Open Access Article
Open Access ArticleBiotemplated synthesis of magnetic filaments
Éva
Bereczk-Tompa
a,
Ferenc
Vonderviszt
 bc,
Barnabás
Horváth
d,
István
Szalai
d and
Mihály
Pósfai
bc,
Barnabás
Horváth
d,
István
Szalai
d and
Mihály
Pósfai
 *a
*a
aDepartment of Earth and Environmental Sciences, University of Pannonia, Egyetem u. 10, 8200 Veszprém, Hungary. E-mail: eva.tompa88@gmail.com; mihaly.posfai@gmail.com
bBio-Nanosystems Laboratory, Research Institute of Biomolecular and Chemical Engineering, University of Pannonia, Egyetem u. 10, 8200 Veszprém, Hungary. E-mail: von007@almos.uni-pannon.hu
cInstitute of Technical Physics and Materials Science, Centre for Energy Research, Konkoly-Thege u. 29-33, 1121 Budapest, Hungary
dInstitute of Physics and Mechatronics, University of Pannonia, Egyetem u. 10, 8200 Veszprém, Hungary. E-mail: bhorvath@almos.uni-pannon.hu; szalai@almos.uni-pannon.hu
First published on 26th September 2017
Abstract
With the aim of creating one-dimensional magnetic nanostructures, we genetically engineered flagellar filaments produced by Salmonella bacteria to display iron- or magnetite-binding sites, and used the mutant filaments as templates for both nucleation and attachment of the magnetic iron oxide magnetite. Although nucleation from solution and attachment of nanoparticles to a pre-existing surface are two different processes, non-classical crystal nucleation pathways have been increasingly recognized in biological systems, and in many cases nucleation and particle attachment cannot be clearly distinguished. In this study we tested the magnetite-nucleating ability of four types of mutant flagella previously shown to be efficient binders of magnetite nanoparticles, and we used two other mutant flagella that were engineered to periodically display known iron-binding oligopeptides on their surfaces. All mutant filaments were demonstrated to be efficient as templates for the synthesis of one-dimensional magnetic nanostructures under ambient conditions. Both approaches resulted in similar final products, with randomly oriented magnetite nanoparticles partially covering the filamentous biological templates. In an external magnetic field, the viscosity of a suspension of the produced magnetic filaments showed a twofold increase relative to the control sample. The results of magnetic susceptibility measurements were also consistent with the magnetic nanoparticles occurring in linear structures. Our study demonstrates that biological templating can be used to produce one-dimensional magnetic nanostructures under benign conditions, and that modified flagellar filaments can be used for creating model systems in which crystal nucleation from solution can be experimentally studied.
Introduction
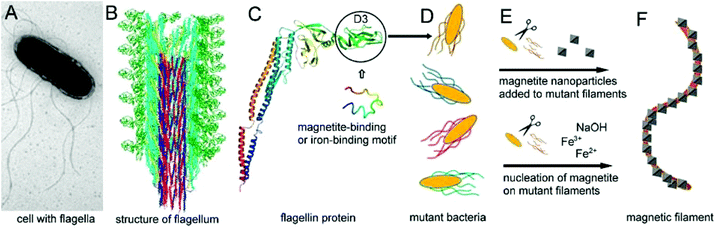
Self-assembling biological systems offer a promising platform for the development of new nanomaterials because they can enable the production of nanoscale materials with more strictly controlled properties than is possible in synthetic systems.1 An example of a natural, self-assembling protein system is represented by the bacterial flagellar filament, which can be used in nanotechnology as a template for nanoparticle arrays or applied as a scaffold to manufacture nanofibers.1,2The use of bacterial filaments in nanofabrication is attractive because their main component – the flagellin (FliC) protein – can be produced in large quantities, exported out of the cell and can self-assemble into long, homogeneous fibers,3 which can be easily purified.4 The length of the filaments can be controlled during in vitro polymerization by changing the conditions, i.e. the applied precipitant or flagellin monomer concentration.5 Using flagellin in nanofabrication offers one more benefit: it can be easily tailored by genetic manipulation. The middle hypervariable region of the FliC gene forms the surface-exposed D3 domain in the filament structure, which can be removed, modified or replaced by foreign proteins without adversely affecting the polymerization ability of flagellin.6 Mutant flagellin variants obtained by genetic engineering can be used as building blocks to add various functionalities to the filament.1,7,8
