Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A DNA bipedal nanowalker with a piston-like expulsion stroke†
Q. Y.
Yeo‡
a,
I. Y.
Loh‡
a,
S. R.
Tee
a,
Y. H.
Chiang
a,
J.
Cheng
a,
M. H.
Liu
a and
Z. S.
Wang
 *ab
*ab
aDepartment of Physics, National University of Singapore, 2 Science Drive 3, Singapore 117542. E-mail: phywangz@nus.edu.sg
bNUS Graduate School of Integrative Sciences and Engineering, National University of Singapore, Singapore
First published on 2nd August 2017
Abstract
Artificial molecular walkers beyond burn-bridge designs are important for nanotechnology, but their systematic development remains difficult. Herein, we have reported a new rationally designed DNA walker–track system and experimentally verified a previously proposed general expulsion regime for implementing non-burn-bridge nanowalkers. The DNA walker has an optically powered engine motif that reversibly extends and contracts the walker via a quadruplex–duplex conformational change. The walker's extension is an energy-absorbing and force-generating process, which drives the walker's leg dissociation off-track in a piston-like expulsion stroke. The unzipping-shearing asymmetry provides the expulsion stroke a bias, which decides the direction of the walker. Moreover, three candidate walkers of different sizes were fabricated. Fluorescence motility experiments indicated two of them as successful walkers and revealed a distinctive size dependence that was expected for these expulsive walkers, but was not observed in previously reported walkers. This study identifies unique technical requirements for expulsive nanowalkers. The present DNA design is readily adapted for making similar walkers from other molecules since the unzipping-shearing asymmetry is common.
Introduction
Track-walking molecular motors are a major form of intracellular machines1 in the field of biology, and man-made counterparts2–12 are key elements in future nanotechnology. The artificial molecular walkers demonstrate various applications such as in automated sequence-dependent synthesis,13,14 nanoscale assembly lines,15 and walker-guided surface patterning.16 However, the majority of reported artificial nanowalkers2–4,6,8,12 gain a direction by damaging or blocking the traversed track (hence rendering it non-reusable). A few walkers5,7,9–11 beyond the burn-bridge approach have been reported, but their systematic development remains difficult to date.In a recent study,10 Loh et al. demonstrated a generally applicable design principle for implementing non-burn-bridge nanowalkers. In the design principle, a symmetric bipedal nanowalker gains a direction on a periodic track by alternately switching between a short length and a long length. Moreover, two regimes for implementing the design principle have been proposed10 according to the force-generating process and resultant direction. In the so-called winding regime, a walker's contraction from long to short length produces the force to drive the walker's leg dissociation off-track. The walker's direction is decided by a dissociation bias between two legs. In the expulsion regime, a walker's extension from short to long length produces the force for leg dissociation. The walker has an opposite direction as the bias for extension-induced dissociation is often reversed from that for contraction-induced dissociation. Hence, the two regimes differ in direction as well as in experimental implementation, especially in the technical requirements for the engine motifs responsible for force generation.
The winding regime was implemented and verified using a rationally designed DNA walker–track system.10,17 In this study, we demonstrated the expulsion regime with an entirely new DNA walker–track system designed to meet the special requirements of this regime. Moreover, the leg-track binding is reduced to a single DNA duplex, and the dissociation bias is derived from a shearing-unzipping asymmetry.18–20 Since this asymmetry is common for polymers, the design of this DNA walker is readily adapted for making similar walkers from peptides21 and synthetic polymers.22
Results and discussion
Walker design and fabrication
As shown in Fig. 1, the DNA walker has two identical single-stranded legs protruding from two ends of a duplex bridge. The track has multiple repeats of single-stranded binding sites separated by duplex segments. A leg binds a site by forming a single duplex. The walker's inter-leg bridge can extend and recover via an engine motif capable of a duplex–quadruplex conformational change. The walker adopts a short length when a 21 nt-long guanine-rich sequence connecting one leg to the bridge forms a compact G-quadruplex structure (see the segment labelled as G4 in Fig. 1A or in state i in Fig. 1D). The walker adopts a long length when this G4 segment forms a duplex with a nearby cytosine-rich overhang (state ii). The walker is switched between two lengths via the light operations of an azo-embedded, guanine-rich fuel strand: it hybridizes with the overhang under visible light to free the G4 segment for the walker's contraction to the short length; however, the fuel-overhang duplex breaks under UV light for the walker's extension to the long length via the overhang-G4 hybridization. The sequences for the G4 segment, cytosine-rich overhang, and guanine-rich fuel have been obtained from ref. 23, in which the quadruplex unfolding by the overhang sequence and re-folding by the fuel sequence have been demonstrated. Similar optical controls of azo-embedded DNA duplexes have been previously demonstrated in nanowalkers.7–10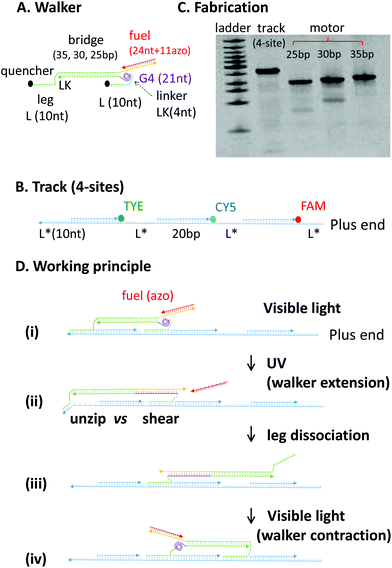 | ||
