis that the dendrimicelles are made by coacervation of the positively and negatively charged building blocks.
The critical salt concentration (CSC), a measure of the salt stability of (dendri)micelles formed from the self-assembly of oppositely charged building blocks, is 1.5 M for NaCl (Fig. S11†), and the critical micelle concentration (CMC) of these dendrimicelles is ∼0.6 mg L−1 total polymer concentration (Fig. S12†). Compared to dendrimicelles made from acid-terminated PAMAM dendrimers,17 both CSC and CMC are slightly improved, and in line with other micelle systems.14 DLS results obtained over time (Fig. S13†) show that both the scattered light intensity as well as the obtained size remained virtually constant over a period of seven days.
To investigate whether the stability of the dendrimicelle as a whole also is representative of the stability of the micelle core, i.e. to see whether the dendrimicelle core is dynamic, we mixed dendrimicelles made from empty PAMAM with dendrimicelles containing Au256DENS. Here cryo-TEM micrographs recorded one day (Fig. S14†) or after 3 months at RT (Fig. 2 and S15/16†) both show dendrimicelles either without any AuDENs, or completely filled with AuDENs. Analysis of the dendrimicelles containing AuDENs shows that these contain 26 ± 8 NPs per dendrimicelle, in agreement with the number of AuNPs per dendrimicelle found before (Fig. 1). Clearly, even at these timescales, no exchange between filled and empty dendrimicelles occurs. This implies that the dendrimer core is (kinetically) trapped and no exchange of dendrimers between the dendrimicelles takes place.
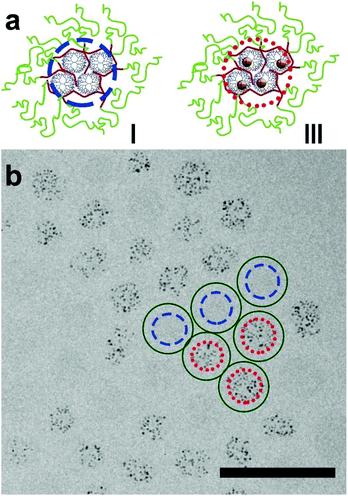 | ||
| Fig. 2 The effect of mixing dendrimicelles formed with empty dendrimers (I) and dendrimicelles formed with AuDENs, i.e. Au-nanoparticle filled dendrimers (III). (a) Schematic representation of the two dendrimicelles that were mixed: the blue, dashed circle is drawn around the core of a selection of three empty dendrimicelles (I) and the red, dotted, circle is drawn around the nanoparticle-filled dendrimer core of another selection of three micelles (III). (b) Cryo-TEM micrograph of a mix of dendrimicelles I and III, containing empty, and Au256-filled dendrimers, respectively; these were separately prepared and consecutively mixed, and measured after 30 days. As can be seen, mixing empty and filled micelles does not result in half-filled micelles, showing the high stability of the dendrimicelles. For visualization purposes, the blue circles are drawn around the micelle-core of three empty dendrimicelles, whereas red-dotted lines are drawn around the cores of three dendrimicelles containing Au-DENs. Additional images can be found in the ESI.† The scale bar is 100 nm. | ||
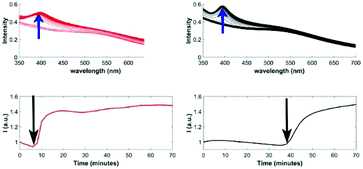
Besides the conceptual value, the advantage of a nanocontainer lies in the fact that properties of the contained particle/molecule are altered, e.g. protected, activated, and selectively accessible. The presented AuDENs in dendrimicelles indeed have properties that are different from AuDENs in solution; we used thiol-induced etching of AuNPs to study the differences.30 Upon adding two molar equivalents of mercaptoethanol (ME) to Au atoms the AuDENs are slowly degraded, the appearance of an absorption band at 395 nm (Fig. 3) being indicative of the formation of Au(I)–thiol complexes.31,32 In the case of the dendrimicelle sample, it takes about six times as long before this peak appears. Additionally, the rate at which the peak appears, i.e. the slope in the intensity vs. time plots, is also higher for the sample with just the Au256NPs than for the dendrimicelle sample. These observations suggest that the dendrimicelle forms a barrier that delays degradation.