In our recent work9 flagellar filaments of Salmonella were engineered to facilitate the formation of 1D magnetic nanostructures under ambient conditions. We constructed four different flagellin mutants displaying magnetite-binding motifs, two of which contained fragments of magnetosome-associated proteins from magnetotactic bacteria (MamI and Mms6), and another two contained synthetic sequences. The self-assembled mutant filaments were used as scaffolds to which separately synthesized magnetite nanoparticles could attach. Even though MamI was previously reported to play a crucial role in magnetite nucleation,10 the mutant filaments containing the fragment of the MamI protein proved to be efficient templates for magnetite nanoparticle attachment. Considering that nucleation and nanoparticle binding are closely related phenomena, in this paper we explore whether the nucleation of magnetic particles could be induced by the modified filaments. Therefore, magnetite was synthesized in the presence of mutant Salmonella bacteria. In addition to the above mentioned sequences (Table 1), we created two additional mutants which were engineered to display known iron-binding motifs. Our goal was to use these motifs to bind iron ions from solution and initiate the nucleation of magnetite particles on the surfaces of the filaments. We compared the magnetite templating and magnetite binding activities of the six different motifs, as well as the products resulting from the two different (iron- and magnetite-binding) processes (Fig. 1).
| Peptide name | Abbreviation | Sequence | Isoelectric point |
|---|---|---|---|
| Loop region of MamI9 | MamI_L | WWWSVTEFLRG | 6.00 |
C-terminal region of Mms6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 11 11 |
Mms6_C | YAYMKSRDIESAQSDEEVELRDALA | 4.19 |
Synthetic magnetite-binding oligopeptide1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12 12 |
SP1 | SGVYKVAYDWQH | 6.74 |
Synthetic magnetite-binding oligopeptide2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 13 13 |
SP2 | TLNKPNRALHFN | 11.00 |
Iron-binding oligopeptide1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 14 14 |
IB1 | DLGEQYFKG | 4.37 |
Iron-binding oligopeptide2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 15 |
IB2 | HREERHKEEKR | 8.60 |
Experimental
Construction of flagellin-based fusion genes, expression and purification of mutant flagellin variants
The iron-binding segments were synthesized by Genscript (Piscataway, NJ), cloned into pUC57 plasmid. Construction of flagellin-based fusion genes in which the D3 domain of FliC was replaced by the iron-binding motifs was performed according to Bereczk-Tompa et al.9 The expression and selection of the fusion protein variants were also carried out as described previously.9 Swimming ability of the bacteria was checked by dark-field microscopy using an Olympus BX50 microscope.Transmission electron microscopy (TEM)
Mutant cells and magnetic nanofibers were visualized using TEM, for which samples were deposited onto Cu grids covered by continuous Formvar/carbon film. In order to enhance image contrast, bare filaments were stained with 2% phosphotungstic acid (pH 7.0). We used unstained filaments in the experiments of magnetite nucleation and attachment. Bright-field images were obtained on a CM20 (200 kV; Philips), high-resolution TEM images on JEM-3010 (300 kV, JEOL) and JEM-2010F (200 kV, JEOL) microscopes.Nucleation experiments
Mutant cells were grown overnight at 37 °C with vigorous agitation, then the samples (500 μL) were subjected to centrifugation (at 3000 rpm, 5 min). The bacteria pellets were suspended in (500 μl) distilled water.Synthesis of magnetite onto surface-modified flagellar filaments was performed by the co-precipitation method: 60 μL of iron chloride solution (66 mM; FeII![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) FeIII = 1
FeIII = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) was added to 500 μL cell suspension and was gently mixed. The pH of the reaction mixture was ∼2.5, and then a base (0.1 M NaOH) was gradually added dropwise to increase the pH until a black precipitate (magnetite) formed. Although reconstructed filaments are stable between pH 4 and 10,16 the distal ends of filaments formed in vivo on bacteria are covered by the pentameric HAP2 cap17 which may help to preserve their stability even at extreme values of pH.