| Fig. 1 (A and B) The DNA walker and track. Lengths for the major components of the walker and track are indicated (bp stands for base pair, nt for nucleotides, and asterisk indicates complementary sequences). The walker shown is under visible light, with the azo-embedded fuel (red) forming a 24 bp duplex with the cytosine-rich overhang (yellow, 27 nt long). Under UV light, this duplex is broken and a new 21 bp duplex forms between the G4 segment and the overhang (with 6 nt exposed at the 3′ end for fuel binding again under visible light, see state ii in panel D). The nucleotide sequences of the DNA strands are given in the ESI (Fig. S1†). (C) Native PAGE (polyacrylamide gel electrophoresis) analysis of the annealed track and three walker candidates with different lengths for the inter-leg bridge. (D) Working principle of the walker based on the unzipping vs. shearing asymmetry. | ||
To achieve expulsion-based dissociation, two requirements must be satisfied. First, the walker at its short length should form a stable two-leg bound state with the track. Second, the walker at its long length should sufficiently destabilize the two-leg state for leg dissociation. The first requirement is fulfilled if the walker's permanent bridge without quadruplex unfolding matches the track's site separation, which is the inter-site duplex spacer plus the leg-site duplex (see state i in Fig. 1D). The second requirement is fulfilled if the walker's bridge extension (i.e., overhang-G4 duplex) is long enough for leg dissociation. The bridge extension destabilizes walker's one leg in an unzipping mode and the other in a shearing mode (see state ii, Fig. 1D). As the threshold force to open a DNA duplex by shearing is ∼two times18 that of unzipping, the unzipped leg will be preferentially dissociated. The number of unzipped bps n can be estimated with the formula: 2n + 2nLK + 5 bp = 21 bp. Herein, nLK is the nucleotide number of the linker between the bridge and either leg (one nucleotide is approximately the equivalent of one bp); 5 bp accounts for the ∼2 nm width of the unzipped leg-site duplex; 21 bp is the length of the overhang-G4 duplex.
We chose a length of 10 nt for legs and sites and 4 nt for linkers. According to the abovementioned formula, the overhang-G4 duplex can unzip the 10 bp-long leg-site duplex by ∼4 bps, leaving the ∼6 bp remainder readily broken via thermal fluctuations. Moreover, the inter-site spacer is chosen to be 20 bp long, yielding an ideal bridge length of 30 bp from the first requirement. Considering the uncertainty in the estimation of DNA lengths, we tested three versions of the walker with a bridge of 25 bp, 30 bp and 35 bp.
The different versions of the walker and the tracks were assembled from their constituent DNA strands via single-pot annealing and characterised using native polyacrylamide gel electrophoresis. Formations of the walker and tracks were confirmed by a single major band for each of the assembled targets (Fig. 1C). The reversible G-quadruplex formation is confirmed by a fluorescence resonance energy transfer (FRET) experiment using a modified version of the walker's G4-containing strand with donor and acceptor dyes chemically labelled at two ends of the G4 sequence within the strand (Fig. S2†). The FRET efficiency swiftly decreases when the modified strand is mixed with an opener strand that is part of the cytosine-rich overhang. This suggests that the quadruplex structure can be opened by the overhang. In addition, the FRET efficiency decreases upon heating the modified strand (without opener) from 25 °C to 75 °C and completely recovers via reverse cooling back to 25 °C. These results suggest that the G4 sequence can reversibly form the quadruple structure when it is not interfered by the cytosine-rich overhang.
The walker works as follows. Under visible light, the walker takes the short length in the presence of fuel, and the two legs are bound to two adjacent sites (see Fig. 1D, state i). In this state, the G4 segment forms a compact quadruplex structure because the cytosine-rich overhang is hybridized with the fuel. When the UV light dissociates the fuel, the overhang opens the quadruplex structure and hybridizes with the G4 segment into a longer duplex. When the walker extends under UV, the unzipped leg is dissociated preferentially over the sheared leg (states ii and iii). The unzipped leg, therefore, fulfils the role of a rear leg, with the other leg being designated as the front leg and pointing to the track's plus end. When the walker contracts to its original length under visible light, the dissociated leg binds the track either forward or backward (state iv). Upon alternating the UV and visible light, the walker drives to the track's plus end on an average in a hand-over-hand manner.
