Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Surface-enhanced hyper Raman hyperspectral imaging and probing in animal cells†
Zsuzsanna
Heiner
 ab,
Marina
Gühlke
b,
Vesna
Živanović
ab,
Fani
Madzharova
b and
Janina
Kneipp
ab,
Marina
Gühlke
b,
Vesna
Živanović
ab,
Fani
Madzharova
b and
Janina
Kneipp
 *ab
*ab
aSALSA School of Analytical Sciences Adlershof, Humboldt-Universität zu Berlin, Albert-Einstein-Str. 5-9, 12489 Berlin, Germany
bDepartment of Chemistry, Humboldt-Universität zu Berlin, Brook-Taylor-Str. 2, 12489 Berlin, Germany. E-mail: janina.kneipp@chemie.hu-berlin.de
First published on 25th May 2017
Abstract
Hyper Raman scattering, that is, spontaneous, two-photon excited Raman scattering, of organic molecules becomes strong when it occurs as surface-enhanced hyper Raman scattering (SEHRS), in the proximity of plasmonic nanostructures. Its advantages over one-photon excited surface-enhanced Raman scattering (SERS) include complementary vibrational information resulting from different selection rules, probing of very small focal volumes, and beneficial excitation with long wavelengths. Here, imaging of macrophage cells by SEHRS is demonstrated, using SEHRS labels consisting of silver nanoparticles and two different molecules, 2-naphthalenethiol and para-mercaptobenzoic acid, that are excited off-resonance. The vibrational signatures of the molecules are discriminated using hyperspectral analysis and provide information about the subcellular localization of the SEHRS probes. The SEHRS based hyperspectral imaging approach presented here uses principal component analysis (PCA) to localize the reporter molecules inside the cells and is augmented by hierarchical cluster analysis (HCA). The high sensitivity of SEHRS spectra with respect to small environmental changes can be utilized for mapping of physiological parameters in the endosomal system of the cells. This is illustrated by discussing the spatial distribution of endosomes of varying pH inside the cytosol.
Introduction
Gold and silver nanoparticles play an increasingly important role in biosensing and theranostics,1,2 mainly because of the tunability of their optical properties, the versatility regarding their functionalization with targeting sequences and active molecules, and the possibility to direct them into complex biomaterials, such as cells and tissues. There exist several microscopic and spectroscopic approaches to visualizing metal nanoparticles or nanoprobes in tissues and cells, ranging from electron3,4 and X-ray microscopies5 over mass spectrometry imaging6 to high-resolution7 and dark field microscopies.8 Surface-enhanced Raman scattering (SERS) is one of the most powerful optical approaches, due to the possibility to both image the plasmonic nanostructures and to characterize the molecular composition and structure of their environment.9 As SERS takes place in the local optical fields of such plasmonic nanoprobes, it can provide chemical and structural information on their surroundings with high sensitivity. Therefore, specifically, fast imaging of intrinsic molecules in cells, usually occurring at concentrations in the nM range is possible with this method.10–13For biological cells and tissues, nonlinear spectroscopy offers several advantages, mainly owing to excitation with lower photon energy and to the reduced interaction volume.14,15 As we and others have shown, surface enhanced hyper Raman scattering (SEHRS), the two photon excited analogue of SERS, is useful for spectroscopic studies of organic molecules and specifically for characterizing their interaction with metal nanostructures.16–21 Due to different selection rules of the hyper Raman process, SEHRS allows to collect complementary vibrational information, revealing infrared-active or even “silent” vibrational modes.22 The surface enhancement of hyper Raman scattering signals benefits specifically from their nonlinear scaling with the excitation field intensity,23–25 and also from an altered chemical enhancement.26,27 By the former, using SEHRS, the low multi-photon cross sections can easily be overcome, and SEHRS cross sections were determined to be up to 10−46–10−45 cm4 s in the non-resonant case,24 which is higher than typical two-photon fluorescence cross-sections. Furthermore, compared to SERS, SEHRS was shown to be more sensitive to changes in the local microenvironment.17,18,20
Here, we use the high signal strengths in SEHRS spectra, along with the possibility to obtain them in small focal volumes,18 to demonstrate SEHRS microscopy on single cells. Using hyperspectral imaging, we localize SEHRS nanoprobes in the endosomal system of macrophage cells. In our experiments, the SEHRS nanoprobes carry different reporter molecules, which make them unequivocally identifiable by their SEHRS (and SERS) spectral fingerprints. One of the molecules, para-mercaptobenzoic acid (pMBA), is responsive to variations in surrounding pH, with higher sensitivity of the SEHRS spectrum18,28 compared to the SERS signature.29,30 Here, we make use of this high sensitivity and demonstrate SEHRS pH mapping to image endosomal pH inside the cells. Building on the recent successful separation of two-photon resonant SEHRS data,31 multivariate signal processing is applied to non-resonant SEHRS imaging datasets now, providing proof that the sensitivity of the new imaging approach is high enough for vibrational imaging of complex biosystems.