2) was added to 500 μL cell suspension and was gently mixed. The pH of the reaction mixture was ∼2.5, and then a base (0.1 M NaOH) was gradually added dropwise to increase the pH until a black precipitate (magnetite) formed. Although reconstructed filaments are stable between pH 4 and 10,16 the distal ends of filaments formed in vivo on bacteria are covered by the pentameric HAP2 cap17 which may help to preserve their stability even at extreme values of pH.
In order to avoid oxidation, all solutions were degassed before use, and the system was kept under argon during the synthesis.
The experiments were performed with three controls that were not supposed to contain iron- or magnetite-binding sites on their flagella: (1) wild-type bacteria, (2) a mutant with its D3 domain removed, and the two ends of D2 connected with a small, uncharged linker (ΔD3_FliC_GNLSA),6 and (3) another mutant in which the D3 domain was replaced by a longer linker (LETGPGEL),8 encoded by a gene cassette containing the recognition sites of the restriction enzymes used in the genetic engineering experiments (ΔD3_FliC_LETGPGEL).
Magnetic measurements
The magnetic properties of magnetic nanofibers were characterized by viscosity and magnetic susceptibility measurements, using the samples prepared with FliC-MamI_L filaments.9The rheological behavior of magnetic nanofibers in an external magnetic field was determined by an Anton Paar MCR 301 rotational rheometer equipped with an MRD 70/1 T magnetorheological accessory. The measurements were conducted at a constant shear rate of 5 s−1 with parallel plate geometry (at T = 20.5 °C). During constant shear the samples were subjected to an external homogeneous magnetic field (20 s long rectangular pulse, B = 800 mT) perpendicular to the shear flow.
The dynamic magnetic response of the nanofibers was determined by a special magnetic susceptibility measurement method, which is the magnetic counterpart of the dielectric response-time measurement technique.18 The sample was placed inside a solenoid, which is the frequency-determining element of an LC oscillator. The frequency was measured by an HP 53310A modulation domain analyzer. A Helmholtz coil pair was used to generate a 50 ms long ramp pulse of a homogeneous magnetic field (25 ms rise time, Bmax = 4.5 mT). The Helmholtz coil was driven by a Labworks PA-138 power amplifier, and the input signal was supplied by a data acquisition card (National Instruments PCI-6052E).
Results and discussion
Development of flagellar filaments displaying iron-binding motifs
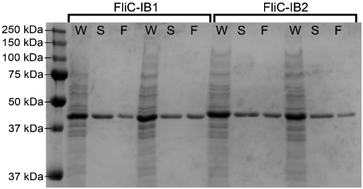
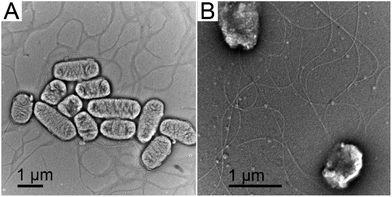
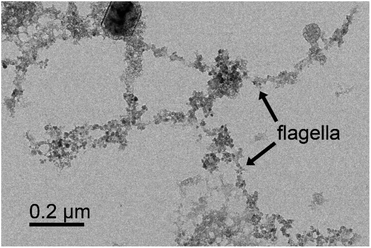
Previously we used bacterial flagellar filaments as templates for the formation of magnetic nanofibers by capturing magnetite nanoparticles from solution. Flagellin subunits were genetically engineered to display magnetite-binding sites on the surface of the filaments. The details of the construction of four mutants were described by Bereczk-Tompa et al.9 In the present work, we constructed two new flagellin mutants displaying iron-binding motifs. One of these peptides, with the sequence DLGEQYFKG, was identified from porcine plasma after hydrolysis with Flavourzyme.14 The other peptide with the sequence HREERHKEEKR was originally proposed as an iron oxide binding motif,15 but we found it to have iron-binding capability. The DNA segments coding for the oligopeptides were synthesized, and ligated into the pKOT-based plasmid containing the D3 deletion mutant FliC gene. After plasmid