Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Properties of in situ generated gold nanoparticles in the cellular context†
D.
Drescher
ab,
H.
Traub
b,
T.
Büchner
a,
N.
Jakubowski
b and
J.
Kneipp
 *ab
*ab
aHumboldt-Universität zu Berlin, Department of Chemistry, Brook-Taylor-Str. 2, 12489 Berlin, Germany. E-mail: janina.kneipp@chemie.hu-berlin.de
bBundesanstalt für Materialforschung und –prüfung (BAM), Richard-Willstätter-Str. 11, 12489 Berlin, Germany
First published on 26th July 2017
Abstract
Gold nanostructures that serve as probes for nanospectroscopic analysis of eukaryotic cell cultures can be obtained by the in situ reduction of tetrachloroauric acid (HAuCl4). To understand the formation process of such intracellularly grown particles depending on the incubation medium, the reaction was carried out with 3T3 fibroblast cells in three different incubation media, phosphate buffer, Dulbecco's Modified Eagle Medium (DMEM), and standard cell culture medium (DMEM with fetal calf serum). The size, the optical properties, the biomolecular corona, and the localization of the gold nanoparticles formed in situ vary for the different conditions. The combination of surface-enhanced Raman scattering (SERS) and laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS) microscopic mapping and transmission electron microscopy (TEM) provides complementary perspectives on plasmonic nanoparticles and non-plasmonic gold compounds inside the cells. While for the incubation with HAuCl4 in PBS, gold particles provide optical signals from the nucleus, the incubation in standard cell culture medium leads to scavenging of the toxic molecules and the formation of spots of high gold concentration in the cytoplasm without formation of SERS-active particles inside the cells. The biomolecular corona of nanoparticles formed in situ after incubation in buffer and DMEM differs, suggesting that different intracellular molecular species serve for reduction and stabilization. Comparison with data obtained from ready-made gold nanoparticles suggests complementary application of in situ and ex situ generated nanostructures for optical probing.
Introduction
The generation of gold and silver nanostructures by living cells of bacteria,1,2 fungi,3,4 plants,5–7 and animals has been discussed and studied in various contexts, ranging from biodiagnostics8 to materials chemistry.9,10 In animal cells and tissues, the interaction of cells with nanomaterials is important with respect to possible diagnostic applications. Gold nanoparticles are formed in cell cultures of animal cell lines and in their supernatant after incubation with tetrachloroauric acid (HAuCl4) in buffer.11–14 Also hybrid structures with graphene oxide could be obtained in this way.14 Even inside epithelial cells of the skin15 and in intact tumor tissues,16 formation of nanoparticles and nanoclusters is observed.Many different biomolecular species were shown to be involved in gold nanoparticle formation and stabilization,17 yielding nanoparticles as well as smaller nanoclusters. These molecular species include amino acids, such as tyrosine,18 different protein side chains,19,20 glutathione,21 phospholipids,22 chitosan and chitin,23 bilirubin,24 and many more.
Upon the in situ reduction of gold and silver salts in or by cells, several excellent plasmonic substrates were reported, and the nanostructures obtained in such in situ syntheses were exploited for optical nanospectroscopy, specifically in surface-enhanced Raman scattering (SERS).1,4,12,13,25
From the perspective of analytical and diagnostic applications, in situ synthesis of metal nanostructures that serve as probes for optical detection enables us to obtain them in direct proximity to their target.1,16,26–28 In order to apply gold nanoparticles that are generated in situ as optical probes inside animal cells, it is crucial to know the biomolecular species involved in their formation, as the biomolecular corona will determine the processing of the nanostructures inside the cells and their physical properties. This is expected from the numerous studies in the area of nanotoxicology carried out with gold nanoparticles synthesized in the absence of biomaterials (ex situ) and added to cells or tissues later.29,30
In the work presented here, we characterize the biomolecular species involved in in situ nanoparticle formation in fibroblast cell cultures, and we discuss the influence of the biosynthesis conditions, as consequence of the composition of the incubation medium, on the surface composition and the subcellular localization of the in situ formed gold nanostructures.
In our experiments, SERS provides detailed information about the nano-bio interaction at the surface of the plasmonic gold nanoparticles formed in live cells. We have demonstrated the feasibility of such an approach in work with plasmonic nanoparticles synthesized outside cells and taken up by endocytosis.31–33 At the same time, SERS can only be used as a probe of nanoparticle formation, as long as nanoparticles with significant enhancement factors are produced. Therefore, we observe the in situ formation of gold nanostructures in cultured cells under varied incubation conditions also by spatially resolved laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS) and by transmission electron microscopy (TEM). LA-ICP-MS is widely used for elemental bioimaging in tissue sections or cells, as it provides high spatial resolution and multielemental detection with high sensitivity.34,35 Capable of quantifying gold in individual eukaryotic cells,36 LA-ICP-MS gives information about the distribution of gold, independent of nanoparticle formation.
As we discuss here, our data indicate that gold nanoparticle formation and the properties and localization of the nanostructures strongly depend on the experimental conditions chosen for the cell culture in the incubation experiment. This is of importance for bioimaging and other diagnostic applications. We compare the in situ formation of the nanostructures by reduction inside the cells with results obtained after incubation of the same cell culture with ready-made 13 nm gold nanoparticles that were prepared by a standard synthesis protocol. Both strategies prove feasible for probing cellular systems and address different cellular substructures for bioanalytical and nanobiophotonic investigations. Furthermore, relevant to potential applications of gold nanoparticles generated in situ, we demonstrate different impact on cell metabolism in the different incubation approaches, resulting in great differences with respect to cytotoxicity.