Materials and methods
Preparation of SEHRS labels
The reporter molecules para-mercaptobenzoic acid (pMBA) and 2-naphthalenethiol (2-NAT) were purchased from Sigma-Aldrich (Munich, Germany). A 10−2 M stock solution was prepared in ethanol and diluted in two steps at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, respectively, with sterilized Millipore water. Silver nitrate (Sigma-Aldrich) and sodium citrate (Merck) were used to synthesize citrate stabilized silver nanoparticles according to ref. 32. SEHRS labels carrying pMBA or 2-NAT as reporter molecules were prepared by mixing the silver nanoparticles with pMBA and 2-NAT, respectively, yielding the respective molecule at a concentration of 10−6 M.
100, respectively, with sterilized Millipore water. Silver nitrate (Sigma-Aldrich) and sodium citrate (Merck) were used to synthesize citrate stabilized silver nanoparticles according to ref. 32. SEHRS labels carrying pMBA or 2-NAT as reporter molecules were prepared by mixing the silver nanoparticles with pMBA and 2-NAT, respectively, yielding the respective molecule at a concentration of 10−6 M.
The silver nanoparticles were characterized by transmission electron microscopy, the average particle diameter was 115 ± 32 nm (Fig. S1a†), and is in accord with data from dynamic light scattering (Fig. S1b†). Absorbance spectra of all nanoparticle suspensions (Fig. S1c†) were recorded before application to the cell cultures. Absorbance spectra were measured using a V-670 double-beam ultraviolet-visible (UV-vis)/near-infrared (NIR) spectrophotometer (JASCO, Gross-Umstadt, Germany). The nanoparticles, the SERS labels, and their suspensions in the cell culture media were also dried on TEM grids and examined with a Tecnai G2 20 S-TWIN electron microscope at 200 kV (ZELMI TU Berlin, Germany).
Application of SEHRS labels in cells
Swiss albino mouse macrophages of cell line J774 (from DSMZ, Braunschweig, Germany) were maintained in Dulbecco's modified eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 1% ZellShield™ (Biochrom AG, Berlin, Germany) and grown under standard conditions (37 °C, 5% CO2).For the imaging experiments, the cells were grown on sterile cover-slips in six-well plates (Thermo Fisher Scientific, Waltham, USA). 120 μL of each SEHRS label were added to 880 μL of culture medium, respectively, resulting in a final nanoparticle concentration of 1.9 pM in culture medium. The cells were incubated with such a mixture for 3 h. When both SEHRS labels were applied, the cells were incubated with 2-NAT SEHRS labels in culture medium for 3 h, and after washing with PBS, they were incubated with pMBA SEHRS labels in the culture medium, also for 3 h. After incubation, the cells were washed thoroughly with PBS buffer, in which they were also kept during the measurements. We have investigated the cytotoxicity of silver nanoparticles33 and of the reporter molecules11 in similar experiments, and found that no toxicity was associated under these experimental conditions with either of them, specifically with the concentrations used here.
Surface enhanced hyper Raman scattering
Hyper Raman experiments were performed using a home-built epi-illuminated Raman micro-spectrometer as described in ref. 18 equipped with a 60× water immersion objective. 1064 nm, 7 ps laser pulses with a peak intensity of 6 × 109 W cm−2 on the sample (average power of 30 mW) were used for excitation of SEHRS in all experiments. Spectra were acquired over duration of 30 s. The focal spot of the laser beam was ∼1.08 μm in diameter. Maps from the cells were acquired with a step size of 2 microns. Bright field images were taken both before and after the measurements to confirm that no visible degradation of the cells occurred. In each condition (after incubation with each of the labels and with both labels, respectively), a minimum of 15 cells in a total of 3 incubation experiments were investigated, yielding a total of ∼45 mapped cells. The number of mapping points from cells incubated with 2-NAT, pMBA and both SEHRS labels was 280, 300 and 160, respectively. The spectral resolution was 3–6 cm−1 in the whole spectral range.Data pre-processing and analysis
Each spectrum was frequency calibrated using the spectrum of a toluene–acetonitrile mixture, and spikes were removed. All SEHRS spectra were analyzed using MatLab (The MathWorks, Inc., Ismaning, Germany) and Cytospec (Cytospec, Inc., Berlin, Germany) software. Hierarchical cluster analysis (HCA) was used to eliminate spectra without SEHRS signals. HCA was carried out using the Euclidean distance method and Ward's algorithm on vector normalized first derivatives over the whole spectral range of 300–2000 cm−1. The group of spectra remaining after this sorting procedure consisted of 180 data points obtained with the SEHRS labels carrying 2-NAT and pMBA, respectively, and spectra of their mixtures.Results and discussion
SEHRS labels in cell culture medium