constructs were introduced into the flagellin-deficient SJW2536 Salmonella strain, the mutant flagellin variants were expressed at a high level. The preserved swimming ability of the bacteria (checked by dark-field microscopy) suggested a well-folded structure of the fusion proteins. The amounts of subunits built into filaments were studied by gel electrophoresis because our earlier observations9 showed that a significant fraction of the flagellin-based fusion proteins was secreted into the culture medium in monomeric form. Therefore, the mutant cells were collected by centrifugation, suspended in PBS and heated to 65 °C to disassemble flagellar filaments, and the depolymerized fractions were analyzed by SDS-PAGE. We observed strong bands at the expected positions for the depolymerized FliC-IB1 and FliC-IB2 fractions (Fig. 2), but comparison of their band intensities to the secreted fractions suggested that again a minor portion of subunits formed filaments on the surfaces of the bacteria. TEM studies demonstrated that IB1 filaments were 7–12 μm and IB2 filaments were 5–8 μm long (Fig. 3).Magnetite precipitation on the surfaces of mutant filaments
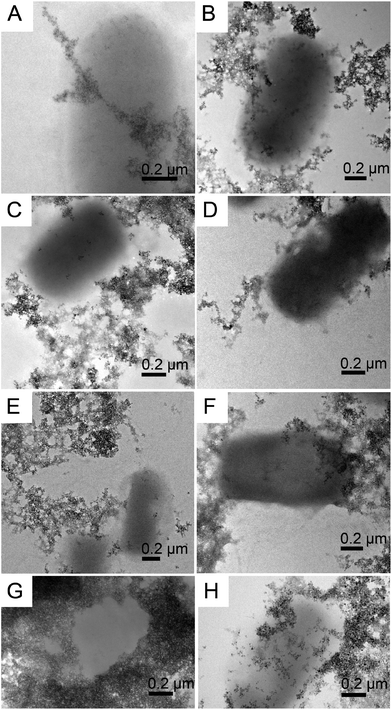
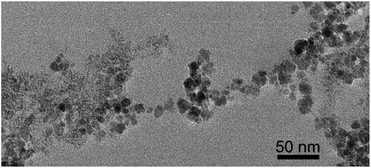
We performed magnetite nucleation experiments with all six mutant flagellin variants (Table 1) and three controls. Magnetite precipitation was initiated by adding a solution containing a mixture of ferrous and ferric chlorides to the suspension of filaments. We expected that iron ions from solution would be bound by the inserted functional motifs and thus localized on the surfaces of filaments. Then we added NaOH in order to initiate the formation of magnetite.The controlled precipitation led to the formation of networks of magnetite chains in all six iron-binding mutants and in one of the controls (ΔD3_FliC_LETGPGEL) (Fig. 4A–F). In contrast, cells in two control samples (WT and ΔD3_FliC_GLNSA) were tightly embedded in magnetite aggregates, but networks of magnetite chains were absent around the cells (Fig. 4G and H). In the case of the mutants producing networks of magnetite-covered filaments, the question arises whether (1) iron from solution was bound by the modified filaments, initiating magnetite crystal formation directly on the surfaces of filaments, or (2) magnetite nanoparticles formed by homogeneous nucleation in the solution and then attached to the filaments. The resulting particle size distribution suggests that process (1) was primarily responsible for the formation of magnetite. We observed particle sizes in the range of 1.5–15 nm showing a bimodal distribution centered around 1.5–3 nm and 10–15 nm (Fig. 5). Magnetite nanoparticles <5 nm (Fig. 6) were observed neither in inorganic co-precipitation experiments in which the same reagents were used (and that yielded 13.5(±6) nm diameter nanoparticles),9 nor in the experiments in which pre-made magnetite particles were attached to mutant filaments.9 Although the homogeneous nucleation of magnetite in the solution and its later attachment to the flagella cannot be entirely excluded, we found in a previous study that the FliC-Mms6_C mutant could not interact with magnetite nanoparticles.9 We also checked the magnetite-binding ability of the IB1 and IB2 variants but failed to observe any binding. The fact that Mms6_C, IB1 and IB2 flagella were not able to interact with pre-existing magnetite surfaces but were efficient in the nucleation experiments (Table 2) also suggests that crystals nucleated directly on these filaments (Fig. 7).