Directional on-track translocation
The walker's motility is detected by measuring the fluorescence of quencher-labelled walkers operated on dye-labelled tracks. The positions of dyes and quenchers are chosen to form highly efficient contact quenching once a binding site is occupied by a leg. Based on literature,10,11 the walker's motion is characterized by a control-calibrated signal, which is the fluorescence obtained from the operation experiments of an operated walker–track mix divided by that of a track-only control experiment under the same light conditions. This control-calibrated fluorescence is a reliable indicator of site occupancy as this quantity removes the influence of the properties of the dyes (e.g., photobleaching).Fig. 2A shows the typical calibrated signal for the walker on a truncated three-site track with dyes labelled at all three sites. The signal decreases for the plus-end dye but increases for the minus-end dye, and the signal changes for the mid-site dye, sandwiched between those for the plus-end and minus-end dyes (Fig. 2B). This fluorescence pattern signifies a hand-over-hand on-track translocation of the walker from the minus end to the plus end, as found in previous studies.10,11
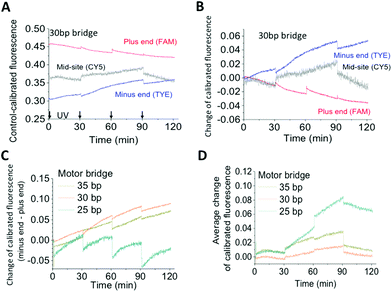 | ||
Fig. 2 Walker operation on a three-site track. The track is the same as the four-site track in Fig. 1B, but with the dye-free site deleted. (A) The control-calibrated fluorescence shown is the ratio of fluorescence obtained from operation experiments over that obtained from a track-only control experiment. For operation experiments, alternating UV and visible light (each 30 minutes long) is applied to a pre-incubated equimolar walker–track mix (10 nM walker/track, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 fuel–walker ratio). The control experiment is conducted with the same amount of the track sample under the same light conditions. The time axis covers only the visible light time during which the fluorescence is obtained (arrows indicating times at which UV light is applied). (B) Change of the calibrated fluorescence relative to the pre-operation values. (C) Direction signal, i.e., the gap between fluorescence changes at the minus and plus ends shown in (B). (D) Dissociation signal, i.e., the average of fluorescence changes of the three dyes in (B). 1 fuel–walker ratio). The control experiment is conducted with the same amount of the track sample under the same light conditions. The time axis covers only the visible light time during which the fluorescence is obtained (arrows indicating times at which UV light is applied). (B) Change of the calibrated fluorescence relative to the pre-operation values. (C) Direction signal, i.e., the gap between fluorescence changes at the minus and plus ends shown in (B). (D) Dissociation signal, i.e., the average of fluorescence changes of the three dyes in (B). | ||
The data shown in Fig. 2 are obtained under alternating UV and visible light that both last for 30 minutes. Shorter light durations were employed, but the yield reduced the signals for directional motion. Moreover, a fuel–walker ratio of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was used at which saturated fuel binding to the walker was already observed.
1 was used at which saturated fuel binding to the walker was already observed.
Direction-dissociation anti-correlation
The walker's direction, as quantified by the gap of fluorescence changes between the minus and plus ends, increases when the walker's bridge varies from 25 bp to 35 bp and 30 bp (Fig. 2C). However, an opposite trend is observed for the walker's overall dissociation off the track, which is quantified by the average fluorescence change for the three dyes (Fig. 2D). Thus, the walker shows a size-dependent performance with a direction-dissociation anticorrelation, reflecting a mechanical mechanism for the walker's direction rectification and coordinated leg motion.A similar anticorrelation was observed for a previous DNA bipedal walker,9,24 but with the decreasing direction and increasing overall dissociation for the elongated bridge size. The reversed size dependence between the present walker and the previous walker reflects their different mechanisms of symmetry breaking. The biased leg dissociation occurs for the present walker at a length beyond two adjacent binding sites (expulsion mode), but occurs for the previous walker at a length within the adjacent sites (winding mode). The expulsion-based dissociation is more effective for a longer walker, rationalizing a better direction for the walkers with a 35 bp and 30 bp bridge than for the walker with a 25 bp bridge. The winding-based dissociation is more effective for a shorter walker; hence, the reversed size dependence of walker performance is observed.
Therefore, the distinct size-dependent performance of the present walkers is an unambiguous indicator that the expulsive regime is experimentally realized as designed.
The observed direction-dissociation anti-correlation is one more piece of evidence for on-track walking. For the walkers with the 30 bp or 35 bp bridge, the direction signal successively increases with more rounds of light operation. However, the overall dissociation at the end of the operation largely recovers the initial values; this suggests a low probability for either walker's entire derailment off-track. The two walkers are successful directional walkers. However, the walker with the 25 bp bridge has a poor direction and unrecoverable dissociation.