In this work, two kinds of SEHRS labels, generated by attaching pMBA and 2-NAT, respectively, to silver nanoparticles, were administered to cultured macrophages by adding them to the cell culture medium. Before the experiments with the cells, the SEHRS labels were characterized using UV/vis spectroscopy, TEM and SEHRS. The absorbance spectra indicate the formation of nanoaggregates, evidenced by a small change in the absorbance above 825 nm when a reporter molecule is present, and when the SEHRS labels are mixed with the cell culture medium (Fig. S1c†). This could cause possible resonances that can be beneficial for the excitation of SEHRS using a wavelength of 1064 nm. The absorbance spectra do not change over time, indicating that the SEHRS labels are stabilized, analogous to similar silver nanoprobes used in cell culture media previously.33 In accord with this, the TEM micrographs (Fig. 1a) show that nanoaggregates of different shapes with diameters of ∼500 nm have formed before the mixture is added to the cells.Fig. 1b shows SEHRS spectra of the pMBA (Fig. 1b, red trace) and the 2-NAT (Fig. 1b, black trace) SEHRS labels in the culture medium obtained under the same experimental conditions as later on in the live cells. All bands in the spectrum of the pMBA SEHRS label in the culture medium are in good agreement with the SEHRS spectrum of pMBA obtained on different silver nanostructures and discussed in detail recently.18 The spectrum of the 2-NAT SEHRS label in culture medium displays bands that are also known from the SERS spectra of 2-NAT34,35 and of the same 2-NAT SERS labels composed of silver nanoparticles11 though with different relative intensities. The SEHRS spectra obtained from the SEHRS probes at the incubation concentration in the culture medium show very weak signals, due to the relatively small number of reporter molecules in this experiment. Even though the space requirements of pMBA,36 and also of 2-NAT on the silver surface yield complete coverage of the silver nanostructures with the molecules, the number of molecules in the ∼2 μm3 focal volume would amount to ∼1080, as on average less than one nanoparticle would reside there.
SEHRS spectral mapping in macrophage cells
After an incubation time of 3 hours with the SEHRS labels, spectra were obtained from the live cells by raster scanning areas with a step size of 2 microns. Fig. 1c shows three representative spectra extracted from mapping data of three individual cells after 3 hours incubation with pMBA (red traces), 2-NAT (black traces) and both SEHRS labels (blue traces), respectively. Compared to the signals obtained from the SEHRS labels in the culture medium, the SEHRS signatures are more enhanced when obtained inside the cells, due to the high concentrations of silver nanoparticles that are yielded inside the focal volume in regions with many endosomal structures9 and the beneficial plasmonic properties that are generated during the formation of different vesicular structures.10 The full coverage of the silver nanostructures with the reporter molecules and their interaction via the thiol group29,34 prevents an interaction of other molecules so that no intrinsic signals from the culture medium (Fig. 1b) or from the cells (Fig. 1c) are observed in the SEHRS spectra. This is different from several SERS hybrid probes that were reported,37,38 including probes containing 2-NAT.11In the spectra of the pMBA SEHRS labels, (Fig. 1c, red traces), the pH-independent18,28 bands at 1076, 1185, and 1585 cm−1 are marked in blue, the intensities that are dependent on pH are marked in red. In agreement with previous discussions,18,28 the example spectra of the pMBA SEHRS labels shown in Fig. 1c indicate a pH of 5.5 in the spectra displayed here, based on the relative intensities of the COO− stretching vibration at 1365 cm−1 and the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration at 1685 cm−1, respectively.18
O stretching vibration at 1685 cm−1, respectively.18
While the SEHRS spectra of pMBA on silver nanoparticles are well known,18 it should be mentioned here that the SEHRS spectrum of 2-NAT (Fig. 1c, black traces) has to the best of our knowledge not been discussed so far. Band positions and assignments in SEHRS spectra of the 2-NAT SEHRS label are shown in Table S1.† All bands in the spectrum of 2-NAT are also found in the one-photon excited SERS spectra.11,34,35 The SEHRS spectrum (Fig. 1c, black spectra) is dominated by the ring stretching vibrational modes at 1569, 1584 and 1624 cm−1. In contrast, in the one-photon excited case we and others have observed more enhanced ring deformation (1068 cm−1) and stretching (1380 cm−1) modes.11,34 The relatively high similarity of the SEHRS spectra of 2-NAT in Fig. 1c and the SERS spectra discussed in ref.11 and 34 suggests that the silver surface strongly lowers the symmetry of the 2-NAT molecule resulting in very similar SERS and SEHRS relative intensities.39 When the macrophages were incubated with both SEHRS labels, first for 3 h with 2-NAT and after washing of the cells with PBS buffer for 3 h with pMBA, the obtained spectral signature (Fig. 1c, blue traces) mainly resembles the spectrum of the 2-NAT label, with which the cells were incubated first. Due to the sequential incubation, at the time of the Raman experiment, the 2-NAT SEHRS labels have been processed in the cells twice as long as the pMBA labels. As discussed above (Fig. 1 and S1†) the SEHRS labels in the culture medium (Fig. 1a) form nanoaggregates of ∼500 nm, corresponding to 5–9 nanoparticles, which can be phagocytosed by the J774 cells. From nanotomography experiments conducted with the same macrophage cell line under very similar incubation conditions for 3 h (ref. 33) and for longer times40 we know that the morphology of silver nanoaggregates changes inside the phagosomes over time. It is very likely, that the longer processing time of the 2-NAT SEHRS labels leads to nanoaggregate geometries that support stronger SEHRS enhancement, yielding higher signals from the 2-NAT labels than from the pMBA labels. Considering that in each focal volume both kinds of SEHRS labels contribute to the spectrum, either contained in different phagosomes or in one vesicular structure,10,11 stronger enhancement of the SEHRS spectrum of 2-NAT will result in the high similarity to the spectrum of the 2-NAT labels (compare blue and black traces in Fig. 1c).