 | ||
| Fig. 7 Bright-field TEM image of a web of magnetic nanofibers synthesized by co-precipitation of Fe(II) and Fe(III) onto mutant FliC-Mms6_C flagella. Since Mms6_C flagella were not able to interact with pre-existing magnetite surfaces,8 the presence of magnetite on these filaments indicates that the crystals nucleated directly on the filaments (the large crystal with dark contrast on the top is NaCl). | ||
| Filament sample | Results of the magnetite-binding experiment | Results of the nucleation experiment |
|---|---|---|
| MamI_L | + | + |
| Mms6_C | − | + |
| SP1 | + | + |
| SP2 | + | + |
| IB1 | − | + |
| IB2 | − | + |
| ΔD3_FliC_LETGPGEL (control) | − | + |
| ΔD3_FliC_GLNSA (control) | − | − |
| Wild-type (control) | − | − |
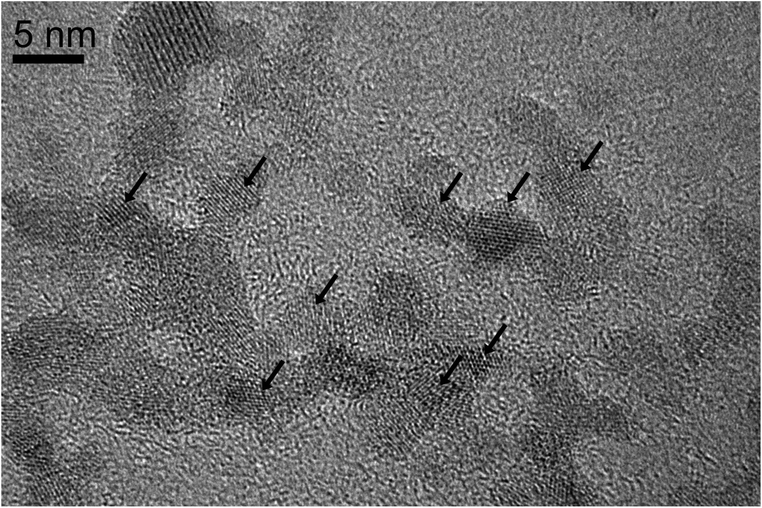
HRTEM images of the nm-sized crystallites and their Fourier transforms indicate that the particles are magnetite. In many systems the formation of amorphous precursors was observed before crystallization of the final product.19 For magnetite, a ferrihydrite precursor was found by cryo-TEM to form in the solution first and then to convert to magnetite.20 Here, we observed cyrstalline, nm-sized magnetite without any evidence for the presence of a precursor. Interestingly, the nm-sized crystallites did not arrange themselves along the protein templates by oriented attachment, and did not fuse into larger single crystals, but were present in random crystallographic orientations along the filaments.
In the present study we tested the magnetite-nucleating ability of six types of mutant flagella, some of which had been previously shown to be efficient binders of magnetite nanoparticles.9 The results of both magnetite nucleation and magnetite attachment experiments are outlined in Table 2. The loop segment of MamI can perform a double role: it may both nucleate and bind magnetite nanoparticles. Previous studies suggested that MamI may be involved in magnetite nucleation in magnetotactic bacteria;21 our experiments provide the first direct evidence for this role. For the C-terminal part of Mms6, both iron- and magnetite-binding capacities have been reported.22,23 However, a recent study found that the 20 amino acid C-terminal residue of Mms6 binds iron ions (especially Fe(II)) rather than magnetite, suggesting that Mms6 is a magnetite-nucleating protein.24 This is confirmed by our experiments in which the C-terminal segment of Mms6 performed as a nucleating agent but was unable to bind pre-made magnetite particles. The apparent contradiction between reports about the magnetite-binding ability of Mms6_C probably results from the same protein adopting different conformations in the various fusion constructs used in the above studies.