Conclusions
Nanoparticles, dendrimers and micelles find wide applications in fields like nanomedicine, responsive materials and catalysis,33 and the controlled assembly strategy presented here opens up a vast horizon of applications. It offers a great variety for generating box-in-a-box structures and nanoparticle/molecule-in-a-box-in-a-box structures. There is, first, the possibility of changing the AuNP size by tuning the metal–dendrimer ratio during the synthesis of the DENs. Second, the use of different dendrimer generations can be exploited. Third, the system allows for making dendrimicelles with different metal NPs or even mixed-metal NPs. Likewise, combinations can be made by mixing different generations, using different types of dendrimers, by encapsulating molecules inside the dendrimer, or even using other macromolecular systems. Finally, coacervate-core micelles form upon charge interactions, but other micellar strategies can be explored as well, e.g. based on hydrophobic or supramolecular interactions.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Jan van Lent and Marcel Giesbers of the Wageningen Electron Microscopy Centre are thanked for their help with (cryo)electron microscopy. JBtH and AHV thank NWO for financial support of the MONALISA project 717.013.006. The research leading to these results has received funding from the European Research Council (ERC) under the European Union's Seventh Framework Program FP7/2007–2013 (Grant No. 2012-306890) and the Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No. 642192.Notes and references
- T. S. Koblenz, J. Wassenaar and J. N. H. Reek, Chem. Soc. Rev., 2008, 37, 247–262 RSC.
- P.-C. Chen, G. Liu, Y. Zhou, K. A. Brown, N. Chernyak, J. L. Hedrick, S. He, Z. Xie, Q.-Y. Lin, V. P. Dravid, S. A. O'Neill-Slawecki and C. A. Mirkin, J. Am. Chem. Soc., 2015, 137, 9167–9173 CrossRef CAS PubMed.
- S. Mirtschin, A. Slabon-Turski, R. Scopelliti, A. H. Velders and K. Severin, J. Am. Chem. Soc., 2010, 132, 14004–14005 CrossRef CAS PubMed.
- B. M. Rosen, C. J. Wilson, D. A. Wilson, M. Peterca, M. R. Imam and V. Percec, Chem. Rev., 2009, 109, 6275–6540 CrossRef CAS PubMed.
- J. Jansen, E. M. M. de Brabander-Van den Berg and E. W. Meijer, Science, 1994, 266, 1226–1229 CAS.
- Z. Zhou, Y. Wang, Y. Yan, Q. Zhang and Y. Cheng, ACS Nano, 2016, 10, 4863–4872 CrossRef CAS PubMed.
- L. Luo, L. Zhang, Z. Y. Duan, A. S. Lapp, G. Henkelman and R. M. Crooks, ACS Nano, 2016, 10, 8760–8769 CrossRef CAS PubMed.
- S. V. Myers, M. G. Weir, E. V. Carino, D. F. Yancey, S. Pande and R. M. Crooks, Chem. Sci., 2011, 2, 1632–1646 RSC.
- R. M. Crooks, M. Zhao, L. Sun, V. Chechik and L. K. Yeung, Acc. Chem. Res., 2001, 34, 181–190 CrossRef CAS PubMed.
- F. Gröhn, B. J. Bauer, Y. A. Akpalu, C. L. Jackson and E. J. Amis, Macromolecules, 2000, 33, 6042–6050 CrossRef.
- V. M. Gomez, J. Guerra, A. H. Velders and R. M. Crooks, J. Am. Chem. Soc., 2009, 131, 341–350 CrossRef PubMed.
- V. M. Gomez, J. Guerra, S. V. Myers, R. M. Crooks and A. H. Velders, J. Am. Chem. Soc., 2009, 131, 14634–14635 CrossRef PubMed.