Using the SEHRS mapping data sets, chemical images were created. Fig. 2 shows selected chemical maps of the macrophages containing pMBA (Fig. 2a–c), 2-NAT (Fig. 2d–f) and both (Fig. 2g–i) SEHRS labels. Each pixel in a map represents one hyper Raman spectrum. Band intensities were calculated from the integral of one or more defined bands of the reporter molecules as indicated in the figure legend of Fig. 2. In the case of pMBA, the integral of the ring stretching band at 1576 cm−1, and in the case of 2-NAT, the integrals of the bands at 1569, 1584, and 1624 cm−1 were used to create chemical maps and to localize both SEHRS labels. Based on such a univariate analysis of the SEHRS spectral maps, it is not possible to identify the distribution of the different reporters in those cells that have internalized both SEHRS labels (Fig. 2g–i). In accord with the individual example spectra discussed above (Fig. 1c, bottom spectra), the chemical images indicate mainly 2-NAT SEHRS spectra in the perinuclear regions of the cells. As the phagosomes/endosomes will move from the periphery towards the nucleus,9,41,42 the amount of older, 2-NAT labels-containing phago/-lysosomes could be higher there, and/or more intense signals are to be expected due to different plasmonic properties/aggregate morphology in the older lysosomal structures. No signals were detected in the regions of the nuclei (Fig. 2, all maps) due to the size restrictions imposed by the nuclear pore complex43 that will not allow the SEHRS labels to pass the nuclear membrane.
 | ||
| Fig. 2 Bright-field micrographs of J774 macrophage cells containing the two SEHRS labels, with overlaid chemical maps obtained from the SEHRS data. (a–c) 2-NAT label, (d–f) pMBA label, (g–i) both labels. The chemical maps show the integrated signals assigned to the ring stretching vibrations of 2-NAT and pMBA, respectively. In the spectra of 2–NAT (a–c and g–i), this corresponds to the integral in the range 1520–1680 cm−1, in pMBA (d–f) this is covered by the integral in the spectral range 1550–1620 cm−1 (baseline corrected), for example spectra compare Fig. 1c. Step width in the SEHRS maps was 2 microns, scale bar: 4 microns. | ||
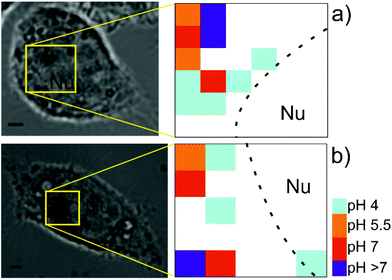
As a specific case of a chemical map, it is also possible to calculate pH maps from the obtained SEHRS spectra of pMBA. Before the experiments in the cells, calibration spectra of the pMBA SEHRS labels were measured for different pH values between 2 and 10. The bands that were used to determine the pH are highlighted with stars in Fig. 1c (red: pH-dependent, blue: pH-independent bands). Calibration curves using the ratio of the bands at 1365 and 1076 cm−1 (Fig. S2a†) as well as at 1365 and 1585 cm−1 (Fig. S2b†), discussed in detail previously regarding their sensitivity towards changes in pH,18 demonstrate the operation range of the SEHRS labels when they are used as pH nanosensors. Acidic pH values can be clearly distinguished, and the SEHRS spectra should be specifically suited for the differentiation of pH values in early to late phagosomes/endosomes and lysosomes. The pH value inside the cells was determined from comparing the two intensity ratios in each spectrum with the calibration curves in Fig. S2.† Each of the maps in Fig. 3a and b shows average pH values determined from the two calibrations using the two different bands. Fig. 3a and b also indicate the positions of the maps in the corresponding bright field images. After incubation for 3 hours, the pH in the macrophages shows a variation between pH 4 and pH 7.5. The highest pH values are denoted as >7. In the white regions of the maps, pH could not be determined due to the absence of spectra with sufficient signal. Examples of corresponding individual SEHRS spectra at different pH are shown in Fig. S2c.† At higher pH in the cells, in all spectra, the band intensities of vibrations associated with the carboxyl group (marked with blue in the pMBA spectra in Fig. 1c) change relative to those of the aromatic ring (marked in red in Fig. 1c). Upon acidification, the intensity of the COO− band at 1365 cm−1 decreases, and the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration at 1685 cm−1 increases, in good agreement with previous discussions.18 The examples in Fig. 3 show clearly that the very low pH values are found in those regions close to the nucleus, which likely contain relatively mature phagosomes and lysosomes, and that regions of less acidic and neutral pH are not in direct proximity of the nucleus. Although positions of varying pH were mapped using pMBA SERS nanosensors in endosomes of other cell lines before,44–46 low pH values could not be discriminated easily,44,45 due to the lower sensitivity of SERS compared to that of SEHRS, specifically in the acidic pH range.18,28 Combining the high sensitivity of the SEHRS spectra within the acidic pH range with mapping as demonstrated in Fig. 3 enables spatially resolved investigations of the acidification process, in this case in the phagolysosomal system, differentiating pH within the perinuclear region.