It is not straightforward to interpret the interactions between the mutant flagellar filaments and iron ions, as well as between filaments and the magnetite nanoparticles. Based on both experimental studies of the interface between magnetite and magnetosome proteins25 and molecular dynamics simulations of interactions between the magnetite surface and acidic amino acids,26 electrostatic forces are thought to dominate. In our experiments, the attachment of magnetite nanoparticles to the mutant filaments was performed at pH 7, where magnetite has a positive surface charge.27 Simple coulombic interactions between iron ions or the magnetite surface and the functional olygopeptides built into the flagellar filaments are insufficient to explain our observations. For example, both SP2 and MamI_L bound pre-made magnetite nanoparticles, even though the isoelectric point of SP2 is at pH 11 (Tables 1 and 2) and MamI_L has a highly hydrophobic character. We have no clear explanation for the observed binding behavior of the applied segments. The process of magnetite nucleation appears to be less specific than magnetite binding, since in addition to the six mutants designed to bind iron, even the ΔD3_FliC_LETGPGEL mutant (intended as a control) was able to nucleate magnetite. In contrast, only three of the mutant filaments could attach to magnetite nanoparticles. Although the ΔD3_FliC_LETGPGEL mutant did not contain the D3 domain of flagellin, two glutamic acid residues were present in the construct. We believe that negatively charged amino acids probably play an important role in binding iron irons from solution and thereby initiating magnetite nucleation.
Even though the atomic scale interactions between the mutant filaments and magnetite remain obscure, the efficiency of the inserted oligopeptide sequences in either nucleating and/or binding magnetite has been clearly established by our experiments.
Magnetic properties of the magnetite-covered filaments
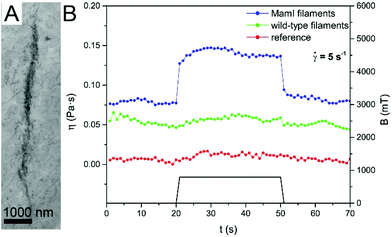
Magnetic nanofibers were prepared by using mutant flagellar filament templates for both magnetite nucleation and attachment. Since the final products obtained by the two different processes did not differ significantly, the magnetic properties of the nanofibers were measured on a selected sample, containing MamI_L filaments with attached magnetite nanoparticles. SAED patterns and HRTEM images clearly identify the nanoparticles as magnetite, the size-dependent magnetic properties of which are well known.28 Therefore, we performed measurements that were designed to shed light on magnetic behavior that could specifically result from the special, filamentous nature of the magnetic nanostructures.To test whether the magnetic nanofibers could be aligned by exposure to a static magnetic field, a drop of suspension containing magnetite-covered MamI_L filaments was placed on a TEM grid and then introduced into a strong magnetic field (up to 800 mT) until the sample dried. TEM images revealed that the magnetic filaments aligned in roughly parallel ropes of several μm long (Fig. 8A), and each rope was composed of bundles of smaller chains of magnetite-covered filaments.
The effect of a magnetic field on the viscosity of a suspension containing magnetite-covered filaments was investigated experimentally. Fig. 8B shows the viscosity versus time of the MamI_L sample and two controls (one containing unmodified filaments with magnetite and the other only magnetite particles), with and without application of an external magnetic field. The viscosity of the sample containing the magnetite-covered filaments increased about twofold upon application of an external magnetic field, and returned to its original value when the magnetic field was switched off. The change of viscosity is caused by the alignment of the magnetic filaments parallel to the applied field (that is perpendicular to the shear exerted by the flow). No viscosity change could be observed in the controls. Apparently, the chains formed from the free magnetite particles under the magnetic field were much less stable than the ordered, linear arrangement of magnetite along the filament templates.
The dynamics of structure formation (alignment of the magnetic filaments) in an external magnetic field was characterized by response time measurements. The structure formation causes a change in the magnetic susceptibility of the nanofiber samples. The response time was extracted from the dynamic magnetic response (change of magnetic susceptibility versus time) as a characteristic time constant. We found that the magnetic nanofiber samples have a response time of τ = 10.2 ms, which is considerably higher than the τ = 6.6 ms response time of the reference sample (magnetite nanoparticles without filaments). The slower response of the magnetic nanofibers is consistent with the magnetite particles being bound to the filaments.