The two longer walkers were confirmed as successful walkers via their operation on a four-site track. The plus-end fluorescence decrease and successive increase of the direction signal are observed for both walkers (Fig. 3). The results suggest that the walkers can operate on longer tracks for real applications, especially when the tracks are embedded in a larger, rigid platform such as a DNA origami scaffold.25,26
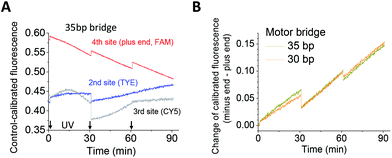 | ||
| Fig. 3 Walker operation on a four-site track. The track design and dye labelling are shown in Fig. 1B, and the experimental procedure is the same as stated in the case of Fig. 2. | ||
Leg binding bias
For the walker's operation on the three-site track, the fluorescence changes at the plus and minus ends during visible light are mainly caused by leg binding from the UV-induced single-leg population located at the middle site. For the walker with the 30 bp bridge, the control-calibrated fluorescence decreases at the plus end, but increases at the minus end during visible light irradiations (Fig. 2B); this suggests a preference for the dissociated leg to bind the front site over the back site. This binding bias is expected as the forward binding places the mobile leg's end ∼20 bp away from the track-bound leg (counting from the bridge-leg junction), whereas the backward binding places it ∼40 bp away (see Fig. 1D, state iv).A similar pattern of fluorescence decrease at the plus-end and increase at the minus end is observed for the walker with the 35 bp bridge (Fig. S2,† panel A); this suggests a forward binding bias for this walker as well. The bias is lost for the walker with the 25 bp bridge as the plus-end fluorescence remains flat or slightly increases during visible light irradiation (Fig. S3,† panel B).
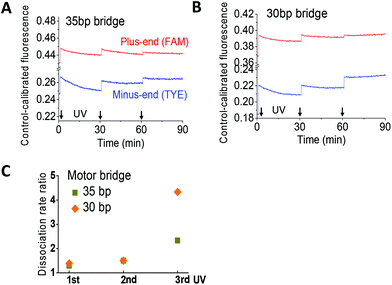
Dissociation bias
Whether or not the walkers attain the designed dissociation bias is tested with a truncated two-site track since its dye-labelled plus and minus ends unambiguously report the leg dissociation events of the same walker in a two-leg bound state. An equimolar walker–track mix plus redundant fuels is incubated to prepare the two-leg bound state. Application of UV extends the walker from its fuel-induced short length, triggering leg dissociation from either the plus end or the minus end. The UV-induced change of fluorescence from the two sites, again calibrated to a track-only control, yields information on the dissociation bias.For the walkers with the 30 bp and 35 bp bridge, the control-calibrated fluorescence is increased by UV light and decreased by visible light (Fig. 4A and B). This is consistent with the UV-induced walker extension and ensuing leg dissociation and with the visible light-induced walker contraction and ensuing leg binding. Based on previous studies,10,11 the UV-induced fluorescence increase yields the rate ratio for leg dissociation from the minus end over the plus end (Fig. 4C). The rate ratio is consistently above one for both walkers, indicating a preference for the rear leg dissociation over the front leg. The dissociation bias for the walker with the 30 bp bridge is higher than that with the 35 bp bridge, consistent with their direction signals (Fig. 2C).
The track-only control shows a slight UV-induced jump of emission from the dye labelled to the plus end (Fig. S4†). However, the fluorescence data obtained for the walker–track mix exhibit a bigger jump for the walkers with 30 bp and 35 bp bridges such that the UV-induced increase in the control-calibrated fluorescence for both walkers is a real signal for leg dissociation.
Operating the walker with the 25 bp bridge on the same two-site track yields a calibrated fluorescence that shows a UV-induced decrease for both dyes at the track's ends (Fig. S5†). These results suggest that the UV-induced extension of this short walker causes leg binding instead of dissociation. One possible interpretation is that this walker at its fuel-induced short length adopts the single-leg bound state more than the two-leg bound state.