O stretching vibration at 1685 cm−1 increases, in good agreement with previous discussions.18 The examples in Fig. 3 show clearly that the very low pH values are found in those regions close to the nucleus, which likely contain relatively mature phagosomes and lysosomes, and that regions of less acidic and neutral pH are not in direct proximity of the nucleus. Although positions of varying pH were mapped using pMBA SERS nanosensors in endosomes of other cell lines before,44–46 low pH values could not be discriminated easily,44,45 due to the lower sensitivity of SERS compared to that of SEHRS, specifically in the acidic pH range.18,28 Combining the high sensitivity of the SEHRS spectra within the acidic pH range with mapping as demonstrated in Fig. 3 enables spatially resolved investigations of the acidification process, in this case in the phagolysosomal system, differentiating pH within the perinuclear region.
SEHRS hyperspectral mapping
To utilize as many as possible of the bands in the SEHRS spectra of pMBA and 2-NAT to generate image contrast, we apply multivariate methods to map the distribution of the different SEHRS labels and their mixtures. First, hierarchical cluster analysis (HCA) was used to eliminate spectra without sufficiently high SEHRS signals from the mapping data sets. Fig. 4 displays an example of a chemical map obtained with 2-NAT SEHRS labels. In the HCA map (Fig. 4b), each spectrum is assigned to either one of two classes. Class average spectra (Fig. 4c) revealed one class per map where no spectrum displayed any signals, e.g., class 2 in the example shown in Fig. 4. The spectra of the other respective class were extracted from each data set and were used for further analysis.In order to account for cell-to-cell variation, spectra from all kinds of data sets (of cells with 2–NAT SEHRS label, with pMBA SEHRS label, and with both labels) were analyzed in one principal component analysis (PCA). In a PCA, the data points are projected into a new, variance-weighted coordinate system, such that the greatest variance by any projection comes to lie on the first coordinate. Thereby, for example, the direction of greatest variance (first principal component), can be used to generate contrast in a SEHRS image, using the whole spectral range from 300 to 1800 cm−1. The scores plot from a PCA with all types of data from many cells (Fig. 5a) indicates the formation of groups of spectra that are similar or dissimilar with respect to the first and the second principal components. Each point in Fig. 5a represents a single SEHRS spectrum from one of all the data sets. The red symbols are spectra from the mapping experiments with the pMBA SEHRS labels, the blue ones from cells containing 2-NAT SEHRS labels, and the green symbols from the data sets of cells that were incubated with both SEHRS labels. The scores of the PCA of the SEHRS spectra (Fig. 5a) yield clear separation of the pMBA spectra (red symbols) from the 2-NAT (blue symbols) and the spectra from cells with two SEHRS labels (green symbols) along PC1 (dashed line in Fig. 5a). The loadings indicate the bands in the spectra that are responsible for the variance in all the combined data sets (Fig. 5b). In accord with the separation of pMBA and 2-NAT along PC1 (Fig. 5a), the loading of PC1 (Fig. 5b, upper trace) contains signals at 845 cm−1 and 1160 cm−1 that originate only from the 2-NAT molecule, and also the features at 1569 and 1624 cm−1 in the loading of PC1 can be assigned to 2-NAT. PC2 (Fig. 5b, lower trace) represents the main variance within the group of data obtained with the pMBA SEHRS labels (red symbols in Fig. 5a). Accordingly, in the loading of PC2, the vibrational mode at 1585 cm−1 is very pronounced, as are also typical pMBA signals at 1076 and 1158 cm−1 (bottom trace in Fig. 5b). The variance in the pMBA spectra that is represented by PC2 is in accord with the sensitivity of the SEHRS spectrum to changes in pH18 and the fact that all different data from early endosomes with high pH and those that are older and have low pH are included in the analysis.
