Open Access Article
Open Access ArticleHost–guest driven ligand replacement on monodisperse inorganic nanoparticles†
B.
Shirmardi Shaghasemi
a,
E. S.
Dehghani
b,
E. M.
Benetti
b and
E.
Reimhult
 *a
*a
aInstitute for Biologically Inspired Materials, Department of Nanobiotechnology, University of Natural Resources and Life Sciences, Vienna, Austria. E-mail: erik.reimhult@boku.ac.at
bLaboratory for Surface Science and Technology, Department of Materials, ETH Zürich, Zürich, Switzerland
First published on 12th June 2017
Abstract
We demonstrate that crown ether-assisted ligand replacement on Fe3O4 NPs using halide salts leads to quantitative stripping of an existing stabilizer shell with unprecedented (complete) efficiency; this allows subsequent re-grafting of functional ligands at maximal surface density. The mechanism of the anion-driven ligand replacement is elucidated by varying the halide salt and the versatility by varying the re-grafted ligand.
Inorganic nanoparticles (NPs) have many applications in different areas including diagnostic,1 multimodal imaging,2 catalysis3 and electronics-optoelectronics.4 Such opportunities have led to the development of methods to synthesize a large variety of nanocrystals of well-defined size and uniform shape.5 The size and morphology of NPs is precisely tunable by applying stabilizing agents (mostly oleic acid (OA)) with high affinity to the NP surface during synthesis.6 These capping molecules are not suitable ligands for most of the subsequent applications, which e.g. require a close access to the inorganic cores (via thin organic7 or inorganic8 shells) for the efficient charge transfer between semiconductor NPs,9 or densely grafted organic shells for colloidal stability in physiological media10 or functional drug delivery assemblies. This has led to an enormous effort to control the grafting and exchange of ligand shells on inorganic nanoparticles.11
Commonly applied routes to efficiently replace the capping oleic acid layer include the use of adsorbates with high affinity to the surface of NPs, e.g. chalcogenide for quantum dots12 or disodium 4,5-dihydroxy-1,3-benzenedisulfonate for Fe3O4 NPs.13 These processes are typically irreversible, with no option for further surface modification, such as the re-grafting of a molecularly tailored surface modifier that presents ideal physico-chemical properties for the application medium to which the NPs subsequently will be subjected. Addressing this problem by directly replacing OA with the desired functional ligand equipped with a high-affinity anchor binding to the core is extremely challenging and requires multi-step protocols to achieve complete functionalization.14 Additionally, the displacement of the existing ligands can be achieved by applying a large excess of anions such as BF4− (using NOBF4 or Et3BF4); this approach exploits the electrostatic interactions between the ions and the NP surface, followed by the final displacement of the BF4− layer by high-affinity ligands featuring the desired chemistry.14,15 The reason for using these exotic salts is that their weak association achieve sufficient reactive free anion to favour surface displacement of strongly adhering ligands such as oleic acid.
We demonstrate that host–guest coordination complexes of crown ethers and common, “laboratory bench” metal–halide salts, such as NaF and KF generate “naked” F− anions driving a quantitative and highly efficient ligand replacement on the surface of Fe3O4 NPs. Nuclear magnetic resonance (NMR), infrared spectroscopy (ATR-FTIR), thermogravimetric analysis (TGA) and X-ray photoelectron spectroscopy (XPS) are used to prove the quantitative removal of oleic acid from the surface of spherical, single-crystalline, superparamagnetic iron oxide nanocrystals synthesized with the method of Hyeon et al.16 The stripped NPs are subsequently re-grafted by organic ligands presenting different physical and chemical properties; this demonstrates how our simple ligand replacement protocol is a powerful new tool for creating monodisperse NPs decorated with precise shell chemistries of highest purity.
15-crown-5 or 18-crown-6 (250 mg) and NaF (50 mg) were dissolved in 1 ml water and added to a mixture of 10 ml hexane solution of NPs (25 mg ml−1) and 7 ml isopropanol. Crown ether strongly coordinates Na+ from added salts to obtain the naked nucleophilic anion (Scheme 1). The naked anion can displace the deprotonated OA at the NP surface; the latter possibly weakly coordinating to the cation-crown ether host–guest complex. Stripped NPs precipitate immediately after gentle shaking of the NP dispersion and can subsequently be spun down via centrifugation after addition of isopropanol to decrease the surface tension of the solvent interface during centrifugation. Precipitated NPs were washed several times with hexane, isopropanol and water, respectively to remove residues of OA, salt and crown ether.
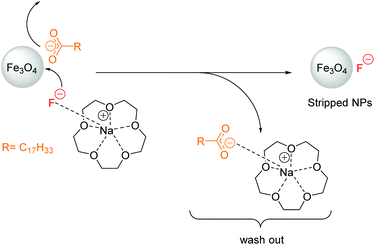 | ||
| Scheme 1 Mechanism of oleic acid stripping demonstrated on Fe3O4 NPs using excess of 15-crown-5 and NaF. | ||
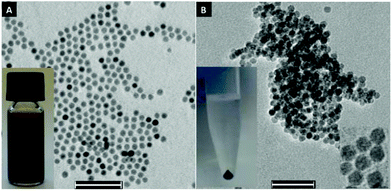
Fig. 1A shows TEM images of NPs as-synthesized. These nanoparticles have a dense shell of OA and are well dispersed on the grid. After stripping by 15-crown-5 and NaF, the particles display no change in shape and size, but the absence of an organic shell leads to dense aggregates after drying on the grid (Fig. 1B).