Experimental
Particle synthesis and characterization
For the synthesis of gold nanoparticles sized 13 nm according to a protocol of Tkachenko et al., 250 mL of 1 mM gold(III)chloride trihydrate solution (99.9%, Sigma-Aldrich, Germany) were boiled for 10 min before 25 mL of 40 mM trisodium citrate dehydrate solution (99%, Merck, Germany) were added. After boiling of the reaction mixture for 10 min, the suspension was stirred for 15 min at room temperature. Extinction spectra were recorded with a UV-vis/NIR double-beam spectrophotometer (JASCO GmbH, Germany). Transmission electron micrographs of gold particles and of HAuCl4 after suspension for 3 hours in PBS and culture medium were obtained with a Tecnai G2 20 S-TWIN microscope (FEI, USA) using an acceleration voltage of 200 kV. Ultrapure water (18 MΩ) and phosphate-buffered saline (PBS; Biochrom AG, Germany) were utilized for all experiments.Cell experiments
Mouse fibroblast cells of cell line 3T3 (DSMZ, Germany) were cultivated in Dulbecco's modified eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 1% ZellShield (all from Biochrom AG, Germany) in a controlled environment at 37 °C and 5% CO2. For the SERS, TEM and LA-ICP-MS experiments, cells were harvested and grown on cover slips (Thermo Fisher Scientific, USA) in sterile culture plates. Under standard cell culture conditions, fibroblast cells were incubated with 1 mM HAuCl4 in PBS, DMEM and DMEM with 10% FCS for 24 hours. For comparison, cells were exposed to 13 nm gold nanoparticles suspended in DMEM with 10% FCS for 1, 3 and 24 hours under the same conditions. After incubation, cells were washed thoroughly with PBS and immediately analyzed (SERS) or fixed (LA-ICP-MS, TEM).XTT cytotoxicity test
Cytotoxicity of tetrachloroauric acid was determined utilizing XTT reagent (2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide, Biozol Diagnostica, Germany) in a procedure described recently.37 In this assay cell viability is reflected as the total mitochondrial activity. After growing in a 96-well culture plate under controlled conditions at 37 °C and 5% CO2, cells were incubated with HAuCl4 in PBS and in DMEM with 10% FCS in a concentration range from 0.01 mM to 1 mM for 24 hours. Fibroblasts not exposed to gold salt served as reference in these experiments. Cell viability is expressed as relation of the experiments with exposed cells to untreated cells under the same conditions (arithmetic mean ± standard deviation of four replicates).SERS experiments
Measurements were conducted with a LabRam HR Raman spectrometer (HORIBA Jobin Yvon GmbH, Germany) equipped with a 60× water immersion objective. An excitation wavelength of 785 nm with an intensity at the sample in the range between 1.9 × 105 W cm−2 and 2.9 × 106 W cm−2 and an acquisition time of 1 s were used. The spectral resolution was ∼2 cm−1.TEM preparation
Ultrathin sections of fixed fibroblast cells were examined with a Zeiss EM 900 electron microscope at 80 kV (Carl Zeiss SMT GmbH, Germany) using a 1 K slow-scan CCD camera (Proscan). As described in the cell Experimental section, fibroblast cells were incubated with 1 mM HAuCl4 in PBS and in DMEM with 10% FCS as well as with 13 nm gold nanoparticles suspended in DMEM with 10% FCS. After an incubation time of 24 hours, the cells were washed twice with PBS and fixed with 2.5% glutaraldehyde in PBS (pH 7.4) for 15 min. In the next step, the cells were postfixed in 1% osmium tetroxide in 0.1 M phosphate buffer (PB, pH 7.4) for 10 min, washed repeatedly in PB and dehydrated in a graded series of ethanol including a block staining with 2% uranyl acetate in 70% ethanol. For embedding the sample in Epon 812 resin, hydroxypropyl methacrylate (HPMA, Fluka, Switzerland) was applied as intermedium. For TEM analysis, ultramicrotome cuts (70 nm) of the aldehyde-fixed and resin embedded cells were prepared and stained with uranyl acetate and lead citrate.Laser ablation ICP-MS
LA-ICP-MS experiments of fixed fibroblast cells were performed with an NWR213 laser ablation system (ESI, USA) equipped with a two-volume cell and coupled to an ICP sector field mass spectrometer (Element XR, Thermo Fisher Scientific, Germany). Details on operating parameters are given in Tables S2 and S3 of the ESI.† Prior to LA-ICP-MS analysis, cells grown on sterile cover-slips (Thermo Fisher Scientific, USA) and exposed to HAuCl4 or nanoparticles were fixed with 4% para-formaldehyde in PBS and dehydrated in a graded series of ethanol/water mixtures. Representative areas of the cell sample were continuously ablated by line scans. High resolution images were obtained by optimizing the laser ablation parameters for high sensitivity at high spatial resolution (see ESI†). To convert each raw data point of the 197Au intensity time profiles to a single pixel in a color-coded intensity plot, Origin 8.5 (Originlab Corporations, USA) was applied. ImageJ software (NIH, USA) was used for data analysis.Results and discussion
Intracellular distribution of in situ biosynthesized nanostructures
In the experiments, fibroblast cells from cell cultures were exposed to tetrachloroauric acid in the presence of DMEM culture medium with and without fetal calf serum (FCS), and PBS buffer, respectively. In order to localize and to quantify the gold, single 3T3 fibroblast cells were investigated by LA-ICP-MS micro-mapping analysis, allowing sensitive high-resolution imaging of the 197Au+ distribution inside cells.36,38 The variability in uptake between individual cells was accounted for by choosing 10–15 cells for each experiment.Fig. 1 shows LA-ICP-MS images of cells that were exposed to 1 mM HAuCl4 for 24 hours in PBS (Fig. 1A) and in DMEM with 10% FCS (Fig. 1B). Both LA-ICP-MS maps show an evenly distributed gold signal in the cytoplasm. Independent of the incubation medium, the 197Au+ intensity indicates high gold concentrations over the whole cells, with the highest intensities (up to 5 × 106 cps) in the regions of the nuclei (Fig. 1). This gives evidence of an interaction of tetrachloroauric acid with components of the cell nucleus. Apart from a possible preferential accumulation in the chemical environment of the nucleus, the high amount of gold in the nucleus may be related to the high number of oxidizable biomolecules per ablation spot due to the greater thickness of the cells in the nucleus region.