 | ||
| Fig. 5 Results of the principal component analysis (PCA) using the SEHRS mapping data sets of 15 macrophage cells, containing 2-NAT or pMBA, or both SEHRS labels, and example of a hyperspectral map of one data set using. The vector-normalized first derivatives of the pre-selected SEHRS spectra (see Fig. 4) were subjected to a PCA using the spectral range 300–1800 cm−1. (a) Scores plot of the first two principal components (PCs). (b) Corresponding loadings and variance that is explained by each PC in percent. (c) Based on the score of the first PC shown in (a), false-color SEHRS maps can be reconstructed for each individual data set, here from a sample containing both 2-NAT and pMBA SEHRS labels. The dashed line in (a) demarcates a threshold score value of the first PC that was used for the differentiation between different types of spectra (for examples see Fig. 1). The colors indicate the different assignments of the spectra, red: pMBA-like, blue: 2-NAT-like. | ||
Based on the first and second PC scores, the spectra of the 2-NAT labels (Fig. 5a, blue symbols) and spectra obtained from cells with both labels (Fig. 5a, green symbols) are indistinguishable. In order to identify those spectra in the data sets with both labels that represent the pMBA signature and hence indicate the presence of phago/-lysosomes containing the pMBA SEHRS labels, the PC1 scores value can be used. Based on the scores of PC1, using the scores of the SEHRS data from the pMBA label experiments as reference, a threshold was defined (Fig. 5a, dashed line). According to this threshold score, all mapping points from the cells containing both labels were assigned along PC1 to ‘pMBA-like’ or ‘2-NAT-like’ spectra. False-color SEHRS maps were generated, an example is shown in Fig. 5c. The low number of ‘pMBA-like’ red pixels in this hyperspectral map is typical and in accord with the resemblance of most of the spectra with 2-NAT as discussed for the univariate analysis of the spectra in the beginning (Fig. 1c). It is important to note that the contrast in the map depends only on the definition of the threshold, and that in the example used here, ‘reference’ spectra from other experiments were available, but that they are not necessary. Here, we aimed at the discrimination of two non-resonant SEHRS labels. Nevertheless, depending on the underlying chemical or biological problem, PCs can be chosen depending on the type of variance they represent, and imaging can also be based on more than one PC,47–49 for example, if additional intrinsic information from the cell can be considered as well, as is the case in some SERS nanoprobes.11
Conclusions
We have shown SEHRS based hyperspectral imaging in live macrophage cells incubated with non-resonant SEHRS labels that carry the reporter molecules 2-NAT and pMBA, respectively. The spectra of both reporter molecules were obtained in the cell culture medium and inside intracellular vesicles after phagocytosis in live cells. Chemical mapping of both reporter signatures indicated an accumulation of the SEHRS labels in the perinuclear region, suggesting their accumulation in phago/lysosomes. The SEHRS spectra of pMBA that were used as pH meter verified the localization in lysosomes close to the nuclei in chemical pH maps. The SEHRS spectra of pMBA from the cellular interior were in good agreement with spectra obtained in solution previously.18 The two-photon excited SEHRS spectrum of 2-NAT obtained inside and outside of cells strongly resembles the one-photon excited SERS spectrum of the molecule.11,34The unambiguous differentiation between changing acidic pH values using two bands in the SEHRS spectrum of the pMBA reporter molecule has only been proposed for individual SEHRS microspectra so far18,28 and was successfully extended to a two-photon imaging approach here that has similar or higher sensitivity than two-photon fluorescence.24 It will enable further studies of uptake and acidification mechanisms at high lateral resolution in live cells, which is of high impact for a large number of biomedical questions, ranging from pathogen interaction50–52 to therapeutic approaches.53,54
Different from recent proof-of-principle of multivariate SEHRS mapping,31 the SEHRS spectra that were used for hyperspectral imaging here were excited off-resonance with electronic transitions in the reporter molecules. This puts SEHRS hyperspectral mapping of cellular biomolecules within reach, as the SEHRS cross sections of the reporters used here18 are very similar to those of typical biomolecules such as nucleobases19,24 or amino acids.20 In conclusion, we have demonstrated multivariate microspectroscopic mapping using two-photon excited SEHRS spectra. SEHRS hyperspectral mapping provides a sensitive tool for the spatially resolved vibrational analysis of complex biological systems, including the characterization of their interaction with nanomaterials, in the fields of bioanalytical chemistry, biomedicine, and biotechnology.
Acknowledgements
We thank Sören Selve (ZELMI, Technical University Berlin) for the TEM measurements, Harald Kneipp for valuable discussions and support in setting up experiments, Daniela Drescher and Tina Büchner for advice regarding cell culture, and Peter Lasch (Cytospec) for providing Cytospec software. Financial Support by ERC Starting Grant no. 259432 (MULTIBIOPHOT) to J. K. and M. G., by DFG (GSC 1013 SALSA) to Z. H. and V. Ž., and an FCI Chemiefonds Fellowship to F.M. is gratefully acknowledged.References
- L. A. Austin, B. Kang and M. A. El-Sayed, Nano Today, 2015, 10, 542–558 CrossRef CAS.