- G. Riess, Prog. Polym. Sci., 2003, 28, 1107–1170 CrossRef CAS.
- I. K. Voets, A. de Keizer and M. A. C. Stuart, Adv. Colloid Interface Sci., 2009, 147–148, 300–318 CrossRef CAS PubMed.
- J. Y. Wang, A. H. Velders, E. Gianolio, S. Aime, F. J. Vergeldt, H. Van As, Y. Yan, M. Drechsler, A. de Keizer, M. A. C. Stuart and J. van der Gucht, Chem. Commun., 2013, 49, 3736–3738 RSC.
- J. Y. Wang, A. Groeneveld, M. Oikonomou, A. Prusova, H. Van As, J. W. M. van Lent and A. H. Velders, Soft Matter, 2016, 12, 99–105 RSC.
- J. Wang, I. K. Voets, R. Fokkink, J. van der Gucht and A. H. Velders, Soft Matter, 2014, 10, 7337–7345 RSC.
- L. Balogh, R. Valluzzi, K. S. Laverdure, S. P. Gido, G. L. Hagnauer and D. A. Tomalia, J. Nanopart. Res., 1999, 1, 353–368 CrossRef CAS.
- R. Esfand and D. A. Tomalia, Drug Discovery Today, 2001, 6, 427–436 CrossRef CAS PubMed.
- R. W. J. Scott, O. M. Wilson and R. M. Crooks, J. Phys. Chem. B, 2005, 109, 692–704 CrossRef CAS PubMed.
- O. M. Wilson, R. W. J. Scott, J. C. Garcia-Martinez and R. M. Crooks, J. Am. Chem. Soc., 2005, 127, 1015–1024 CrossRef CAS PubMed.
- Y. Niu, L. Sun and R. M. Crooks, Macromolecules, 2003, 36, 5725–5731 CrossRef CAS.
- L. Ruiz-Pérez, A. Pryke, M. Sommer, G. Battaglia, I. Soutar, L. Swanson and M. Geoghegan, Macromolecules, 2008, 41, 2203–2211 CrossRef.
- D. G. Mullen, A. Desai, M. A. van Dongen, M. Barash, J. R. Baker and M. M. Banaszak Holl, Macromolecules, 2012, 45, 5316–5320 CrossRef CAS PubMed.
- M. A. van Dongen, A. Desai, B. G. Orr, J. R. Baker and M. M. B. Holl, Polymer, 2013, 54, 4126–4133 CrossRef CAS PubMed.
- Y. Zheng, Y.-Y. Won, F. S. Bates, H. T. Davis, L. E. Scriven and Y. Talmon, J. Phys. Chem. B, 1999, 103, 10331–10334 CrossRef CAS.
- C. J. Fee and J. M. Van Alstine, Bioconjugate Chem., 2004, 15, 1304–1313 CrossRef CAS PubMed.
- Y. Zheng, Z. Lin, J. L. Zakin, Y. Talmon, H. T. Davis and L. E. Scriven, J. Phys. Chem. B, 2000, 104, 5263–5271 CrossRef CAS.
- S. Labeit and B. Kolmerer, Science, 1995, 270, 293–296 CAS.
- C.-Y. Ke, T.-H. Chen, L.-C. Lu and W.-L. Tseng, RSC Adv., 2014, 4, 26050–26056 RSC.
- Y. Shichibu, Y. Negishi, H. Tsunoyama, M. Kanehara, T. Teranishi and T. Tsukuda, Small, 2007, 3, 835–839 CrossRef CAS PubMed.
- T. G. Schaaff and R. L. Whetten, J. Phys. Chem. B, 1999, 103, 9394–9396 CrossRef CAS.
- H. Ye and R. M. Crooks, J. Am. Chem. Soc., 2005, 127, 4930–4934 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental section and additional figures. See DOI: 10.1039/c7nr06773a |
| ‡ These authors contributed equally to the work. |
| This journal is © The Royal Society of Chemistry 2017 |