Simulation study
To further understand the dissociation bias, we carried out a realistic simulation of the DNA-walker systems using the oxDNA package.27 Specifically, we obtained the free-energy profiles for dissociation of the front and rear leg during the walker's elongation at the experimental temperature (25 °C). For the walkers with the 35 bp or 30 bp bridge, the profiles have a higher dissociation barrier for the front leg than that for the rear leg; this suggests a bias favouring the rear leg dissociation (Fig. S6 and S7, ESI†). For the walker with the 25 bp bridge, the free-energy profiles have virtually equal barriers for the rear and front legs; this suggests no dissociation bias (Fig. S8†). Thus, the simulation predicts a dissociation bias for the two longer walkers, but not for the 25 bp bridge walker, which is in line with the experimental data.Size sensitivity
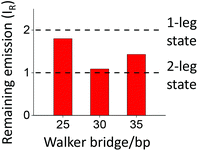
Among the three walkers tested herein, the walker with the intermediate bridge size (30 bp) shows the optimal performance, i.e. the highest direction and biases, but the lowest off-track derailment. Elongation of the bridge by 5 bp reduces the performance, whereas its shortening by 5 bp loses the direction/biases and amplifies derailment. Thus, a size change of less than 2 nm is able to drastically change the walker's performance. This size sensitivity is remarkable, which is sharper than previously observed for a DNA bipedal walker9 operating in the winding regime.Similar size sensitivity exists in the three walkers’ binding on the three-site track, as deduced from the observed quenching of this fully labelled track (Fig. 5). Total percentage of the remaining emission from the track's three dyes, IR, is given by the sum of control-calibrated fluorescence over the three dyes. For an equimolar walker–track mix, IR = 1 if all the walkers (each labelled with two quenchers) form a two-leg bound state with the track, and IR = 2 if all the walkers form a single-leg bound state. The IR value obtained from the incubated equimolar mixes for fuel-bound walkers (hence at their short lengths) is ∼1.80, 1.09, and 1.43 for the walkers with the 25 bp, 30 bp, and 35 bp bridge, respectively. The results suggest that the 30 bp-bridge walker forms the two-leg state, but the 25 bp-bridge walker mostly forms the single-leg state, with a mix of both states for the 35 bp-bridge walker.
As a consequence, the first requirement for expulsion-based dissociation is best fulfilled by the 30 bp-bridge walker, but violated by the 25 bp-bridge walker. These findings explain the sharp size sensitivity of the walkers’ performance. The binding data are consistent with the previous size consideration that predicts the ideal walker-sites match for the 30 bp bridge. The binding data for the 25 bp-bridge walker suggest that this short walker is likely near the winding regime. This explains the walker's lost biases and slightly reversed direction (Fig. 2C).
General guidelines for implementing expulsive nanowalkers
The study of three DNA walkers yields generally applicable guidelines for implementing nanowalkers within the expulsion regime. First, it is necessary to have an asymmetric leg-site binding in the sense that pulling the leg backward along the track requires a lower force than pulling it forward. This asymmetric leg-site binding is the equivalent of a binding potential that has two edges with different slopes (a steeper edge corresponds to a higher force against the leg's pulling). This type of binding potential can be realized using the unzipping-shearing asymmetry, as demonstrated by the DNA present walkers and schematically illustrated in Fig. 6A.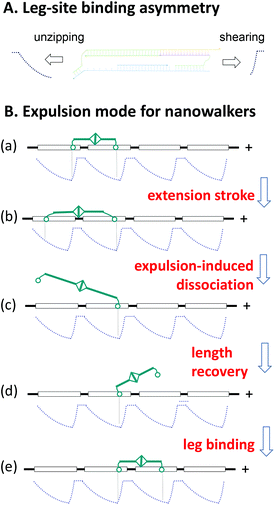 | ||
| Fig. 6 A generic scheme for designing expulsive nanowalkers. Panel A shows how the unzipping-shearing asymmetry of the present DNA walker results in an asymmetric leg-site binding potential with two edges of differing slopes. The flat part of either edge corresponds to a fully dissociated leg. Panel B illustrates how a symmetric bipedal nanowalker (cyan) gains a direction on a track with periodic binding sites (horizontal square, each supporting an asymmetric potential) by expulsion-based dissociation of the walker's legs (circles). The flow of walker–track states shows the general working principle of expulsive walkers (Fig. 1D is a special case for the present DNA walker, with the states i, ii, iii, and iv corresponding to the states a, b, c, and e, respectively). | ||
Second, expulsive nanowalkers can be designed following the generic scheme shown in Fig. 6B. For generality, the walker may be a symmetric biped and the track may host identical asymmetric binding sites in a periodic array. The walker, initially at a short length, matches the separation between two adjacent sites to form a stable two-leg bound state (i.e., the two legs settle down to the bottom of the binding potentials, see state a in Fig. 6B). The walker's extension to a long length generates an intra-walker expelling force, which lifts the two legs from the potential bottom along different edges, therefore dissociating the leg along the less steep edge preferentially over the other leg (states b and c). The dissociated leg diffuses back and forth, and upon recovery of the walker's short length, it will bind to a site before or behind the track-bound leg (states d and e). The forward binding completes a directional step for the walker, whereas the backward binding results in a futile step (but not a backward step). On average, the walker with alternatingly long and short lengths will move hand-over-hand towards a direction dictated by the bias of expulsion-based dissociation, even for equal chance of forward or backward binding.