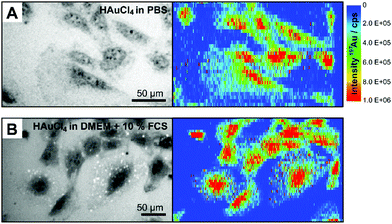 | ||
| Fig. 1 Bright field images and the corresponding intensity distribution of 197Au+ in single fibroblast cells after 24 h-exposure to 1 mM tetrachloroauric acid (A) in PBS and (B) in DMEM with 10% FCS determined by LA-ICP-MS. Enlarged figures of the LA-ICP-MS maps are shown in the ESI (Fig. S1†). Parameters: Laser spot size 4 μm, line distance 6 μm, scan speed 5 μm s−1, frequency 10 Hz, pixel size 6 × 1 μm, fluence 0.7 J cm−2. | ||
The LA-ICP-MS images reveal differences in gold accumulation with respect to the incubation medium. When DMEM with 10% fetal calf serum (FCS) is used for the incubation of HAuCl4, single pixels of high 197Au+ intensity are observed in the cytoplasm of the fibroblast cells (Fig. 1B, for enlarged maps see Fig. S1†). The corresponding bright field image also indicates small bright structures in the cytoplasm (Fig. 1B), which do not occur upon incubation in PBS (Fig. 1A). Some of the LA-ICP-MS signals co-localize with them. As will be discussed below, other evidence, such as the absence of larger particles in TEM micrographs and lack of plasmonic enhancement in SERS, suggests, that these structures must be small, non-plasmonic gold clusters generated either in the exposure medium during incubation or inside the cells. It is possible that such small clusters are stabilized immediately after their formation by biological molecules,7,19,20 in this case components of the cell culture medium, e.g., proteins or amino acids. Such small gold clusters were shown to have very beneficial properties for theranostic applications.39,40
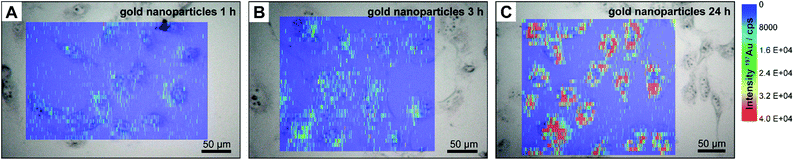
For comparison, we also conducted experiments with small gold nanoparticles of a size of 13 nm generated by citrate reduction. Fig. 2 shows the intensity distribution of 197Au+ in 3T3 fibroblast cells after incubation of these gold nanoparticles for one, three, and 24 hours, respectively. While upon incubation with tetrachloroauric acid, either biosynthesized particles, gold clusters, or excess gold salt could be present in the cell, especially localized in the region of the cell nucleus (Fig. 1), the gold nanoparticles accumulate in the cytoplasm (Fig. 2). Comparison of the LA-ICP-MS maps in Fig. 2A–C reveals differences in the amount and distribution of 197Au+ in the cellular compartments, that depend on the incubation time (for details see enlarged maps in Fig. S2†): after an exposure of one hour (Fig. 2A), the gold nanoparticles are evenly distributed in the cytoplasm, while after three hours (Fig. 2B) aggregates of particles are already formed in the perinuclear region in the course of endosomal maturation and multivesicular fusion.30,41 With longer exposure time of 24 hours (Fig. 2C), the signal intensity of 197Au+ rises, indicating an increase in the number of particles per sampled spot and particle aggregation. Importantly, for all incubation times, only low 197Au+ signal intensities are determined in the nucleus region, which originate from particles above and below the cell nucleus and maybe a few particles that entered the nucleus. The increase of the intracellular nanoparticle concentration observed in the LA-ICP-MS mapping of the single cells was also confirmed by ICP-MS experiments after digestion of the cells (see ESI Fig. S3†). The number of gold nanoparticles per cell is estimated to amount to 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 after 1 hour, and increasing to 227
000 after 1 hour, and increasing to 227![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nanoparticles after 24 hours.
000 nanoparticles after 24 hours.
For the cases of the incubation with HAuCl4 under the experimental conditions shown in Fig. 1, also the intensity profiles of representative LA-ICP-MS line scans are very different (Fig. 3). The region of the cell nucleus (marked by bar symbols) can be clearly differentiated from the cytoplasm and the background signal, and the 197Au+ signals differ in absolute intensity (Fig. 3A). While only low signal intensities are obtained in the cell nucleus after incubation with gold nanoparticles (Fig. 3B, compare also Fig. 2), the highest intensities are measured in this region after tetrachloroauric acid is incubated with the cells (Fig. 3A, compare also Fig. 1). Under all experimental conditions, separated, narrow signals occur and indicate local accumulations of 197Au+ in the form of gold clusters and/or nanoparticles. Especially for the incubation of tetrachloroauric acid in cell culture medium with 10% FCS (blue and red trace in Fig. 3A), narrow signals with very high intensities are observed in the cytoplasm, representing localized areas of very high gold concentrations. As a first conclusion, the LA-ICP-MS micro-mapping experiments of the cells after the incubation of tetrachloroauric acid confirm the presence of 197Au+ especially in the cell nuclei. This is different from incubation with gold nanoparticles and would allow optical investigations of the nucleus.
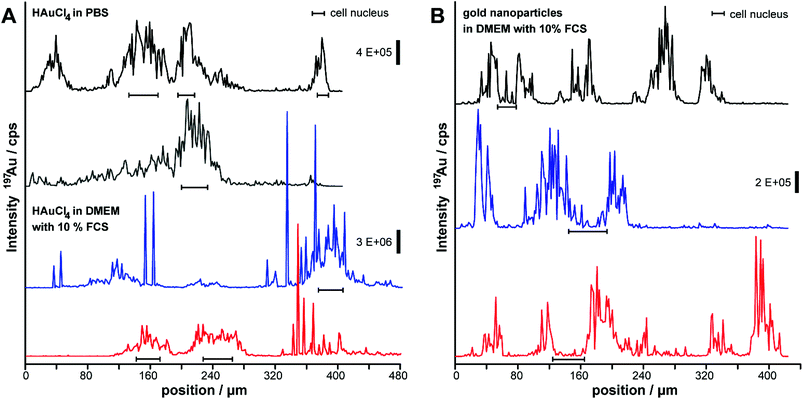 | ||
| Fig. 3 Representative line scans of the LA-ICP-MS images in Fig. 1 and 2 of the 197Au+ intensity distribution inside fibroblasts. Cells were incubated with (A) gold nanoparticles sized 13 nm and (B) tetrachloroauric acid in PBS and in DMEM with 10% FCS for 24 hours. Parameters: Laser spot size 4 μm, line distance 6 μm, scan speed 5 μm s−1, frequency 10 Hz, fluence 0.7 J cm−2. | ||
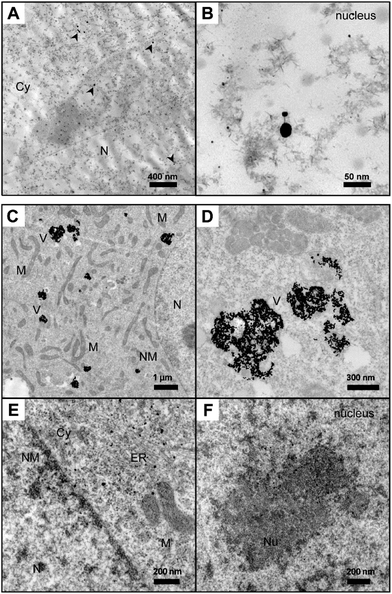
Also the TEM micrographs obtained from the cells vary greatly (Fig. 4). Owing to the high spatial resolution, TEM allows the localization of nanoparticles in cellular substructures of ultramicrotome sections, so that the presence of in situ biosynthesized nanoparticles or of gold nanoparticles taken up by the cells can be proven. The TEM micrographs in Fig. 4 show fibroblast cells after exposure to tetrachloroauric acid (Fig. 4A and B) and the intracellular distribution of gold nanoparticles with a diameter of 13 nm (Fig. 4C–F). TEM images of fibroblast cells incubated with 1 mM tetrachloroauric acid in PBS for 24 hours (Fig. 4A and B) provide evidence of gold nanoparticles inside the cellular ultrastructure. The in situ biosynthesized nanoparticles are characterized by a spherical shape and sizes ranging from a few nanometers to 30 nm. The particles are distributed over the whole cell. The TEM images (Fig. 4A and B; for more details see enlarged image in Fig. S4†) demonstrate that also in the nucleus, numerous particles are formed. This is in accord with the data shown in Fig. 3, where high 197Au+ intensities are found in the nucleus region when PBS is used as incubation medium for the tetrachloroauric acid (black and grey traces in Fig. 3A). The formation of nanoparticles inside the cells can be explained by the reduction of HAuCl4 by naturally occurring biomolecules.18,19,22 As evidenced by the absence of a plasmon band in the absorbance spectrum (Fig. S5A†), HAuCl4 remains unchanged during the incubation in PBS buffer. Therefore, we exclude that a reduction of Au3+ can take place already in the incubation medium PBS. Different particle sizes are observed in the TEM micrographs of the cells (Fig. 4A and B). Apart from very small nanostructures (<5 nm in diameter), only a few larger particles are found. The variation in size could be associated with the respective biomolecules that act as reducing agent and naturally vary within the surrounding cellular environment. Comparing the results with those of previous experiments on the quantification of gold nanostructures inside cells36,38 the number of nanoparticles that is visible in the ultramicrotome sections used for TEM is not in agreement with the high amount of 197Au+ found in the LA-ICP-MS experiments (compare Fig. 1A and 3A). Since the high 197Au+ content cannot be explained by the presence of nanoparticles visible in the TEM micrographs, it must be due to the presence of small gold clusters or unreacted gold salt in the cell.