Conclusions
In this study mutant flagella were used as templates for both magnetite nucleation and attachment. Both experimental approaches resulted in filamentous structures covered by randomly oriented magnetite nanoparticles. The obtained magnetic filaments can be aligned in an external magnetic field, showing a highly ordered arrangement. Our results demonstrate that one-dimensional biological templates can be built that either trigger the nucleation of magnetite from solution or bind magnetite nanoparticles from their aqueous suspension, with both processes leading to the formation of filamentous magnetic nanostructures. The produced magnetic nanofibers can be potentially used in various applications in the future, including magnetic nanoparticle imaging and the preparation of new soft intelligent materials.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Dr Aleksander Rečnik for the use of the TEM facility at the Jožef Stefan Institute in Ljubljana, Slovenia. This research was supported by NKFIH grants ERA-CHEMISTRY NN117642 and M.ERA-NET NN117849, and the BIONANO_GINOP-2.3.2-15-2016-00017 project.References
- M. T. Kumara, B. C. Tripp and S. Muralidharan, Chem. Mater., 2007, 19, 2056–2064 CrossRef CAS.
- M. T. Kumara, S. Muralidharan and B. C. Tripp, J. Nanosci. Nanotechnol., 2007, 7, 2260–2272 CrossRef CAS PubMed; W. R. Hesse, L. Luo, G. Zhang, R. Mulero, J. Cho and M. J. Kim, Mater. Sci. Eng., 2009, 29, 2282–2286 CrossRef; W. Jo, K. J. Freedman, D. K. Yi and M. J. Kim, Nanotechnology, 2012, 23, 055601 CrossRef PubMed; D. Li, B. Mathew and C. Mao, Small, 2012, 8, 3691–3697 CrossRef PubMed.
- K. Namba and F. Vonderviszt, Q. Rev. Biophys., 1997, 30, 1–65 CrossRef CAS PubMed.
- F. Vonderviszt, S. Kanto, S.-I. Aizawa and K. Namba, J. Mol. Biol., 1989, 209, 127–133 CrossRef CAS PubMed.
- S. Asakura, Adv. Biophys., 1970, 1, 99–155 Search PubMed; S. Asakura, G. Eguchi and T. Lino, J. Mol. Biol., 1964, 10, 42–56 CrossRef CAS PubMed.
- A. Muskotál, C. Seregélyes, A. Sebestyén and F. Vonderviszt, J. Mol. Biol., 2010, 403, 607–615 CrossRef PubMed.
- K. Deplanche, R. D. Woods, I. P. Mikheenko, R. E. Sockett and L. E. Macaskie, Biotechnol. Bioeng., 2008, 101, 873–880 CrossRef CAS PubMed; Á. Klein, B. Tóth, H. Jankovics, A. Muskotál and F. Vonderviszt, Protein Eng., Des. Sel., 2012, 25, 153–157 CrossRef PubMed.
- V. Szabó, A. Muskotál, B. Tóth, M. Mihovilovic and F. Vonderviszt, PLoS One, 2011, 6, e25388 Search PubMed.
- É. Bereczk-Tompa, M. Pósfai, B. Tóth and F. Vonderviszt, ChemBioChem, 2016, 17, 2075–2082 CrossRef PubMed.
- A. Lohße, S. Borg, O. Raschdorf, I. Kolinko, É. Tompa, M. Pósfai, D. Faivre, J. Baumgartner and D. Schüler, J. Bacteriol., 2014, 196, 2658–2669 CrossRef PubMed.
- L. Wang, T. Prozorov, P. E. Palo, X. Liu, D. Vaknin, R. Prozorov, S. Mallapragada and M. Nilsen-Hamilton, Biomacromolecules, 2012, 13, 98–105 CrossRef PubMed.
- M. Widdrat, PhD thesis, Mathematisch-Naturwissenschaftlichen Fakultät der Universität Potsdam, 2015.