However, expulsive walkers can have a biased forward binding as well. This binding bias occurs when the walker's short length restricts the dissociated leg to such an extent that it accesses the less steep rear edge of the binding potential at the front site, but not the steep front edge at the back site (see the states d and e). This is the case for the present DNA walker. Hence, the expulsive walkers can integrate two biases: one for dissociation and one for binding, which are necessary for making high-fidelity28–30 nanowalkers.
As a characteristic of the expulsion regime, the expulsion-based dissociation is the predominant force-generating process in the walker's entire working cycle. This is because leg dissociation from the potential bottom is the most force-demanding transition. The walker's length recovery in the resultant single-leg state occurs under a vanishing intra-walker force, and the ensuing leg binding is a downhill transition in terms of energy change. The expulsion-based dissociation is also an energy-absorbing process, which powers the walker's uphill transition from a low-energy two-leg state to a high-energy single-leg state. Thus, the walker's expulsion plays the double roles of force generation and energy intake, essentially resembling the steam-driven stroke of a piston in heat engines.
Hence, an important technical requirement for implementing an expulsion nanowalker is to design a molecular engine motif that absorbs the supplied energy to generate a high force upon the energy-driven extension. Comparatively, nanowalkers operated in the winding regime, as demonstrated in ref. 10, have the predominant force-generating process in the contraction-induced leg dissociation (e.g., via shortening of the walker in state a, Fig. 6). As a consequence, expulsive walkers and winding walkers have different technical requirements for implementation, especially for the engine motifs. Moreover, the two types of walkers need to be optimized in different ways because they follow opposite trends in their size-performance relations as the present DNA walkers show.
Conclusions
We have demonstrated a general expulsion regime for implementing nanowalkers beyond burn-bridge approaches. The present demonstration is carried out with DNA, but similar expulsive nanowalkers can be made with other building blocks following the common principle outlined in Fig. 6. The strategy of realizing asymmetric leg-site binding from the shearing-unzipping asymmetry likely works for other molecular systems too as this asymmetry is common (e.g., in the β sheets of peptides,19 in parallel binding between nucleotides, and man-made analogues20). Indeed, the design of the present DNA walker–track can be recast into a new walker with a different DNA engine or a hybrid walker with a peptide engine covalently linked to the DNA legs, or into a new hybrid track with the same DNA sites but hosted on a backbone of peptides or synthetic polymers. Implementation of a similar walker–track design with only peptides or synthetic polymers is also possible. In addition, the present walker requires a DNA track that has no extruding single-stranded sticky ends at each binding site. For comparison, the previously reported walkers mostly require binding sites with one or two sticky ends. A track free of sticky ends requires a smaller space for the walker's operation, and it is less difficult to integrate this track into large DNA origami scaffolds. Future applications of the present walker can be envisioned in light-controlled transport, molecular synthesis, or nanoscale assembly lines based on compact DNA origami platforms.Experimental
Walker–track design and fabrication
The nucleotide sequences for the DNA strands were first selected using the NuPack server (http://www.nupack.org). Secondary structures were manually checked and eliminated by sequence adjustment. The chosen sequences were also tested using the oxDNA package27 to see if the correct walker–track binding could be formed or not. The DNA strands were purchased from Nihon Techno Service Co., Ltd (for azobenzene-tethered fuel strand, high-performance liquid chromatography purification) and from Integrated DNA Technologies Pt. Ltd (for all other strands). For any purchased DNA strand, its purity was checked using gel electrophoresis to see if it was a single band at the right molecular weight. To assemble the walkers and tracks, the strands were stoichiometrically mixed in a TE buffer (10 mM tris, 1 mM EDTA, 150 mM NaCl, pH 8.0) and heated at 98 °C for 30 minutes and then slowly cooled down to 25 °C over 24 hours. The annealing products were analysed in a 10% native PAGE (polyacrylamide gel electrophoresis) against a low molecular weight DNA Ladder (purchased from New England BioLabs, Inc.). The buffer and annealing procedure largely follow those optimized in previous studies7,10,11 for the self-assembly of DNA walkers and tracks. The salt concentration for this study was chosen to balance two requirements. On the one hand, the salt concentration cannot be too low as it is necessary for the charge screening effect, stabilizing DNA duplexes, and providing cations, stabilizing the G-quadruplex structure.31 On the other hand, a salt concentration that is too high causes DNA aggregation that is often signified by almost complete quenching of fluorescent dyes. The salt concentration was selected in this study through trial and error by checking the dye quenching for the upper limit of salt concentration and checking the walker's integrity and mobility for the lower limit.Fluorescence motility experiments
For each operation experiment, the annealed walker was incubated with the fuel at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 ratio over 24 hours to ensure saturation of the fuel binding to the walker. The sample was then incubated with the annealed track for another 10 hours before the operation experiment was started. The incubations and later operations (and controls) were conducted at 25 °C in a TE buffer. The operation buffer had a pH value of 8.0, which was known from previous studies7,10,11 to ensure stability of the DNA duplexes, vital for the walker/track integrity, and sufficient fluorescent signals (low dye emission32 at low pH). As suggested by the FRET experiment (Fig. S2†), the G-quadruplex structure also formed although it was most favoured at a lower pH value.31 The pH value in this study has been chosen to favour the duplex formation of the G4 sequence with the cytosine-rich overhang over the quadruplex structure because the duplex formation drives the force-generating expulsion stroke, and the quadruplex formation is not against a force. The walker/track concentration was kept low (∼10 nM) throughout this study to suppress track crosslinking by a walker. The irradiation operation and fluorescence measurement were conducted using an RF-5301PC spectrophotometer (150 W Xenon lamp, Shimadzu Corp.). For each round of irradiation operation, the walker–track sample was first irradiated by UV (300–400 nm wavelength; 3 nm excitation slit width) followed by visible light (kinetic mode scanning wavelengths 495 nm, 549 nm, and 648 nm for FAM, TYE and Cy5, respectively). The fluorescence was obtained during the visible irradiation (10 nm emission slit width; wavelength 520 nm, 563 nm, and 668 nm for FAM, TYE, and Cy5, respectively).