When standard cell culture medium (DMEM with 10% FCS) is used for the incubation of HAuCl4 instead of PBS, no particulate nanostructures can be identified in the TEM images of fibroblast cells (data not shown). Also here, the absence of visible nanoparticles suggests that very small gold clusters or excess gold salt must be present in very confined spots that lead to the high 197Au+ intensities detected by mass spectrometry (compare Fig. 1B and 3A). Compared to the incubation of HAuCl4 in PBS (dotted spectrum in Fig. S5A†), in the case of tetrachloroauric acid in DMEM (dashed spectrum in Fig. S5A†) the absorbance is increased due to the reduction of the gold by medium constituents, e.g., by amino acids and glucose.42,43 In the presence of serum proteins (DMEM with 1% FCS), a broad plasmon band with a maximum around 580 nm appears (solid line in Fig. S5A†), which can be explained by the reduction of HAuCl4 in particular by the serum components. The broad absorbance indicates the presence of gold nanoparticles with a wide size distribution and the formation of nanoaggregates in this incubation medium. Several studies already describe the successful application of amino acids and proteins for the reduction of metal salts.18,19,24 From the absorbance spectra and TEM micrographs, we conclude the formation of very small gold nanostructures already in the DMEM incubation medium and their stabilization by serum proteins. Therefore, the uptake of gold clusters by the cells, rather than of tetrachloroauric acid is very likely, even though they cannot be resolved by TEM. As will be discussed below, to assess these effects further, Raman experiments were conducted for all incubation conditions.
When cells were incubated with gold nanoparticles, a large number of particles are localized in endosomal vesicles in the vicinity of the nuclei (Fig. 4C and D). After an incubation time of 24 hours, nanoparticle aggregates occur inside the vesicular structures, differing in size by several hundred particles. Also, individual particles are associated with membrane structures of the endoplasmic reticulum (Fig. 4E) and have passed the nuclear membrane (Fig. 4F) after they must have undergone endo-lysosomal escape.44 Interestingly, several of these particles are separated from each other, embedded in the nucleoli.
Nano-bio interaction inside the fibroblast cells
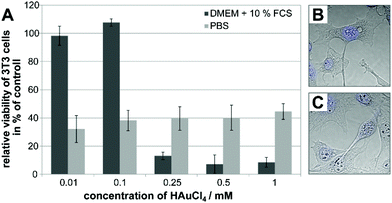
An important step in delineating the processes associated with in situ nanoparticle formation inside the cell is an assessment of cell viability, and a characterization of the newly formed metal nanoparticles’ interaction with the biomolecular surroundings. Therefore, the influence of the HAuCl4 incubation medium on the viability of 3T3 fibroblast cells was studied, and SERS spectra were obtained from the cells.The toxic action of tetrachloroauric acid regarding the metabolic activity of the cells was determined in an XTT assay. Mouse fibroblast cells were incubated with varying concentrations of tetrachloroauric acid (0.01 mM to 1 mM) in PBS and in standard cell culture medium (DMEM with 10% FCS), respectively, for 24 hours. In Fig. 5A, the relative viability in the presence of tetrachloroauric acid is shown as a function of concentration and of the incubation medium. The data discussed in Fig. 1–4 were obtained upon incubation of 1 mM HAuCl4 in the respective medium, representing the highest concentration applied in the experiments. In both PBS and standard cell culture medium, for 1 mM tetrachloroauric acid, the relative viability of fibroblast cells (Fig. 5A) is significantly reduced. In PBS (light gray bars in Fig. 5A), it is approximately 40% in the whole concentration range of 0.01 to 1 mM HAuCl4. The low stability of the nanoparticles formed in DMEM without FCS does not allow incubation experiments with cells for the cytotoxicity investigations.
Micrographs of the fibroblast cells (Fig. 5B and C) as well as the TEM images (Fig. 4A and B) reveal changes of the cell morphology compared to control cells. As observed from the lack of further cell division, the cell cycle is impaired and the cell structure is irreversibly damaged due to the oxidative action of Au3+, similar to observations reported for erythrocytes after incubation of tetrachloroauric acid.45 When DMEM with 10% FCS is used as incubation medium for tetrachloroauric acid (dark gray bars in Fig. 5A), further decrease in the cell viability is observed compared to the exposure in PBS, at the concentration used for the experiments discussed above. Interestingly, different from PBS, the use of this standard cell culture medium leads to a concentration-dependent toxicity of the tetrachloroauric acid (dark gray bars in Fig. 5A). While lower concentrations (≤0.1 mM) have no impact on the viability of the fibroblast cells, for concentrations ≥0.25 mM HAuCl4, the metabolic activity of the cells is significantly reduced (relative cell viability <15%, see Fig. 5A).