- J. Kneipp, ACS Nano, 2017, 11, 1136–1141 CrossRef CAS PubMed.
- P. Nativo, I. A. Prior and M. Brust, ACS Nano, 2008, 2, 1639–1644 CrossRef CAS PubMed.
- A. W. Sanders, K. M. Jeerage, C. L. Schwartz, A. E. Curtin and A. N. Chiaramonti, ACS Nano, 2015, 9, 11792–11799 CrossRef CAS PubMed.
- C. Hagen, S. Werner, S. Carregal-Romero, A. N. Malhas, B. G. Klupp, P. Guttmann, S. Rehbein, K. Henzler, T. C. Mettenleiter, D. J. Vaux, W. J. Parak, G. Schneider and K. Grunewald, Ultramicroscopy, 2014, 146, 46–54 CrossRef CAS PubMed.
- D. Drescher, C. Giesen, H. Traub, U. Panne, J. Kneipp and N. Jakubowski, Anal. Chem., 2012, 84, 9684–9688 CrossRef CAS PubMed.
- A. G. Tkachenko, H. Xie, Y. L. Liu, D. Coleman, J. Ryan, W. R. Glomm, M. K. Shipton, S. Franzen and D. L. Feldheim, Bioconjugate Chem., 2004, 15, 482–490 CrossRef CAS PubMed.
- A. K. Oyelere, P. C. Chen, X. Huang, I. H. El-Sayed and M. A. El-Sayed, Bioconjugate Chem., 2007, 18, 1490–1497 CrossRef CAS PubMed.
- D. Drescher and J. Kneipp, Chem. Soc. Rev., 2012, 41, 5780–5799 RSC.
- J. Kneipp, H. Kneipp, M. McLaughlin, D. Brown and K. Kneipp, Nano Lett., 2006, 6, 2225–2231 CrossRef CAS PubMed.
- A. Matschulat, D. Drescher and J. Kneipp, ACS Nano, 2010, 4, 3259–3269 CrossRef CAS PubMed.
- J. Ando, K. Fujita, N. I. Smith and S. Kawata, Nano Lett., 2011, 11, 5344–5348 CrossRef CAS PubMed.
- M. Aioub and M. A. El-Sayed, J. Am. Chem. Soc., 2016, 138, 1258–1264 CrossRef CAS PubMed.
- W. R. Zipfel, R. M. Williams and W. W. Webb, Nat. Biotechnol., 2003, 21, 1369–1377 CrossRef CAS PubMed.
- K. König, J. Microsc., 2000, 200, 83–104 CrossRef.
- T. Itoh, Y. Ozaki, H. Yoshikawa, T. Ihama and H. Masuhara, Appl. Phys. Lett., 2006, 88, 084102 CrossRef.
- J. C. Hulteen, M. A. Young and R. P. Van Duyne, Langmuir, 2006, 22, 10354–10364 CrossRef CAS PubMed.
- M. Guhlke, Z. Heiner and J. Kneipp, Phys. Chem. Chem. Phys., 2015, 17, 26093–26100 RSC.
- F. Madzharova, Z. Heiner, M. Guhlke and J. Kneipp, J. Phys. Chem. C, 2016, 120, 15415–15423 CAS.
- F. Madzharova, Z. Heiner and J. Kneipp, J. Phys. Chem. C, 2017, 121, 1235–1242 CAS.
- F. Madzharova, Z. Heiner and J. Kneipp, Chem. Soc. Rev., 2017 10.1039/c7cs00137a.
- V. N. Denisov, B. N. Mavrin and V. B. Podobedov, Phys. Rep., 1987, 151, 1–92 CrossRef.
- H. Kneipp, K. Kneipp and F. Seifert, Chem. Phys. Lett., 1993, 212, 374–378 CrossRef CAS.
- J. Kneipp, H. Kneipp and K. Kneipp, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 17149–17153 CrossRef CAS PubMed.
- J. Mullin, N. Valley, M. G. Blaber and G. C. Schatz, J. Phys. Chem. A, 2012, 116, 9574–9581 CrossRef CAS PubMed.
- J. T. Golab, J. R. Sprague, K. T. Carron, G. C. Schatz and R. P. V. Duyne, J. Chem. Phys., 1988, 88, 7942–7951 CrossRef CAS.
- N. Valley, L. Jensen, J. Autschbach and G. C. Schatz, J. Chem. Phys., 2010, 133, 054103 CrossRef PubMed.
- J. Kneipp, H. Kneipp, B. Wittig and K. Kneipp, Nano Lett., 2007, 7, 2819–2823 CrossRef CAS PubMed.
- A. Michota and J. Bukowska, J. Raman Spectrosc., 2003, 34, 21–25 CrossRef CAS.