4 ratio over 24 hours to ensure saturation of the fuel binding to the walker. The sample was then incubated with the annealed track for another 10 hours before the operation experiment was started. The incubations and later operations (and controls) were conducted at 25 °C in a TE buffer. The operation buffer had a pH value of 8.0, which was known from previous studies7,10,11 to ensure stability of the DNA duplexes, vital for the walker/track integrity, and sufficient fluorescent signals (low dye emission32 at low pH). As suggested by the FRET experiment (Fig. S2†), the G-quadruplex structure also formed although it was most favoured at a lower pH value.31 The pH value in this study has been chosen to favour the duplex formation of the G4 sequence with the cytosine-rich overhang over the quadruplex structure because the duplex formation drives the force-generating expulsion stroke, and the quadruplex formation is not against a force. The walker/track concentration was kept low (∼10 nM) throughout this study to suppress track crosslinking by a walker. The irradiation operation and fluorescence measurement were conducted using an RF-5301PC spectrophotometer (150 W Xenon lamp, Shimadzu Corp.). For each round of irradiation operation, the walker–track sample was first irradiated by UV (300–400 nm wavelength; 3 nm excitation slit width) followed by visible light (kinetic mode scanning wavelengths 495 nm, 549 nm, and 648 nm for FAM, TYE and Cy5, respectively). The fluorescence was obtained during the visible irradiation (10 nm emission slit width; wavelength 520 nm, 563 nm, and 668 nm for FAM, TYE, and Cy5, respectively).
Simulation study
The oxDNA package27 was used to obtain the free energy profiles for leg dissociation through Monte Carlo simulations (at 25 °C). The walker was simulated in its fuel-free extended state on the two-site track (see Fig. S1D† for the track's design), with the duplex on each end truncated to 10 bp to accelerate the simulations. To efficiently sample the configuration space, which includes a high energy barrier, umbrella sampling33 was used to obtain the free energy as a function of the reaction coordinate, which was chosen as the number of base pairs formed between the walker leg and target binding site before leg dissociation or the leg-site separation after the dissociation (measured by the minimum distance of the 10 complementary pairs of nucleotides in the leg and the binding site). Herein, three or more independent Monte Carlo runs producing a free energy profile were performed, each lasting 107–108 steps. The convergence was tested, and error bars were estimated from the runs.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Ministry of Education of Singapore under grants R-144-000-320-112, R-144-000-325-112 and R-144-000-372-114.Notes and references
- M. Schliwa and G. Woehlke, Nature, 2003, 422, 759–765 CrossRef CAS PubMed.
- P. Yin, H. Yan, X. G. Daniell, A. J. Turberfield and J. H. Reif, Angew. Chem., Int. Ed., 2004, 43, 4906–4911 CrossRef CAS PubMed.
- Y. Tian, Y. He, Y. Chen, P. Yin and C. Mao, Angew. Chem., Int. Ed., 2005, 44, 4355–4358 CrossRef CAS PubMed.
- J. Bath, S. J. Green and A. J. Turberfield, Angew. Chem., Int. Ed., 2005, 44, 4358–4361 CrossRef CAS PubMed.
- S. J. Green, J. Bath and A. J. Turberfield, Phys. Rev. Lett., 2008, 101, 238101 CrossRef CAS PubMed.
- T. Omabegho, R. Sha and N. C. Seeman, Science, 2009, 324, 67–71 CrossRef CAS PubMed.
- J. Cheng, S. Sreelatha, R. Z. Hou, A. Efremov, R. C. Liu, J. R. van der Maarel and Z. S. Wang, Phys. Rev. Lett., 2012, 109, 238104 CrossRef PubMed.