The SERS spectra discussed now (Fig. 6) are therefore characteristic of the nano-bio interaction during toxic action of the tetrachloroauric acid in the live fibroblast cells. At the same time, as SERS relies on the plasmonic properties of the nanostructures,46 the possibility to obtain SERS spectra from the cells is a sensitive indicator that such plasmonic nanoparticles must have formed. Concentrations up to 0.01 mM did not lead to the formation of plasmonic nanoparticles that provided sufficient SERS enhancement. In the case of incubation with HAuCl4 in DMEM with FCS, SERS signals were observed neither for non-toxic (up to 0.1 mM) nor for toxic concentrations (from 0.25 mM). The samples for the SERS experiments were prepared parallel to those for the LA-ICP-MS (Fig. 1 and 3) and the TEM experiments (Fig. 4A and B). The TEM images show in situ generated nanoparticles in the cell nucleus, which in principle can serve as SERS substrates for in vivo investigations of the nucleus and its components.
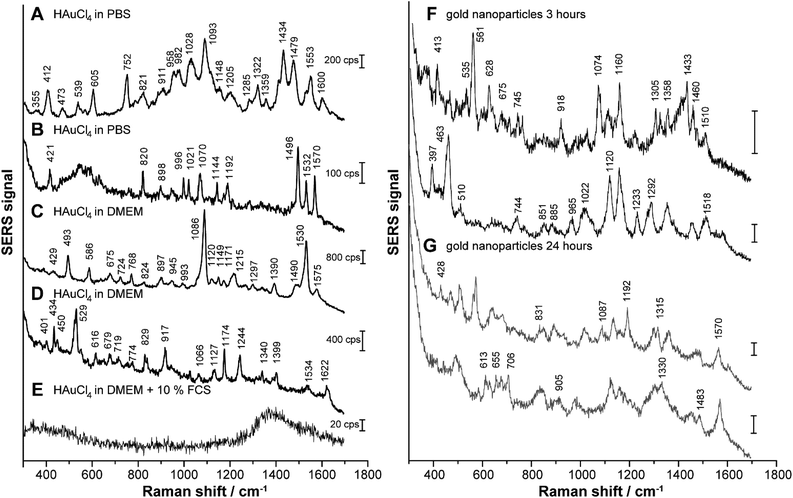
To follow the formation of SERS active nanoparticles inside the cells, 3T3 fibroblast cells were incubated with HAuCl4 in PBS (Fig. 6A and B), in DMEM (Fig. 6C and D) and in DMEM with 10% FCS (Fig. 6E), respectively, for 24 hours. In Fig. 6F and G, representative spectra of fibroblast cells exposed to gold nanoparticles that were synthesized ex situ prior to incubation for 3 hours (Fig. 6F) and 24 hours (Fig. 6G) are displayed for comparison. In accord with previous work,38 they give evidence that incubation time influences the SERS spectral fingerprint. After 24 h-exposure to HAuCl4 in the different cell culture-relevant media, there are significant differences in the spectral fingerprint but also in the number of bands and their average signal intensity, depending on the incubation medium (for examples refer to Fig. 6A–E). The assignments of the Raman bands are summarized in Table S1.† When the cells are incubated with 1 mM HAuCl4 in PBS (Fig. 6A and B) or in DMEM (Fig. 6C and D) many signals are identified in the spectra that indicate the presence of amino acids and proteins: as examples, we identify the symmetrical ring breathing vibration of tryptophan at 760 cm−1, the NH2 rocking and the C–N stretching vibrations at 1070 cm−1, the C–C stretching vibration of tyrosine, as well as the C–N stretching vibration of proteins at 1145 cm−1 and the ring vibrations of phenylalanine and tyrosine at 1590 cm−1 (see also Table S1†).46,47 Furthermore, when PBS is used as incubation medium, the bands around 540 cm−1 and 980 cm−1, which can be assigned to the S–S stretching vibration, to skeletal modes and to the C–C stretching vibration, characteristic of proteins46,48 are frequent (Fig. 6A and B). These vibrational modes indicate the presence of proteins in the immediate vicinity of the biosynthesized gold nanostructures. Most of these bands also occur at lower concentrations of tetrachloroauric acid in PBS buffer (Fig. S6†).
After the incubation of HAuCl4 in DMEM (Fig. 6C and D), the signals at 680 cm−1 (C–S stretching vibration of cysteine and tyrosine side chain), 830 cm−1 (tyrosine), 945 cm−1 (C–C and C–COO− stretching vibrations of amino acids), ∼1000 cm−1 (aromatic ring vibration of phenylalanine), 1170 cm−1 (CH3 rocking vibration of threonine) and 1215 cm−1 (ring breathing of tryptophan) are frequently found (see also Table S1†).47,48 All these bands can be assigned to the vibrational modes of amino acids, suggesting their important role in the reduction of HAuCl4 as components of the DMEM medium. The importance of the amino acids in the formation of the gold nanoparticles during incubation with DMEM is supported by the corresponding absorbance spectrum (dashed line in Fig. S5A†), as well as by the TEM image of HAuCl4 in DMEM (Fig. S5B†), both indicating the presence of nanoparticles already in the incubation medium alone, in the absence of the cells. The signals around 730 cm−1 and 1590 cm−1 (Fig. 6C and D) can also be assigned to adenine and guanine (Table S1†).47 In case of an incubation of HAuCl4 in DMEM, the signal intensities are higher than in PBS buffer (compare scale bars in Fig. 6A–D). This indicates that a larger number of SERS-active particles and/or nanoaggregates with a higher enhancement factor are present.49
In contrast, when tetrachloroauric acid is incubated in standard cell culture medium (DMEM with 10% FCS), no signals occur in the spectra (Fig. 6E), which suggests that no SERS-active nanostructures can have formed. This is in accord with the lack of nanoparticles observed in the TEM of the cells (data not shown) as discussed above.