- S. W. Bishnoi, C. J. Rozell, C. S. Levin, M. K. Gheith, B. R. Johnson, D. H. Johnson and N. J. Halas, Nano Lett., 2006, 6, 1687–1692 CrossRef CAS PubMed.
- M. Guhlke, Z. Heiner and J. Kneipp, Phys. Chem. Chem. Phys., 2016, 18, 14228–14233 RSC.
- P. C. Lee and D. Meisel, J. Phys. Chem., 1982, 86, 3391–3395 CrossRef CAS.
- D. Drescher, P. Guttmann, T. Büchner, S. Werner, G. Laube, A. Hornemann, B. Tarek, G. Schneider and J. Kneipp, Nanoscale, 2013, 5, 9193–9198 RSC.
- R. A. Alvarez-Puebla, D. S. Dos Santos Jr. and R. F. Aroca, Analyst, 2004, 129, 1251–1256 RSC.
- R. A. Alvarez-Puebla, E. Arceo, P. J. G. Goulet, J. J. Garrido and R. F. Aroca, J. Phys. Chem. B, 2005, 109, 3787–3792 CrossRef CAS PubMed.
- Y. Yu, S. Handa, T. Yajima and M. Futamata, Chem. Phys. Lett., 2013, 560, 49–54 CrossRef CAS.
- J. Kneipp, H. Kneipp, A. Rajadurai, R. W. Redmond and K. Kneipp, J. Raman Spectrosc., 2009, 40, 1–5 CrossRef CAS.
- J. Kneipp, H. Kneipp, W. L. Rice and K. Kneipp, Anal. Chem., 2005, 77, 2381–2385 CrossRef CAS PubMed.
- W.-hui Yang, J. Hulteen, G. C. Schatz and R. P. V. Duyne, J. Chem. Phys., 1996, 104, 4313–4323 CrossRef CAS.
- D. Drescher, I. Zeise, H. Traub, P. Guttmann, S. Seifert, T. Büchner, N. Jakubowski, G. Schneider and J. Kneipp, Adv. Funct. Mater., 2014, 24, 3765–3775 CrossRef CAS.
- J. Huotari and A. Helenius, EMBO J., 2011, 30, 3481–3500 CrossRef CAS PubMed.
- S. D. Conner and S. L. Schmid, Nature, 2003, 422, 37–44 CrossRef CAS PubMed.
- C. M. Feldherr and D. Akin, J. Cell Biol., 1990, 111, 1–8 CrossRef CAS PubMed.
- C. E. Talley, L. Jusinski, C. W. Hollars, S. M. Lane and T. Huser, Anal. Chem., 2004, 76, 7064–7068 CrossRef CAS PubMed.
- J. Kneipp, H. Kneipp, B. Wittig and K. Kneipp, J. Phys. Chem. C, 2010, 114, 7421–7426 CAS.
- A. Jaworska, L. E. Jamieson, K. Malek, C. J. Campbell, J. Choo, S. Chlopicki and M. Baranska, Analyst, 2015, 140, 2321–2329 RSC.
- P. Lasch and D. Naumann, Cell. Mol. Biol., 1998, 44, 189–202 CAS.
- J. Kneipp, P. Lasch, E. Baldauf, M. Beekes and D. Naumann, Biochim. Biophys. Acta, Mol. Basis Dis., 2000, 1501, 189–199 CrossRef CAS.
- M. Hedegaard, C. Matthäus, S. Hassing, C. Krafft, M. Diem and J. Popp, Theor. Chem. Acc., 2011, 130, 1249–1260 CrossRef CAS.
- J. R. Schnell and J. J. Chou, Nature, 2008, 451, 591–U512 CrossRef CAS PubMed.
- A. Di, M. E. Brown, L. V. Deriy, C. Y. Li, F. L. Szeto, Y. M. Chen, P. Huang, J. K. Tong, A. P. Naren, V. Bindokas, H. C. Palfrey and D. J. Nelson, Nat. Cell Biol., 2006, 8, 933–U952 CrossRef CAS PubMed.
- L. M. Shaughnessy, A. D. Hoppe, K. A. Christensen and J. A. Swanson, Cell. Microbiol., 2006, 8, 781–792 CrossRef CAS PubMed.
- J. Su, F. Chen, V. L. Cryns and P. B. Messersmith, J. Am. Chem. Soc., 2011, 133, 11850–11853 CrossRef CAS PubMed.
- S. P. Parihar, R. Guler, R. Khutlang, D. M. Lang, R. Hurdayal, M. M. Mhlanga, H. Suzuki, A. D. Marais and F. Brombacher, J. Infect. Dis., 2014, 209, 754–763 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Characterization of the SEHRS labels, determination of pH values, and band assignments of 2-NAT SEHRS label. See DOI: 10.1039/c7nr02762a |
| This journal is © The Royal Society of Chemistry 2017 |