- J. Baumgartner, M. A. Carillo, K. M. Eckes, P. Werner and D. Faivre, Langmuir, 2014, 30, 2129–2136 CrossRef CAS PubMed.
- S.-H. Lee and K. B. Song, Process Biochem., 2009, 44, 378–381 CrossRef CAS.
- A. Briel and E. Blazek, US Pat, 2009/0208420A1, 2009 Search PubMed.
- S. Asakura, Adv. Biophys., 1970, 1, 99–155 CAS.
- Z. Diószeghy, P. Závodszky, K. Namba and F. Vonderviszt, FEBS Lett., 2004, 568, 105–109 CrossRef PubMed.
- B. Horváth and I. Szalai, Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top., 2012, 86, 061403 CrossRef PubMed; B. Horváth and I. Szalai, Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top., 2015, 92, 042308 CrossRef PubMed.
- E. M. Pouget, P. H. H. Bomans, J. A. C. M. Goos, P. M. Frederik, G. de With and N. A. J. M. Sommerdijk, Science, 2009, 323, 1455–1458 CrossRef CAS PubMed; Q. Hu, M. H. Nielsen, C. L. Freeman, L. M. Hamm, J. Tao, J. R. I. Lee, T. Y. J. Han, U. Becker, J. H. Harding, P. M. Dove and J. J. De Yoreo, Faraday Discuss., 2012, 159, 509–523 RSC; A. Dey, P. H. H. Bomans, F. A. Müller, J. Will, P. M. Frederik, G. de With and N. A. J. M. Sommerdijk, Nat. Mater., 2010, 9, 1010–1014 CrossRef PubMed.
- J. Baumgartner, G. Morin, N. Menguy, T. Perez Gonzalez, M. Widdrat, J. Cosmidis and D. Faivre, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 14883–14888 CrossRef CAS PubMed.
- A. Lohße, S. Borg, O. Raschdorf, I. Kolinko, É. Tompa, M. Pósfai, D. Faivre, J. Baumgartner and D. Schüler, J. Bacteriol., 2014, 196, 2658–2669 CrossRef PubMed.
- H. Nudelman, C. Valverde-Tercedor, S. Kolusheva, T. Perez Gonzalez, M. Widdrat, N. Grimberg, H. Levi, O. Nelkenbaum, G. Davidov, D. Faivre, C. Jimenez-Lopez and R. Zarivach, J. Struct. Biol., 2016, 194, 244–252 CrossRef CAS PubMed.
- K. Ma, H. Zhao, X. Zheng, H. Sun, L. Hu, L. Zhu, Y. Shen, T. Luo, H. Dai and J. Wang, J. Mater. Chem. B, 2017, 5, 2888–2895 RSC.
- A. E. Rawlings, J. P. Bramble, A. M. Hounslow, M. P. Williamson, A. E. Monnington, D. J. Cooke and S. S. Staniland, Chem. – Eur. J., 2016, 22, 7885–7894 CrossRef CAS PubMed; S. M. Bird, A. E. Rawlings, J. M. Galloway and S. S. Staniland, RSC Adv., 2016, 6, 7356–7363 RSC.
- A. E. Rawlings, J. P. Bramble, A. M. Hounslow, M. P. Williamson, A. E. Monnington, D. J. Cooke and S. S. Staniland, Chem. – Eur. J., 2016, 22, 7885–7894 CrossRef CAS PubMed.
- A. Bürger, U. Magdans and H. Gies, J. Mol. Model., 2013, 19, 851–857 CrossRef PubMed.
- E. Tombácz, A. Majzik, Zs. Horvát and E. Illés, Rom. Rep. Phys., 2006, 58, 281–286 Search PubMed.
- M. Kumari, A. Hirt, R. Uebe, D. Schüler, É. Tompa, M. Pósfai, W. Lorenz, F. Ahrentorp, C. Jonasson and C. Johansson, Geochem., Geophys., Geosyst., 2015, 16, 1739–1752 CrossRef.
| This journal is © The Royal Society of Chemistry 2017 |