- M. You, Y. Chen, X. Zhang, H. Liu, R. Wang, K. Wang, K. R. Williams and W. Tan, Angew. Chem., Int. Ed., 2012, 51, 2457–2460 CrossRef CAS PubMed.
- M. H. Liu, R. Z. Hou, J. Cheng, I. Y. Loh, S. Sreelatha, J. N. Tey, J. Wei and Z. S. Wang, ACS Nano, 2014, 8, 1792–1803 CrossRef CAS PubMed.
- I. Y. Loh, J. Cheng, S. R. Tee, A. Efremov and Z. S. Wang, ACS Nano, 2014, 8, 10293–10304 CrossRef CAS PubMed.
- M. H. Liu, J. Cheng, S. R. Tee, S. Sreelatha, I. Y. Loh and Z. S. Wang, ACS Nano, 2016, 10, 5882–5890 CrossRef CAS PubMed.
- T. G. Cha, J. Pan, H. Chen, J. Salgado, X. Li, C. Mao and J. H. Choi, Nat. Nanotechnol., 2014, 9, 39–43 CrossRef CAS PubMed.
- Y. He and D. R. Liu, Nat. Nanotechnol., 2010, 5, 778–782 CrossRef CAS PubMed.
- B. Lewandowski, G. De Bo, J. W. Ward, M. Papmeyer, S. Kuschel, M. J. Aldegunde, P. M. E. Gramlich, D. Heckmann, S. M. Goldup, D. M. D'Souza, A. E. Fernandes and D. A. Leigh, Science, 2013, 339, 189–193 CrossRef CAS PubMed.
- H. Gu, J. Chao, S. J. Xiao and N. C. Seeman, Nature, 2010, 465, 202–206 CrossRef CAS PubMed.
- K. Lund, A. J. Manzo, N. Dabby, N. Michelotti, A. Johnson-Buck, J. Nangreave, S. Taylor, P. Pei, M. N. Stojanovic, N. G. Walter, E. Winfree and H. Yan, Nature, 2010, 465, 206–210 CrossRef CAS PubMed.
- R. Z. Hou, I. Y. Loh, H. Li and Z. S. Wang, Phys. Rev. Appl., 2017, 7, 024020 CrossRef.
- S. K. Kufer, E. M. Puchner, H. Gumpp, T. Liedl and H. E. Gaub, Science, 2008, 319, 594–596 CrossRef CAS PubMed.
- B. Jagannathana, P. J. Elmsa, C. Bustamante and S. Marquseea, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 17820–17825 CrossRef PubMed.
- J. P. Berezney and O. A. Saleh, Nucleic Acids Res., 2014, 42, e150 CrossRef PubMed.
- S. Niman, M. J. Zuckermann, M. Balaz, J. O. Tegenfeldt, P. M. G. Curmi, N. R. Forde and H. Linke, Naonscale, 2014, 6, 15008–15019 RSC.
- M. von Delius, E. M. Geertsema and D. A. Leigh, Nat. Chem., 2010, 2, 96–101 CrossRef CAS PubMed.
- P. Alberti and J. Mergny, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 1569–1573 CrossRef CAS PubMed.
- J. Cheng, S. Sreelatha, I. Y. Loh, M. Liu and Z. S. Wang, Methods, 2014, 67, 227–233 CrossRef CAS PubMed.
- S. F. J. Wickham, M. Endo, Y. Katsuda, K. Hidaka, J. Bath, H. Sugiyama and A. J. Turberfield, Nat. Nanotechnol., 2011, 6, 166–169 CrossRef CAS PubMed.
- M. Liber, T. E. Tomov, R. Tsukanov, Y. Berger and E. Nir, Small, 2015, 11, 568–575 CrossRef CAS PubMed.
- P. Šulc, F. Romano, T. E. Ouldridge, L. Rovigatti, J. P. K. Doye and A. A. Louis, J. Chem. Phys., 2012, 137, 135101 CrossRef PubMed.
- A. Efremov and Z. S. Wang, Phys. Chem. Chem. Phys., 2011, 13, 5159–5170 RSC.
- A. Efremov and Z. S. Wang, Phys. Chem. Chem. Phys., 2011, 13, 6223–6233 RSC.
- Z. S. Wang, R. Z. Hou and A. Efremov, J. Chem. Phys., 2013, 139, 035105 CrossRef PubMed.
- A. T. Phan and J.-L. Mergny, Nucleic Acids Res., 2002, 30, 4618–4625 CrossRef CAS PubMed.
- R. A. J. Darby, M. Sollogoub, C. McKeen, L. Brown, A. Risitano, N. Brown, C. Barton, T. Brown and K. R. Foxa, Nucleic Acids Res., 2002, 30, e39 CrossRef PubMed.
- G. M. Torrie and J. P. Valleau, J. Comput. Phys., 1977, 23, 187–199 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7nr03809g |
| ‡ These authors contribute equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |