Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Nanotopography mediated osteogenic differentiation of human dental pulp derived stem cells†
Akash
Bachhuka‡
 ab,
Bahman
Delalat‡
ab,
Bahman
Delalat‡
 a,
Soraya Rasi
Ghaemi
a,
Soraya Rasi
Ghaemi
 a,
Stan
Gronthos
a,
Stan
Gronthos
 c,
Nicolas H.
Voelcker
c,
Nicolas H.
Voelcker
 *adefg and
Krasimir
Vasilev
*adefg and
Krasimir
Vasilev
 *ah
*ah
aFuture Industries Institute, University of South Australia, Mawson Lakes, Adelaide, SA 5095, Australia. E-mail: krasimir.vasilev@unisa.edu.au; Fax: +(61)8 8302 5689; Tel: +(61)8 8302 5697
bARC Centre of Excellence for Nanoscale Bio Photonics, Institute for Photonics and Advanced Sensing, School of Physical Sciences, The University of Adelaide, Adelaide, SA 5005, Australia
cAdelaide Medical School, Faculty of Health and Medical Sciences, University of Adelaide, Adelaide, 5005, SA, Australia
dMelbourne Centre for Nanofabrication, Victorian Node of the Australian National Fabrication Facility, 151 Wellington Road, Clayton, Victoria 3168, Australia. E-mail: nicolas.voelcker@monash.edu; Tel: +(61)3 9903 9320
eDrug Delivery, Disposition and Dynamics, Monash Institute of Pharmaceutical Sciences, Monash University, 381 Royal Parade, Parkville, Victoria 3052, Australia
fCommonwealth Scientific and Industrial Research Organisation (CSIRO), Clayton, Victoria 3168, Australia
gINM-Leibniz Institute for New Materials, Campus D2 2, Saarbrücken, 66123, Germany
hSchool of Engineering, University of South Australia, Adelaide, SA 5000, Australia
First published on 8th September 2017
Abstract
Advanced medical devices, treatments and therapies demand an understanding of the role of interfacial properties on the cellular response. This is particularly important in the emerging fields of cell therapies and tissue regeneration. In this study, we evaluate the role of surface nanotopography on the fate of human dental pulp derived stem cells (hDPSC). These stem cells have attracted interest because of their capacity to differentiate to a range of useful lineages but are relatively easy to isolate. We generated and utilized density gradients of gold nanoparticles which allowed us to examine, on a single substrate, the influence of nanofeature density and size on stem cell behavior. We found that hDPSC adhered in greater numbers and proliferated faster on the sections of the gradients with higher density of nanotopography features. Furthermore, greater surface nanotopography density directed the differentiation of hDPSC to osteogenic lineages. This study demonstrates that carefully tuned surface nanotopography can be used to manipulate and guide the proliferation and differentiation of these cells. The outcomes of this study can be important in the rational design of culture substrates and vehicles for cell therapies, tissue engineering constructs and the next generation of biomedical devices where control over the growth of different tissues is required.
Introduction
Understanding the interactions of biological matter with interphases is of paramount importance for creating advanced medical treatments, diagnostics and therapies.1–4 The integration and function of medical devices ranging from heart valves to dental implants is dependent on the response of tissue to material surface.5–8 The precision of many medical diagnostic technologies requires adequate response to biological fluids. The field of cell therapies, which is considered by many as the future of medical treatments, can be realized only if we have the capacity to control cell proliferation, differentiation and expansion on synthetic materials, and provide vehicles for the delivery of these cell.9–11 Thus, it is not surprising that substantial research efforts have focussed on unravelling the processes and phenomena occurring between cells and material surfaces.Recently, dental pulp derived stem cells (DPSC) have attracted interest as potent source of autologous cells in a number of area of regenerative medicine, including cell therapies.12–14 These cells have mesenchymal character and similar to bone marrow stromal cells (BMSC) have the capacity to differentiate into a wide range of other cells.14,15 It was also shown that DPSC can be a source of induced pluripotent stem (IPS) cells.16,17 In addition to tooth regeneration and bone reconstruction, other areas such as neuroregeneration,18 angiogenesis19 and vasculogenesis, and endocrinology20 can benefit from DPSC. The attractive sides of these cells is their relative ease of isolation from the pulp of discarded or removed teeth.14 It was also shown that DPSC can be cultured for periods as long as 6 months without changes in morphology or the expression of stem cell markers.21 These studies demonstrate that DPSC can be maintained and expanded to high numbers, which is a prerequisite for cell therapies and other regenerative medicine applications.
For DPSC and other stem cells to become accessible medical therapies, it is essential to acquire the capacity to control cell differentiation and expansion in an efficient and cost effective manner.22,23 Current strategies to control stem cell differentiation is via soluble factors and chemical formulations.24,25 However, this approach is rather expensive. Furthermore, some of the soluble factors are of animal origin and can cause regulatory problems because of the danger of transmission of diseases. Emerging cost effective and efficient strategies involve the targeted modification of the culture substrate or deliver vehicle via tuning the surface chemistry, ligand type and density, stiffness or topography.26–30 In this context, surface nanotopography has attracted particular attention since it may provide a tool to mimic the physical microenvironment where cells naturally reside.31,32
Surface nanotopography has been extensively investigated and documented to affect the behaviour of a range of cells including dermal,33 inflammatory,34 tumor35 and stem cells.29,36–43 There are a number of phenomena that takes place when cells interact with nanotopography. When a material is placed into a biological fluid, proteins adhere to its surface within minutes. The amount and conformation of adhered proteins can have a significant influence on the subsequent cell attachment, proliferation, and differentiation.44,45 In the presence of nanotopography, surface area is generally significantly increased (relative to a smooth surface) which may result in adsorption of greater amount of protein. Another important role that surface topographical features may play is altering the conformation of absorbed proteins. Recent work carried out in solution have demonstrated that the conformation of fibrinogen can be dramatically altered by the curvature of nanoparticles.46,47 The consequence of fibrinogen unfolding was exposing protein sequences that are shielded in the protein natural conformation. Although this work was carried out in solution, it is reasonable to assume that topographical surface feature at the nanoscale may lead to the same consequence. On the other hand, mechanotransduction due to nanotopography exerts forces on cells which can alter biochemical signalling and induce adaptive functional changes.48 Furthermore, nanotopography is reported to alter focal adhesion, integrin clustering and cytoskeletal organization of cultured cells.49,50
In this study, we explore for the first time the role of controlled nanotopography and tailored outermost surface chemistry on the attachment, proliferation and differentiation of hDPSC. The hypothesis underpinning this work was that carefully designed surface nanotopography can be used to control the behavior of hDPSC. This hypothesis was substantiated by the similarity of hDPSC to MSC, which have been demonstrated to be strongly influenced by surface topography.29,51 To elucidate the role of nanotopographical features in terms of height and lateral spacing, we generated nanoparticle density gradients via a method which we reported earlier.52,53 To create surface topography at the nanoscale we used nearly monodispersed gold nanoparticles (AuNP) of diameters of 16, 38 or 68 nm. The benefit of using gradients is that studies can be conducted using minimum number of substrates thus avoiding errors associated with the use of multiple uniformly modified samples. In addition, the gradient format speeds up analysis, reduces the number of cells used in the experiments and decreases the experimental variability by directly comparing the data obtained using the same cell population. An issue inherent of many published studies, where nanotopography was used to examine various phenomena, is heterogeneous surface chemistry. We tackled this problem by tailoring the outermost surface chemistry by depositing a 5 nm thin plasma polymer coating on top of the nanoparticles from allylamine vapor. hDPSC were cultured on these model surfaces for periods of up to three weeks and analyzed for proliferation and osteogenic differentiation.
Results and discussion
In order to generate gradients of AuNP, 13 mm round coverslips were first coated with a 20 nm thin layer of plasma polymerized allylamine (AApp). These films carry a population of amine groups which protonate in aqueous medium below pH = 8 and produce a positive surface charge.54,55 The modified coverslips were then immersed in a rate controlled fashion in a solution of AuNP of three different sizes (16, 38 or 68 nm) using a dip coating method as we have previously reported.56 Using this technique, we were successfully able to control the time of contact of each region of the AApp coated surface and the gold nanoparticle solution, which resulted in nanoparticle density gradients across the surface. In addition of controlled spacing between nanoscale features, the predetermined size of the immobilized nanoparticles i.e. 16, 38 or 68 nm also allowed us to generate surface nanotopography of controlled height. However, these substrates present not only variation in nanotopography but also have mixed chemistry. The underlying AApp coating is amine/nitrogen rich while gold nanoparticles carry carboxyl acid surface groups. We were able to uniquely tailor the outermost surface chemistry by overcoating the nanoparticle density gradients with a 5 nm thin layer of AApp. We have demonstrated in several published studies that AApp films of this thickness are continuous and pinhole free and thus provide uniform surface chemistry across the surface.57These surface overcoatings also change surface energy further leading to the change in surface wettability.53,58 Using this approach, the stem cells were presented only with changes in nanotopography. Fig. 1a shows AFM images on six equally spaced regions across the nanoparticles density gradients having nanoparticle size of 16 nm (top), 38 nm (middle) and 68 nm (bottom). The nanoparticles were randomly distributed with an increase in nanoparticle density from region 3 mm to region 12 mm and did not form aggregates. The root mean square roughness (RMS) increased with the greater nanoparticle size and density (shown in Fig. 1b). The AFM images were further analyzed to calculate number of particles per μm2, % increase in surface area, % surface coverage and interparticle distance at different regions of the surface. The number of surface bound nanoparticles per μm2 increased from 17 ± 2 at region 3 mm to 121 ± 5 at 12 mm in the case of 16 nm AuNP. For nanoparticles of 38 nm diameter, the number of particles increased from 12 ± 3 to 30 ± 3 across the gradient and for 68 nm nanoparticles from 8 ± 2 to 30 ± 3 (Fig. 1c). The increasing number of nanoparticles across the surface resulted in greater surface area and surface coverage as shown in Fig. 1d and e, respectively. The interparticle distance decreased with the increase in density across the gradients for all nanoparticles. The interparticle distance decreased from 196 nm to 83 nm for the 16 nm AuNP, from 228 nm to 155 nm for the 38 nm nanoparticles and from 268 nm to 156 nm for the 68 nm AuNP (Fig. 1f).