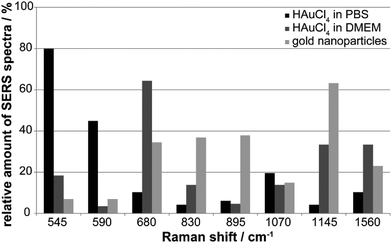
The variation in the nano-bio interaction of the in situ generated nanostructures in PBS and DMEM, and the gold nanoparticles is summarized in Fig. 7. The relative number of SERS spectra with a specific Raman signal can provide an idea of the typical spectral fingerprint that is characteristic of each of the different gold nanostructures (Fig. 7). The SERS signals at 545 cm−1 (skeletal modes and S–S stretching vibration, Table S1†) and 590 cm−1 occur predominately for the incubation of tetrachloroauric acid in PBS (Fig. 7, black bars). While the first band is found in 80% of all spectra, the latter only occurs with a frequency of ∼45% (Fig. 7, black bars). The S–S stretching vibration at 545 cm−1 is also present in ∼20% of the spectra of the HAuCl4 incubation in DMEM, suggesting the formation of disulfide bonds by oxidation of cysteine by tetrachloroauric acid (Fig. 7, dark gray bars and black bars). Since this band is only rarely detected in the case of gold nanoparticles synthesized before incubation (Fig. 7, light gray bars), it seems likely that there, access to the disulfide bonds by the nanoparticles is hindered, possibly by steric conflicts with other stabilizing species on these nanoparticles.
In DMEM (Fig. 7, dark gray bars), in contrast to the HAuCl4 incubation in PBS, the bands at 680 cm−1 (65%) and 1560 cm−1 (35%) occur frequently (Fig. 7, dark gray bars). They are attributed to the amino acid side chains of cysteine and tyrosine, as well as to the amide II band of proteins.47 The particles interact with extracellular and intracellular proteins and lipids in the course of their formation in the culture medium and later, during endocytotic uptake. This is supported by the fact that these bands are also found in every third and fifth spectrum, respectively, of fibroblast cells incubated with gold nanoparticles (Fig. 7, light gray bars). A band at 1145 cm−1 that is characteristic of the C–N stretching vibration of proteins and lipids47 is also observed for both in situ in DMEM formed nanoparticles and typical gold nanoparticles synthesized outside cells (Fig. 7, light gray and dark gray bars at 1145 cm−1). It occurs with a frequency of 65% in the case of the gold nanoparticles and 35% for the incubation of HAuCl4 in DMEM, which further supports the above-mentioned conclusion of an endocytotic uptake of extracellularly formed particles.
For the incubation of gold nanoparticles the bands at 830 cm−1 (tyrosine) and 895 cm−1 (C–C stretching vibration of amino acids)48 can be found in every second spectrum, suggesting the interaction of these amino acid side chains with the particle surface (Fig. 7, light gray bars). The SERS spectra of all gold nanoparticle types display the C–N stretching and NH2 rocking vibrations around 1070 cm−1,47 which occur with a frequency of 12–20%.
Nanoparticle formation in situ
The qualitative differences in the spectral fingerprints during incubation in PBS (Fig. 6A and B) and DMEM (Fig. 6C and D) account for a different mechanism of particle formation and a different composition of the molecular environment. While in the presence of PBS, the nanostructures must form inside the cells (see absorbance spectrum in Fig. S5A† and the TEM micrographs Fig. 4A and B), the DMEM medium causes the formation of particles already outside the cells, as evidenced by both absorbance spectra (Fig. S5A†) and TEM data (Fig. S5B†) taken in the absence of cells. Later on, the nanoparticles formed outside the cells can be taken up into endo-lysosomes, or interact with other cell compartments such as the cell nucleus.In the course of nanoparticle formation by the reduction of tetrachloroauric acid, the culture medium and/or molecules in the cells will be oxidized depending on the incubation medium. In addition to the oxidation of amino acid side chains, e.g., of cysteine, methionine or threonine, also fragmentation of the polypeptide chain or cross-links between proteins can occur.50,51 As an estimate, more than half of the proteinogenic amino acids can be altered due to oxidative stress and react intra- or intermolecularly via reactive intermediates.51 Defective proteins can accumulate in the cell and cause severe cell damage. In particular, high molecular weight compounds from cross-linking of cysteine to disulfide bridges or of tyrosine to di-tyrosine, do partly not undergo proteolytic degradation. The formation of reactive oxygen species as a result of oxidative stress caused by the tetrachloroauric acid can further lead to lipid peroxidation.52,53 In fact, several signals in the SERS spectra can be assigned to vibrational modes of lipids. They include e.g., the C–C and O–P–O stretching vibration at 1089 cm−1 and the C–C stretching vibration at 1120 cm−1 (Fig. 6A and B).54,55
As can be concluded from the qualitative changes in the SERS spectra after incubation of ex situ formed gold nanoparticles (examples shown in Fig. 6F and G) that were obtained for comparison, the spectra differ from those measured after incubation with tetrachloroauric acid. Assignments are also provided in Table S1.† The assignments of the bands to amino acids suggest a stable interaction of the gold nanoparticles with amino acid side chains of the protein corona over the entire time course of endosomal maturation. In addition, we observed Raman bands indicating the presence of nucleotides. The signals around 1087 cm−1 and 1300 cm−1 can be assigned to the symmetric PO2 stretching vibration and to the nucleotides, adenine and cytosine.47,56 This is in accord with the presence of nanoparticles inside the cell nucleus, as revealed by the TEM images above, showing particles associated with structures of the nucleolus (Fig. 4E and F). In addition, the high particle quantities in the endosomal vesicles (Fig. 4C and D), where nucleotides also occur as degradation product, can be the origin of most of these bands.
The analysis of the SERS spectral fingerprints (Fig. 7) shows that the similarity of the biomolecular environment is greater between ex situ synthesized gold nanoparticles and nanoparticles generated in situ with DMEM as incubation medium, than for incubation of tetrachloroauric acid in PBS. SERS spectra obtained by incubation with lower HAuCl4 concentrations in PBS (Fig. S6†) show a greater similarity with those of nanoparticle-incubated cells (Fig. 6F and G).
Conclusions
In the work reported here, to study the in situ formation of gold nanoparticles in cell cultures, 3T3 fibroblast cells were incubated with tetrachloroauric acid in different media, and spatially resolved LA-ICP-MS, Raman microscopy, cytotoxicity, and TEM data were obtained from parallel samples. For comparison, the experiments were also performed with gold nanoparticles synthesized before incubation ex situ outside cells.LA–ICP–MS microscopic mapping indicates different 197Au distribution and uptake pathways for in situ reduction of tetrachloroauric acid, compared to gold nanoparticles synthesized before incubation with cells. In agreement with high gold concentrations found in the regions of the nuclei by LA-ICP-MS mapping, the TEM micrographs confirm the presence of in situ generated gold nanoparticles in the nuclei of the cells. While for the incubation with HAuCl4 in PBS, gold particles of sizes up to 30 nm are found in the entire cellular area, including the nucleus, the incubation of HAuCl4 in DMEM with FCS leads to the reduction and stabilization of the particles already in the incubation medium. The latter prevents both the formation of SERS-active particles inside the cells and the uptake of plasmonic gold nanostructures.