XPS was used to analyze the elemental chemical composition across the gradients of nanoparticle density. The survey spectra on 6 regions along the coverslip modified with 16 nm AuNP is shown in Fig. 2a. The data showing the elemental composition of the corresponding spectra for nanoparticle density gradients of 38 and 68 nm are presented in the ESI Fig. S1 and S2.† The atomic percentage of chemical elements on different regions across the gradient are presented in Table S1 of the ESI.†
Fig. 2a shows the increase in the intensity of the Au 4f peak towards region 12 mm which corresponds in increasing number of surface bound nanoparticles. The nitrogen N 1s peak was detected due to the initial underlying layer of allylamine, which aids in electrostatic immobilization of different sized gold nanoparticles. The surface also contained some amount of oxygen which was due to the carboxyl functionalities on the surface of the nanoparticles and some partial oxidation of the surface typical for this type of plasma polymers.55,57 Survey spectra of nanoparticle density gradient after overcoating with a 5 nm thin layer of plasma polymerized allylamine (AApp) are shown in Fig. 2b. The intensity of the nitrogen N 1s peak increased while gold atomic concentration decreased (Table S1†). However, gold signal was still detectable as XPS sampling depth of ca. 10 nm, which is greater than the thickness of the AApp overcoating. A detailed information of the chemical composition across the nanotopography gradients of different sizes, is shown in ESI Table S1.†
To demonstrate the effect of surface nanotopography on cell attachment, hDPSC were cultured on the gradients of different nanotopography scale and AApp control for 12 h.
Fig. 3 shows the fluorescence microscopy images of cells attached on different regions across the gradients. Nuclei (blue) staining was used to quantify the number of attached cells. The images clearly show an increase in the number of adherent cells toward the greater nanoparticle density (region 1 to region 5). In addition, there were greater number of attached cells on the nanotopography gradients compared to a control coverslip coated with the smooth AApp coating only. The data suggest that nanotopography enhances cell adhesion compared to a smooth surface with the same chemistry.
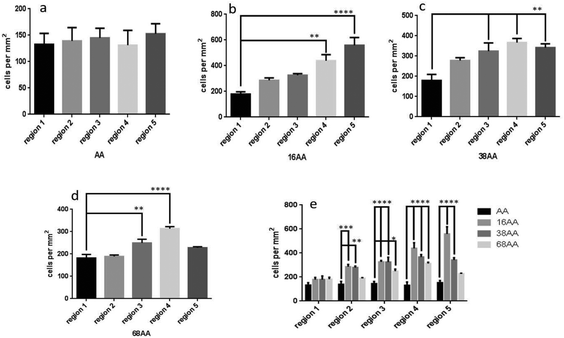
Fig. 4 shows quantitative analysis of cell numbers on smooth AApp surface and across gradients of nanotopography (16, 38 and 68 nm). There was no significant difference in cell number across the control coverslip coated with AApp only (Fig. 4a). In the case of 16 nm nanotopography, cell number per mm2 significantly increased on regions 4 (p < 0.01) and 5 (p < 0.0001) compared to region 1 (Fig. 4b). This shows that smaller nanotopography with an interparticle distance of 97 nm or less plays an important role in cell adhesion. When nanotopography was 38 nm, cell number per mm2 significantly increased on regions 3, 4 and 5 (p < 0.01) compared to region 1 (Fig. 4c), suggesting that features of 38 nm high and interparticle distance below 200 nm is favorable for cell attachment. On gradients of 68 nm nanoparticles, cell number was greatest on regions 3 (p < 0.01) and 4 (p < 0.001) having interparticle distance between 185 and 167 nm, respectively (Fig. 4d). There was no significant difference in cell numbers on regions 2 and 5 compared to region 1. Further increase in nanoparticle density had no effect on the number of adhered cells. The data indicate that both height and spacing between the nanoparticles are strong arbiters for cell adhesion. This is clearly evident in Fig. 4e where the data of the three different nanotopography scales is collectively presented, further suggesting the smaller scale of nanotopography may preferentially encourage cell attachment.
Fig. 5 shows fluorescence microscopy images of cell proliferation on uniformly coated AApp coverslip and across the nanotopography gradients. It can be clearly seen that cell proliferation was same across the uniformly coated AApp control. In case of 16 nm, 38 nm and 68 nm nanotopography, surface density. An increase in proliferation of cells was observed on gradients of higher nanotopography compared to gradients of smaller nanotopography.
Fig. 6 shows quantitative analysis of cell proliferation on AApp coated control and across the gradients of nanotopography. The AApp control had the same level of cell proliferation across the entire surface (Fig. 6a). As a general trend, cell proliferation increased towards greater density of nanotopography features for all gradients. In the case of nanotopography of 16 nm, the increase was statistically significant on regions 3 (p < 0.01), 4 (p < 0.01), and 5 (p < 0.001) relative to region 1 (Fig. 6b). The trends were similar on nanotopography of 38 nm and 68 nm where statistically significant increase in cell proliferation was seen on regions 3, 4 and 5. Interestingly, when nanotopography height was 68 nm, small drop in cell proliferation was observed in region 5 relative to region 4 (Fig. 6d), a trend consistent with the data for cell attachment (Fig. 4d). The comparative representation of cell proliferation on gradients of different nanotopography height is shown in Fig. 6e. While increased nanosize feature surface density stimulated cell proliferation on all gradients, it is evident that cells proliferated fastest on the nanotopography of greatest height.
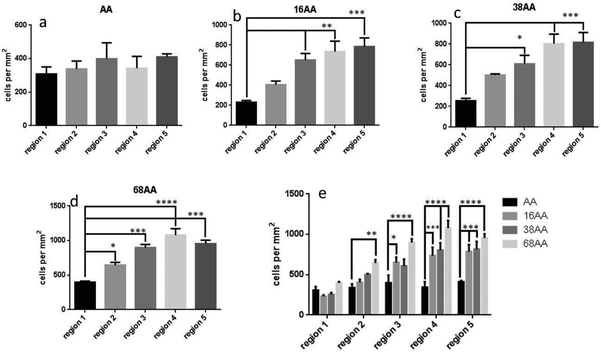
The alkaline phosphatase (AP) activity was monitored across the gradients of nanotopography and AApp control to determine the differentiation of hDPSC into osteogenic lineages. AP was selected as a marker since is well accepted and extensively documented indicator of osteoblasts.59Fig. 7 shows fluorescence microscopy images of the cells stained for AP after 21 d in culture. The cells on AApp coated control showed little or no AP activity, similar to region 1 on the gradients where the nanotopography was same as on the control. This is mainly due to the lack of any surface topography and exogenous biomolecule factors on the control group. However, the images clearly show the increase in AP staining intensity (green) towards increasing density on nanoparticles. The trend was the same for all nanotopography heights but most marked on the smallest nanotopography of 16 nm.
Fig. 8 shows quantitative analysis of cell differentiation on the gradients of surface nanotopography and AApp control. The data is expressed as AP staining intensity per individual cells. Consistent with the images in Fig. 7, the levels of differentiation to osteogenic lineages was small on the smooth AApp film. When nanotopography was presented on the surface, a statistically significant trend to stronger osteogenic differentiation towards greater density of nanotopography features was observed on all gradients. In agreement with the images in Fig. 7, the smaller nanotopography of 16 nm appeared to be the strongest driver to osteogenic differentiation (Fig. 8e). The data presented in Fig. 7 and 8 demonstrate that controlled surface nanotopography is capable to promote differentiation of dental pulp stem cells (hDPSC). The cells were grown in a standard DMEM culture medium, which does not favor differentiation in particular direction.
To confirm differentiation towards osteogenic lineages, the cells were cultured in osteogenic differentiation medium (StemPro® osteogenesis differentiation kit contains components such as dexamethasone, L-glutamine, ascorbate, mesenchymal cell growth supplement and beta-glycerophosphate). Images across the nanotopography gradients and AApp control are shown in ESI Fig. S3 and S4.† The staining intensity of AP followed the same tends as reported in Fig. 7 and 8 above, supporting the finding of self-differentiation caused by surface nanotopography.
This study demonstrates that surface nanotopography can induce differentiation of dental pulp derived stem cells into osteoblasts without addition of any biochemical cues. Gradients of nanoparticle density were used to provide controlled surface nanotopography having engineered and characterized lateral spacing between the nanoscale features. The height of surface nanotopography was tuned by involving nanoparticles of different diamantes i.e. 16, 38 or 68 nm. The outermost surface chemistry was tailored by addition of a 5 nm thin amine rich layer of AApp. The gradient strategy allowed us to interrogate cellular responses with a minimum number of substrates thus avoiding errors associated with the use of multiple uniformly modified samples. Furthermore, the use of gradients speeds up analysis, reduces the number of cells used in the experiments and decreases the experimental variability by directly comparing the data obtained using the same cell population.
As a general trend, greater surface nanotopography (in terms of surface feature density) promoted cell attachment. One possible explanation could be the increase in surface area associated with greater surface nanotopography. This in turn facilitates adsorption of larger amounts of cell adhesion proteins and thus provides greater number of integrin binding sites for the cells to attach.4 This hypothesis seems to hold well for nanotopography of 16 nm where increasing density of surface bound nanoparticles leads to greater number of attached cells. However, when the scale of nanotopography increased to 38 nm and 68 nm, despite similar or greater surface area, the number of adhered cells was smaller compared to 16 nm. Recent studies have suggested protein unfolding when attached to curved surface of nanoparticles. The phenomenon leads to exposure of hidden binding sites that may change cell adhesion dynamics.47,60 Although, these studies were carried out with nanoparticles in solution, it is reasonable to assume that the same phenomenon occurs when nanoparticles are immobilized to solid surfaces. It may be possible that in this scenario nanoparticles of 38 nm and 68 nm lead to some degree on protein unfolding and thus present lower number of integrin binding domains relative to the increased surface area caused by nanotopography.
After two weeks in culture, the cell number was significantly greater on regions of the gradients with higher density of nanotopographical features. Larger nanoparticles of 38 nm and 68 nm promoted stronger cell proliferation. Interrogation of cell proliferation rate across the gradients, presented in Fig. S5,† revealed that in the case of nanotopography of 16 nm the increase in cell numbers towards higher nanoparticle density was merely due to the greater number of initially adhered cells. There was no statistically significant difference in the rate of cell proliferation across the 16 nm nanoparticle density gradient. However, a statistically significant increase in the rate of proliferation was seen when nanotopography increases to 38 nm and 68 nm. The data suggest that nanotopography of 68 nm is the strongest driver of cell proliferation. It should be taken into consideration that the nanotopography of 16 nm was such a strong promoter of cell attachment that space for further proliferation may not have been available to the cells. To determine the exact role of surface nanotopography on cell proliferation rate, studies with lower seeding densities need to be carried out. However, the primary objective of this study was to determine how surface nanotopography affects the differentiation of hDPSC.
Cell differentiation was much more effective on nanotopographically modified surfaces in comparison to smooth surfaces. Differentiation towards a particular type of cell in the absence of any inducing agents points to the capacity of surface nanotopography to directly alter the fate of stem cells. Nanotopography directed differentiation of hDPSC to osteoblasts observed in our studies is consistent with what has already been reported for MSC.30,61 Thus, taking into consideration that hDPSC are of mesenchymal phenotype, the reported results are not surprising. However, our studies allow us to examine the differentiation process as a function of both density and size of surface nanotopographical features. The evident trend associated with all three sizes of nanotopography is an increase in osteogenic differentiation with increasing surface feature density. The greater differentiation to osteoblasts towards higher nanotopography density seems to correlate with the increased number of cells shown in Fig. 3 and 4. However, while cell density reached maximum in region 4 of the gradients the differentiation increases throughout the sample. The later suggests that cell surface number and density are not sufficient to explain the phenomena that were observed. Another interesting observation points to the increase in surface area. It appears that the greater surface area on samples of 16 nm nanotopography (Fig. 1) induced the strongest alkaline phosphatase staining intensity per individual cell (Fig. 7 and 8).
The complex interplay between biological cells and biointerfaces is yet to be fully uncovered. This study and a number of others unambiguously point to the important role that surface nanotopography plays in modulating cellular responses. A large number of parameters come into play in the interactions between the surface and the cells. The attachment of proteins in terms of amount, types, orientation and unfolding are all significant in determining cell fate. Furthermore, the effect of physical cues in terms of physical and mechanical modulation of cell membrane and cytoskeleton have recently been attributed critical importance.23,48,62 The differentiation of hDPSC to osteoblasts on rough surfaces can also be related to the natural response of these cells when needed to repair bone tissue, which is inherently rougher. The results of this study can be important in the rational design of implantable devices and tissue engineering scaffolds where the modulation of specific tissue growth is important. Furthermore, this study can instruct the design of tools and devices such as bioreactors, culture substrates and vehicles needed for the growing field of cell therapies, which are considered by many as the future of medical treatment, pointing to the possibility to control stem cell fate without the use of soluble factors such as growth factors and cytokines. The latter is important because it can make cell therapies more affordable as surface engineering is more cost effective and safer than the use soluble factors.
Experimental
Materials
Allylamine (AA) (98%, Aldrich), hydrogen tetrachloroaurate (99.9985%, ProSciTech), trisodium citrate (99%, BHD Chemicals, Australia Pty. Ltd), 2-mercaptosuccinic acid (97%, Aldrich), were used as received.Plasma polymerization
Plasma polymerization was carried out in a custom-built reactor with a 13.56 MHz plasma generator.63 Deposition of allylamine plasma polymer (AApp) was carried out at a pressure of 0.2 mbar for 2 min. Power used for deposition was 40 W. Using these conditions, a polymer film of thickness 23 nm was obtained. To achieve an overcoating of 5 nm, the time of deposition was 20 s. Before deposition, all substrates were cleaned by applying air plasma for 2 min at 50 W.Synthesis of gold nanoparticles (AuNP)
AuNP were synthesized using hydrogen tetrachloroaurate (HAuCl4). Particles of 16, 38 and 68 nm diameter were synthesized by varying the amount of 1% trisodium citrate from 1 ml to 0.3 ml, respectively.64 Surface modification of these nanoparticles was performed by using 2-mercaptosuccinic acid.65Gradient formation
Allylamine coated surfaces were immersed in a solution of gold nanoparticles in a time controlled manner. The speed of immersion was controlled using Zaber T-LSR series, a computer controlled motorised linear slide using Zaber's software. For 16, 38 and 68 nm nanoparticles the speed of immersion was 10, 30 and 75 s mm−1 respectively. After immersion, all samples were thoroughly washed using Milli-Q water to remove the weakly bound nanoparticles.Atomic force microscopy (AFM)
An NT-MDT NTEGRA SPM atomic force microscope (AFM) was used in non-contact mode to provide topographical images. Silicon nitride non-contact tips coated with Au on the reflective side (NT-MDT, NSG03) were used and had resonance frequencies between 65 and 100 kHz. The amplitude of oscillation was 10 nm, and the scan rate for 5 μm × 5 μm images was 0.5 Hz. All images were scaled relative to the maximum height for like-sized AuNP immobilized surfaces.X-ray photoelectron spectroscopy
XPS analysis was used to determine the surface composition of the plasma polymer and the deposited AuNP. XPS spectra were recorded on a Specs SAGE XPS spectrometer using Al Kα radiation source (hν = 1253.6 eV) operated at 10 kV and 20 mA. Elements present in a sample surface were identified from the survey spectrum recorded over the energy range 0–1000 eV at pass energy of 100 eV and a resolution of 0.5 eV. The areas under selected photoelectron peaks in a wide scan spectrum were used to calculate percentage atomic concentrations (excluding hydrogen). High-energy resolution (0.1 eV) spectra were then recorded for pertinent photoelectron peaks at pass energy of 20 eV to identify the possible chemical binding environments for each element. All the binding energies (BEs) were referenced to the C 1s neutral carbon peak at 285 eV, to compensate for the effect of surface charging. The XPS analysis area was circular with a diameter of 0.7 mm. The processing and curve-fitting of the high-energy resolution spectra were performed using Casa XPS software.Cell culture
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 in PBS) as a osteo lineage marker. After incubation with the primary antibodies for overnight at 4 °C and washing three times with PBS, the secondary antibody, FITC-conjugated donkey anti rabbit (Santa Cruz, diluted 1
200 in PBS) as a osteo lineage marker. After incubation with the primary antibodies for overnight at 4 °C and washing three times with PBS, the secondary antibody, FITC-conjugated donkey anti rabbit (Santa Cruz, diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in PBS) was added for 1 h at room temperature. Negative controls were carried out by eliminating the primary antibody labelling step from the procedure, which in all cases resulted in a complete loss of signal from fluorescence-labelled secondary antibodies. Finally, the nuclei were counterstained with 2 μg ml−1 Hoechst 33342 in PBS for 10 min, rinsed with PBS. Gradients were divided into five equal regions of gradually increasing AuNP as previously described, and cell behavior was compared between each zone. Images were obtained on a full automated inverted fluorescence microscope (Operetta) equipped with Harmony imaging and analysing software. Immunofluorescence analysis was repeated in three independent experiments.