As indicated by the results of an XTT assay, HAuCl4 incubated in PBS buffer is cytotoxic and impairs the metabolic activity of the cells in the concentration ranges that have been reported for incubation experiments with live cells so far,11–13 and also at lower concentration. In contrast, if administered in standard cell culture medium, HAuCl4 does not influence mitochondrial function in concentrations below 0.1 mM. This stands in agreement with the lack of intracellular nanoparticles, implying that molecules in the culture medium must scavenge HAuCl4 before it enters the cells. Nevertheless, our data from LA-ICP-MS indicate small spots of high local gold concentration inside the cytoplasm, which suggests the formation of non-plasmonic gold clusters.
The SERS spectra indicate a different way of how the nanostructures are formed in the different incubation media. For incubation with PBS and DMEM, SERS-active structures form inside the cells. In addition to amino acids and proteins in the surroundings of the in situ biosynthesized nanoparticles, the SERS spectra clearly reveal the presence of nucleotides in the proximity of the nanostructures, in accordance with the observation of nanoparticles interacting with the nucleus by TEM and LA-ICP-MS. The SERS fingerprints indicate similarity of nanoparticles formed upon incubation in DMEM and of standard gold nanoparticles delivered into the cells, and are very different from in situ generated gold nanoparticles in PBS. The possibility of different localization of gold nanostructures, depending on the incubation medium during their in situ formation, together with the application of gold nanoparticles that are formed prior to incubation enables complementary targeting of different cellular compartments for in situ nanoanalytics and for theranostic applications. The results of this work provide a better understanding of the conditions needed in order to optimize such applications.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Dr. G. Laube (Charité Berlin) and S. Selve (ZELMI, TU Berlin) for support in TEM experiments of cells and nanoparticles, respectively. Technical support by A. Cossmer (BAM) is gratefully acknowledged. We thank R. Calheiros Cruz for help during initial experiments. The work was funded by ERC starting grant 259432 MULTIBIOPHOT.References
- S. Efrima and B. V. Bronk, J. Phys. Chem. B, 1998, 102, 5947–5950 CrossRef CAS.
- T. Klaus, R. Joerger, E. Olsson and C. G. Granqvist, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 13611–13614 CrossRef CAS.
- J. Xie, J. Y. Lee, D. I. C. Wang and Y. P. Ting, J. Phys. Chem. C, 2007, 111, 16858–16865 CAS.
- M. A. Prusinkiewicz, F. Farazkhorasani, J. J. Dynes, J. Wang, K. M. Gough and S. G. W. Kaminskyj, Analyst, 2012, 137, 4934–4942 RSC.
- N. C. Sharma, S. V. Sahi, S. Nath, J. G. Parsons, J. L. Gardea-Torresdey and T. Pal, Environ. Sci. Technol., 2007, 41, 5137–5142 CrossRef CAS PubMed.
- J. L. Gardea-Torresdey, J. G. Parsons, E. Gomez, J. Peralta-Videa, H. E. Troiani, P. Santiago and M. J. Yacaman, Nano Lett., 2002, 2, 397–401 CrossRef CAS.
- M. E. Palanco, K. B. Mogensen, M. Gühlke, Z. Heiner, J. Kneipp and K. Kneipp, Beilstein J. Nanotechnol., 2016, 7, 834–840 CrossRef CAS PubMed.
- R. M. Jarvis, N. Law, L. T. Shadi, P. O'Brien, J. R. Lloyd and R. Goodacre, Anal. Chem., 2008, 80, 6741–6746 CrossRef CAS PubMed.
- M. F. Lengke, B. Ravel, M. E. Fleet, G. Wanger, R. A. Gordon and G. Southam, Environ. Sci. Technol., 2006, 40, 6304–6309 CrossRef CAS PubMed.
- F. Reith, B. Etschmann, C. Grosse, H. Moors, M. A. Benotmane, P. Monsieurs, G. Grass, C. Doonan, S. Vogt, B. Lai, G. Martinez-Criado, G. N. George, D. H. Nies, M. Mergeay, A. Pring, G. Southam and J. Brugger, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 17757–17762 CrossRef CAS PubMed.
- A. Anshup, J. S. Venkataraman, C. Subramaniam, R. R. Kumar, S. Priya, T. R. S. Kumar, R. V. Omkumar, A. John and T. Pradeep, Langmuir, 2005, 21, 11562–11567 CrossRef PubMed.
- A. Shamsaie, M. Jonczyk, J. Sturgis, J. P. Robinson and J. Irudayaraj, J. Biomed. Opt., 2007, 12, 020502 CrossRef PubMed.
- H. Huang, W. W. Chen, J. J. Pan, Q. S. Chen, S. Y. Feng, Y. Yu, Y. P. Chen, Y. Su and R. Chen, Spectroscopy, 2011, 26, 187–194 CrossRef CAS.
- Z. M. Liu, C. F. Hu, S. X. Li, W. Zhang and Z. Y. Guo, Anal. Chem., 2012, 84, 10338–10344 CrossRef CAS PubMed.
- E. Larios-Rodriguez, C. Rangel-Ayon, S. J. Castillo, G. Zavala and R. Herrera-Urbina, Nanotechnology, 2011, 22, 355601 CrossRef CAS PubMed.
- J. L. Wang, G. Zhang, Q. W. Li, H. Jiang, C. Y. Liu, C. Amatore and X. M. Wang, Sci. Rep., 2013, 3, 1–6 Search PubMed.
- N. T. K. Thanh and L. A. W. Green, Nano Today, 2010, 5, 213–230 CrossRef CAS.
- W. L. Fu, S. J. Zhen and C. Z. Huang, Analyst, 2013, 138, 3075–3081 RSC.
- J. P. Xie, Y. G. Zheng and J. Y. Ying, J. Am. Chem. Soc., 2009, 131, 888–889 CrossRef CAS PubMed.
- S. Chattoraj and K. Bhattacharyya, J. Phys. Chem. C, 2014, 118, 22339–22346 CAS.
- S. Chattoraj, M. A. Amin, S. Mohapatra, S. Ghosh and K. Bhattacharyya, ChemPhysChem, 2016, 17, 61–68 CrossRef CAS PubMed.
- P. He and X. Y. Zhu, Mater. Res. Bull., 2007, 42, 1310–1315 CrossRef CAS.
- Z. D. Mu, X. W. Zhao, Z. Y. Xie, Y. J. Zhao, Q. F. Zhong, L. Bo and Z. Z. Gu, J. Mater. Chem. B, 2013, 1, 1607–1613 RSC.
- S. P. Shukla, M. Roy, P. Mukherjee, A. K. Tyagi, T. Mukherjee and S. Adhikari, J. Nanopart. Res., 2012, 14, 981 CrossRef.
- V. Joseph, F. Schulte, H. Rooch, I. Feldmann, I. Dörfel, W. Österle, U. Panne and J. Kneipp, Chem. Commun., 2011, 47, 3236–3238 RSC.
- X. Wu, X. X. He, K. M. Wang, C. Xie, B. Zhou and Z. H. Qing, Nanoscale, 2010, 2, 2244–2249 RSC.
- Y. Su, L. Qi, X. Y. Mu and M. L. Wang, Anal. Methods, 2015, 7, 684–689 RSC.
- H. B. Zhou, Q. Q. Wang, D. T. Yuan, J. Y. Wang, Y. Huang, H. H. Wu, J. Y. Jian, D. T. Yang, N. Huang, C. Haisch, Z. J. Jiang and S. Z. Chen, Analyst, 2016, 141, 4293–4298 RSC.
- A. M. Alkilany and C. J. Murphy, J. Nanopart. Res., 2010, 12, 2313–2333 CrossRef CAS PubMed.
- N. Khlebtsov and L. Dykman, Chem. Soc. Rev., 2011, 40, 1647–1671 RSC.
- J. Kneipp, H. Kneipp, M. McLaughlin, D. Brown and K. Kneipp, Nano Lett., 2006, 6, 2225–2231 CrossRef CAS PubMed.
- D. Drescher, P. Guttmann, T. Büchner, S. Werner, G. Laube, A. Hornemann, B. Tarek, G. Schneider and J. Kneipp, Nanoscale, 2013, 5, 9193–9198 RSC.
- D. Drescher, I. Zeise, H. Traub, P. Guttmann, S. Seifert, T. Büchner, N. Jakubowski, G. Schneider and J. Kneipp, Adv. Funct. Mater., 2014, 3765–3775 CrossRef CAS.
- J. S. Becker, U. Kumtabtim, B. Wu, P. Steinacker, M. Otto and A. Matusch, Metallomics, 2012, 4, 284–288 RSC.
- S. J. M. Van Malderen, E. Vergucht, M. De Rijcke, C. Janssen, L. Vincze and F. Vanhaecke, Anal. Chem., 2016, 88, 5783–5789 CrossRef CAS PubMed.
- D. Drescher, C. Giesen, H. Traub, U. Panne, J. Kneipp and N. Jakubowski, Anal. Chem., 2012, 84, 9684–9688 CrossRef CAS PubMed.
- D. Drescher, G. Orts-Gil, G. Laube, K. Natte, R. W. Veh, W. Österle and J. Kneipp, Anal. Bioanal. Chem., 2011, 400, 1367–1373 CrossRef CAS PubMed.
- T. Büchner, D. Drescher, H. Traub, P. Schrade, S. Bachmann, N. Jakubowski and J. Kneipp, Anal. Bioanal. Chem., 2014, 406, 7003–7014 CrossRef PubMed.
- H. H. Wang, C. A. J. Lin, C. H. Lee, Y. C. Lin, Y. M. Tseng, C. L. Hsieh, C. H. Chen, C. H. Tsai, C. T. Hsieh, J. L. Shen, W. H. Chan, W. H. Chang and H. I. Yeh, ACS Nano, 2011, 5, 4337–4344 CrossRef CAS PubMed.
- S. Chattoraj, A. Amin, B. Jana, S. Mohapatra, S. Ghosh and K. Bhattacharyya, ChemPhysChem, 2016, 17, 253–259 CrossRef CAS PubMed.
- W. Stoorvogel, G. J. Strous, H. J. Geuze, V. Oorschot and A. L. Schwartz, Cell, 1991, 65, 417–427 CrossRef CAS PubMed.
- P. Raveendran, J. Fu and S. L. Wallen, Green Chem., 2006, 8, 34–38 RSC.
- N. Wangoo, S. Kaur, M. Bajaj, D. V. S. Jain and R. K. Sharma, Nanotechnology, 2014, 25, 435608 CrossRef PubMed.
- I. Canton and G. Battaglia, Chem. Soc. Rev., 2012, 41, 2718–2739 RSC.
- M. Suwalsky, R. Gonzalez, F. Villena, L. F. Aguilar, C. P. Sotornayor, S. Bolognin and P. Zatta, Coord. Chem. Rev., 2009, 253, 1599–1606 CrossRef CAS.
- R. F. Aroca, R. A. Alvarez-Puebla, N. Pieczonka, S. Sanchez-Cortez and J. V. Garcia-Ramos, Adv. Colloid Interface Sci., 2005, 116, 45–61 CrossRef CAS PubMed.
- F. S. Parker, Applications of Infrared, Raman, and Resonance Raman Spectroscopy in Biochemistry, Plenum Press, New York and London, 1983 Search PubMed.
- S. Stewart and P. M. Fredericks, Spectrochim. Acta, Part A, 1999, 55, 1641–1660 CrossRef.
- J. Kneipp, H. Kneipp and K. Kneipp, Chem. Soc. Rev., 2008, 37, 1052–1060 RSC.
- V. I. Lushchak, Biochemistry, 2007, 72, 809–827 CAS.
- E. R. Stadtman and R. L. Levine, Ann. N. Y. Acad. Sci., 2000, 899, 191–208 CrossRef CAS PubMed.
- A. E. Nel, L. Madler, D. Velegol, T. Xia, E. M. V. Hoek, P. Somasundaran, F. Klaessig, V. Castranova and M. Thompson, Nat. Mater., 2009, 8, 543–557 CrossRef CAS PubMed.
- Y. Omata, J. B. Lewis, P. E. Lockwood, W. Y. Tseng, R. L. Messer, S. Bouillaguet and J. C. Wataha, Toxicol. in Vitro, 2006, 20, 625–633 CrossRef CAS PubMed.
- D. F. H. Wallach, Chem. Phys. Lipids, 1972, 8, 347–354 CrossRef CAS PubMed.
- J. L. Lippert, L. E. Gorczyca and G. Meiklejohn, Biochim. Biophys. Acta, 1975, 382, 51–57 CrossRef CAS.
- C. Otto, T. J. J. Vandentweel, F. F. M. Demul and J. Greve, J. Raman Spectrosc., 1986, 17, 289–298 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: LA-ICP-MS maps and TEM image of fibroblasts; quantification of the number of gold particles in cells; UV-vis absorbance spectra of HAuCl4 in cell media; influence of HAuCl4 concentration and incubation time on the SERS information; band assignments of the cell spectra; and laser parameters in the LA-ICP-MS experiments. See DOI: 10.1039/c7nr04620k |
| This journal is © The Royal Society of Chemistry 2017 |