100 in PBS) was added for 1 h at room temperature. Negative controls were carried out by eliminating the primary antibody labelling step from the procedure, which in all cases resulted in a complete loss of signal from fluorescence-labelled secondary antibodies. Finally, the nuclei were counterstained with 2 μg ml−1 Hoechst 33342 in PBS for 10 min, rinsed with PBS. Gradients were divided into five equal regions of gradually increasing AuNP as previously described, and cell behavior was compared between each zone. Images were obtained on a full automated inverted fluorescence microscope (Operetta) equipped with Harmony imaging and analysing software. Immunofluorescence analysis was repeated in three independent experiments.
Conclusions
This study reveals the role of surface nanotopography on the attachment, proliferation and differentiation of hDPSC. We designed and utilized gradients of nanotopography generated from three different sized gold nanoparticles (i.e. 16, 38 and 68 nm) which had uniformly tailored outermost surface chemistry of plasma polymerised allylamine. hDPSC adhered in greater numbers and proliferated faster on the regions of the gradients with higher density of nanotopography features. Nanoparticles diameters of 16 nm were most effective in enhancing cell attachment with those of 38 nm and 68 nm showing greater proliferation rates. Moreover, we found that greater surface nanotopography density directed the differentiation of hDPSC to osteogenic lineages and the 16 nm sized nanotopography being the strongest driver. In addition to being the first work to explore the effect of surface roughness on hDPSC, this study demonstrates that carefully tuned surface nanotopography can be used to manipulate and guide the proliferation and differentiation of these cells. The outcomes of this study can be important in the rational design of the next generation of biomedical devices and tissue engineering constructs where control over the growth of different tissues is required. These results are also important for the growing field of cell therapies where control of stem cell fate is an essential requirement.Conflicts of interest
There are no conflicts to declare.Acknowledgements
KV thanks ARC (DP15104212) and NHMRC Fellowship (APP1122825) and NV thanks NHMRC (APP595901) for their support.References
- F. Variola, J. B. Brunski, G. Orsini, P. T. de Oliveira, R. Wazen and A. Nanci, Nanoscale, 2011, 3, 335 RSC.
- L. Tang, P. Thevenot and W. Hu, Curr. Top. Med. Chem., 2008, 8, 270 CrossRef.
- M. S. Castro Raucci, M. S. Francischini, L. N. Teixeira, E. P. Ferraz, H. B. Lopes, P. T. de Oliveira, M. Q. Hassan, A. L. Losa and M. M. Beloti, J. Cell. Biochem., 2016, 7, 1718 CrossRef PubMed.
- M. J. P. Biggs, R. G. Richards and M. J. Dalby, Nanomed. Nanotechnol., 2010, 6, 619 CrossRef CAS PubMed.
- S. Brody, T. Anilkumar, S. Liliensiek, J. A. Last, C. J. Murphy and A. Pandit, Tissue Eng., 2006, 12, 413 CrossRef PubMed.
- S. J. Liliensiek, J. A. Wood, J. Yong, R. Auerbach, P. F. Nealey and C. J. Murphy, Biomaterials, 2010, 31, 5418 CrossRef CAS PubMed.
- C. N. Elias and L. Meirelles, Expert Rev. Med. Devices, 2010, 7, 241 CrossRef PubMed.
- E. Bressan, L. Sbricoli, R. Guazzo, I. Tocco, M. Roman, V. Vindigni, E. Stellini, C. Gardin, L. Ferroni and S. Sivolella, Int. J. Mol. Sci., 2013, 14, 1918 CrossRef CAS PubMed.
- M. J. Dalby, N. Gadegaard, R. Tare, A. Andar, M. O. Riehle, P. Herzyk, C. D. W. Wilkinson and R. O. C. Oreffo, Nat. Mater., 2007, 6, 997 CrossRef CAS PubMed.
- A. Klymov, L. Prodanov, E. Lamers, J. A. Jansen and X. F. Walboomers, Biomater. Sci., 2013, 1, 135 RSC.
- P. Decuzzi and M. Ferrari, Biomaterials, 2010, 31, 173 CrossRef CAS PubMed.
- K. Sakai, A. Yamamoto, K. Matsubara, S. Nakamura, M. Naruse, M. Yamagata, K. Sakamoto, R. Tauchi, N. Wakao and S. Imagama, J. Clin. Invest., 2012, 122, 80 CAS.
- P. D. Potdar and Y. D. Jethmalani, World J. Stem Cells, 2015, 7, 839 CrossRef PubMed.
- M. Tatullo, M. Marrelli, K. M. Shakesheff and L. J. White, J. Tissue Eng. Regener. Med., 2015, 9, 1205 CrossRef PubMed.
- S. Gronthos, M. Mankani, J. Brahim, P. G. Robey and S. Shi, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 13625 CrossRef CAS PubMed.
- X. Yan, H. Qin, C. Qu, R. S. Tuan, S. Shi and G. T.-J. Huang, Stem Cells Dev., 2010, 19, 469 CrossRef CAS PubMed.
- N. Tamaoki, K. Takahashi, T. Tanaka, T. Ichisaka, H. Aoki, T. Takeda-Kawaguchi, K. Iida, T. Kunisada, T. Shibata, S. Yamanaka and K. Tezuka, J. Dent. Res., 2010, 89, 773 CrossRef CAS PubMed.
- B. C. Kim, H. Bae, I. K. Kwon, E. J. Lee, J. H. Park, A. Khademhosseini and Y. S. Hwang, Tissue Eng., Part B, 2012, 18, 235 CrossRef CAS PubMed.
- C. Gandia, A. Arminan, J. M. García-Verdugo, E. Lledo, A. Ruiz, M. D. Minana, J. Sanchez-Torrijos, R. Paya, V. Mirabet and F. Carbonell-Uberos, Stem Cells, 2008, 26, 638 CrossRef PubMed.
- R. Patil, B. M. Kumar, W. J. Lee, R. H. Jeon, S. J. Jang, Y. M. Lee, B. W. Park, J. H. Byun, C. S. Ahn, J. W. Kim and G. J. Rho, Exp. Cell Res., 2014, 320, 92 CrossRef CAS PubMed.
- N. F. Lizier, A. Kerkis, C. M. Gomes, J. Hebling, C. F. Oliveira, A. I. Caplan and I. Kerkis, PLoS One, 2012, 7, e39885 CAS.
- M. P. Lutolf, P. M. Gilbert and H. M. Blau, Nature, 2009, 462, 433 CrossRef CAS PubMed.
- S. R. Ghaemi, F. J. Harding, B. Delalat, S. Gronthos and N. H. Voelcker, Biomaterials, 2013, 34, 7601 CrossRef PubMed.
- D. Woodbury, E. J. Schwarz, D. J. Prockop and I. B. Black, J. Neurosci. Res., 2000, 61, 364 CrossRef CAS PubMed.
- L. Qian and W. M. Saltzman, Biomaterials, 2004, 25, 1331 CrossRef CAS PubMed.
- B. Delalat, A. Mierczynska, S. R. Ghaemi, A. Cavallaro, F. J. Harding, K. Vasilev and N. H. Voelcker, Adv. Funct. Mater., 2015, 25, 2737 CrossRef CAS.
- X. Liu, Q. Feng, A. Bachhuka and K. Vasilev, ACS Appl. Mater. Interfaces, 2014, 6, 9733 CAS.
- F. Harding, R. Goreham, R. Short, K. Vasilev and N. H. Voelcker, Adv. Healthcare Mater., 2013, 2, 585 CrossRef CAS PubMed.
- R. J. McMurray, N. Gadegaard, P. M. Tsimbouri, K. V. Burgess, L. E. McNamara, R. Tare, K. Murawski, E. Kingham, R. O. Oreffo and M. J. Dalby, Nat. Mater., 2011, 10, 637 CrossRef CAS PubMed.
- M. J. Dalby, N. Gadegaard and R. O. Oreffo, Nat. Mater., 2014, 13, 558 CrossRef CAS PubMed.
- E. K. Yim and K. W. Leong, Nanomed. Nanotechnol. Biol. Med., 2005, 1, 10 CrossRef CAS PubMed.
- V. Vogel and M. P. Sheetz, Curr. Opin. Cell Biol., 2009, 21, 38 CrossRef CAS PubMed.
- A. Bachhuka, J. Hayball, L. E. Smith and K. Vasilev, ACS Appl. Mater. Sci., 2015, 7, 23767 CrossRef CAS PubMed.
- S. N. Christo, A. Bachhuka, K. R. Diener, A. Mierczynska, J. D. Hayball and K. Vasilev, Adv. Healthcare Mater., 2016, 8, 956 CrossRef PubMed.
- Y. Khung, G. Barritt and N. Voelcker, Exp. Cell Res., 2008, 314, 789 CrossRef CAS PubMed.
- M. J. Dalby, D. McCloy, M. Robertson, H. Agheli, D. Sutherland, S. Affrossman and R. O. Oreffo, Biomaterials, 2006, 27, 2980 CrossRef CAS PubMed.
- A. S. Curtis and C. D. Wilkinson, J. Biomater. Sci., Polym. Ed., 1998, 9, 1313 CrossRef CAS PubMed.
- J.-Y. Jang, S. W. Lee, S. H. Park, J. W. Shin, C. Mun, S.-H. Kim, D. H. Kim and J.-W. Shin, BioMed. Res. Int., 2011, 860652 Search PubMed.
- K. S. Brammer, C. Choi, C. J. Frandsen, S. Oh and S. Jin, Acta Biomater., 2011, 7, 683 CrossRef CAS PubMed.
- L. R. Clements, P.-Y. Wang, W.-B. Tsai, H. Thissen and N. H. Voelcker, Lab Chip, 2012, 12, 1480 RSC.
- P. Y. Wang, L. R. Clements, H. Thissen, A. Jane, W. B. Tsai and N. H. Voelcker, Adv. Funct. Mater., 2012, 22, 3414 CrossRef CAS.
- P.-Y. Wang, L. R. Clements, H. Thissen, S.-C. Hung, N.-C. Cheng, W.-B. Tsai and N. H. Voelcker, RSC Adv., 2012, 2, 12857 RSC.
- P.-Y. Wang, L. R. Clements, H. Thissen, W.-B. Tsai and N. H. Voelcker, Biomater. Sci., 2013, 1, 924 RSC.
- C. J. Wilson, R. E. Clegg, D. I. Leavesley and M. J. Pearcy, Tissue Eng., 2005, 11, 1 CrossRef CAS PubMed.
- F. Guilak, D. M. Cohen, B. T. Estes, J. M. Gimble, W. Liedtke and C. S. Chen, Cell Stem Cell, 2009, 5, 17 CrossRef CAS PubMed.
- Z. J. Deng, M. Liang, M. Monteiro, I. Toth and R. F. Minchin, Nat. Nanotechnol., 2011, 6, 39 CrossRef CAS PubMed.
- Z. J. Deng, M. Liang, I. Toth, M. J. Monteiro and R. F. Minchin, ACS Nano, 2012, 6, 8962 CrossRef CAS PubMed.
- M. Schernthaner, B. Reisinger, H. Wolinski, S. D. Kohlwein, A. Trantina-Yates, M. Fahrner, C. Romanin, H. Itani, D. Stifter and G. Leitinger, Acta Biomater., 2012, 8, 2953 CrossRef CAS PubMed.
- T. P. Kunzler, T. Drobek, M. Schuler and N. D. Spencer, Biomater., 2007, 28, 2175 CrossRef CAS PubMed.
- E. K. Yim, E. M. Darling, K. Kulangara, F. Guilak and K. W. Leong, Biomater., 2010, 31, 1299 CrossRef CAS PubMed.
- R. Ravichandran, S. Liao, C. C. Ng, C. K. Chan, M. Raghunath and S. Ramakrishna, World J. Stem Cells, 2009, 1, 55 CrossRef PubMed.
- R. V. Goreham, A. Mierczynska, L. E. Smith, R. Sedev and K. Vasilev, RSC Adv., 2013, 3, 10309 RSC.
- M. Ramiasa-MacGregor, A. Mierczynska, R. Sedev and K. Vasilev, Nanoscale, 2016, 8, 4635 RSC.
- A. Mierczynska, A. Michelmore, A. Tripathi, R. V. Goreham, R. Sedev and K. Vasilev, Soft Matter, 2012, 8, 8399 RSC.
- J. C. Ruiz, S. Taheri, A. Michelmore, D. E. Robinson, R. D. Short, K. Vasilev and R. Förch, Plasma Processes Polym., 2014, 11, 888 CrossRef CAS.
- R. V. Goreham, A. Mierczynska, M. Pierce, R. D. Short, S. Taheri, A. Bachhuka, A. Cavallaro, L. E. Smith and K. Vasilev, Thin Solid Films, 2013, 528, 106 CrossRef CAS.
- K. Vasilev, A. Michelmore, P. Martinek, J. Chan, V. Sah, H. J. Griesser and R. D. Short, Plasma Processes Polym., 2010, 7, 824 CrossRef CAS.
- Z. Bo, Y. Tian, Z. J. Han, S. Wu, S. Zhang, J. Yan, K. Cen and K. K. Ostrikov, Nanoscale Horiz., 2017, 2(2), 89–98 RSC.
- M. Elsafadi, M. Manikandan, M. Atteya, J. A. Hashmi, Z. Iqbal, A. Aldahmash, M. Alfayez, M. Kassem and A. Mahmood, Stem Cells Int., 2016, 2016, 9378081 Search PubMed.
- Z. J. Deng, M. T. Liang, M. Monteiro, I. Toth and R. F. Minchin, Nat. Nanotechnol., 2011, 6, 39 CrossRef CAS PubMed.
- S. Oh, K. S. Brammer, Y. J. Li, D. Teng, A. J. Engler, S. Chien and S. Jin, Proc.Natl. Acad. Sci. U. S. A., 2009, 106, 2130 CrossRef CAS PubMed.
- O. Mashinchian, L.-A. Turner, M. J. Dalby, S. Laurent, M. A. Shokrgozar, S. Bonakdar, M. Imani and M. Mahmoudi, Nanomed., 2015, 10, 829 CrossRef CAS PubMed.
- K. Vasilev, A. Michelmore, P. Martinek, J. Chan, V. Sah, H. J. Griesser and R. D. Short, Plasma Processes Polym., 2010, 7, 824 CrossRef CAS.
- R. V. Goreham, R. D. Short and K. Vasilev, J. Phys. Chem. C, 2011, 115, 3429 CAS.
- T. Zhu, K. Vasilev, M. Kreiter, S. Mittler and W. Knoll, Langmuir, 2003, 19, 9518 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Supplementary figures showing XPS characterization of surfaces and Fluorescence images of cell differentiation. See DOI: 10.1039/c7nr03131a |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2017 |