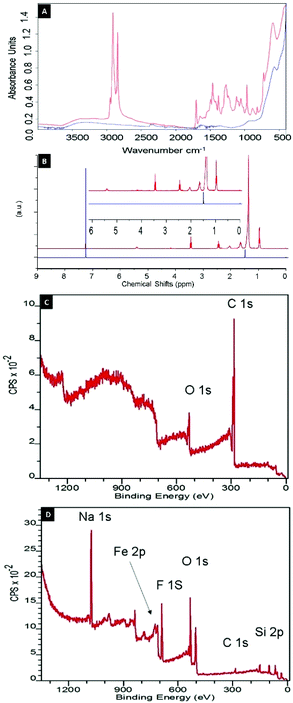
The complete removal of OA represents a crucial step for NP functionalization, since precisely knowing and controlling the ligand stoichiometry is a fundamental requirement for many applications.17 The complete removal of OA from Fe3O4 NPs was confirmed by infrared spectroscopy (ATR-FTIR), nuclear magnetic resonance spectroscopy (NMR), thermogravimetric analysis (TGA) and X-ray photoelectron spectroscopy (XPS). FTIR spectra of as-synthesized and stripped NPs in Fig. 2A show that the C–H stretching vibrations at 2800–3000 cm−1 and the characteristic peaks18 of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O at 1600–1715 cm−1 disappeared completely after treatment with 15-crown-5/NaF.
O at 1600–1715 cm−1 disappeared completely after treatment with 15-crown-5/NaF.
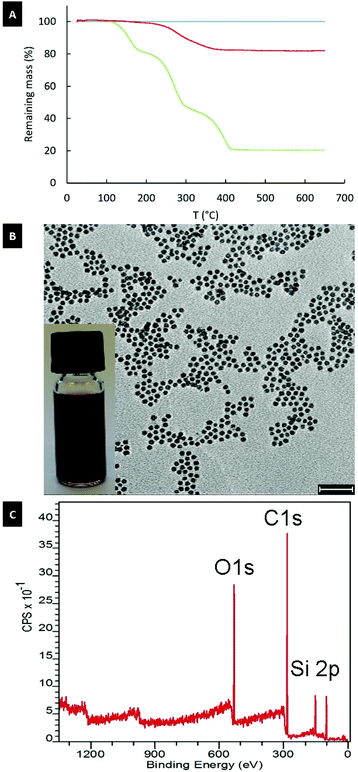
Since superparamagnetic Fe3O4 nanoparticles change the relaxation time and broaden the NMR spectrum, particles were dissolved in concentrated HCl and the shell material was extracted with chloroform. The resulting 1H-NMR profiles neither showed a signal from OA nor from crown ether, which confirms the quantitative removal of the OA ligand and of the excess host–guest complex (Fig. 2B). Finally, the TGA data in Fig. 3 (blue line) shows that no organic content could be found in the stripped NP sample.
The stripping process was tested with different anions including F−, Cl−, Br− and I−. All salts lead to precipitation of NPs in the aqueous phase in the presence of crown ether. The phase transfer and precipitation is caused by the change of solubility of the NPs upon removal of the OA shell. However, complete removal of organics was uniquely observed by TGA for F− as anion (Fig. S1 and Table S1†) and precipitation of the NPs was complete already after 30 seconds. In contrast, even after 24 hours of incubation, detectable traces of organic ligand were recorded by TGA, FTIR and NMR on NPs treated with Cl−, Br− and I− analogues (Fig. S2 and Table S2†).
The chemical composition of the nanoparticle coating was confirmed by XPS after each modification step (Tables S3–S5†). As shown in Fig. 2C, the XPS spectrum of OA-coated NPs exhibits a single C 1s peak at 285 eV, which was attributed to carbon atoms of OA, whereas the C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) O ratio agrees well with the chemical composition of this ligand. After stripping of OA-capped NPs with 15-crown-5/NaF, the intensity of the C 1s peak in the XPS spectrum (Fig. 2D) becomes negligible, confirming the observations derived from NMR, ATR-FTIR and TGA analyses, and further demonstrating the complete removal of OA from the surface of the NPs. Simultaneously, the appearance of the characteristic signals of Na 1s and F 1s peaks at 1071 and 684 eV, respectively, indicate the presence of salt from the stripping process. However, a F
O ratio agrees well with the chemical composition of this ligand. After stripping of OA-capped NPs with 15-crown-5/NaF, the intensity of the C 1s peak in the XPS spectrum (Fig. 2D) becomes negligible, confirming the observations derived from NMR, ATR-FTIR and TGA analyses, and further demonstrating the complete removal of OA from the surface of the NPs. Simultaneously, the appearance of the characteristic signals of Na 1s and F 1s peaks at 1071 and 684 eV, respectively, indicate the presence of salt from the stripping process. However, a F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Na stoichiometric ratio of 2
Na stoichiometric ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 suggests the presence of excess fluoride anions covering the NP cores (Table S4†). Importantly, the stripping did not result in a change to the nanoparticle cores as demonstrated by X-ray diffraction (XRD) measurements (Fig. S3†). XRD showed the presence of Fe2O3/Fe3O4 with the same diffraction peak positions and widths before and after the stripping process.
1 suggests the presence of excess fluoride anions covering the NP cores (Table S4†). Importantly, the stripping did not result in a change to the nanoparticle cores as demonstrated by X-ray diffraction (XRD) measurements (Fig. S3†). XRD showed the presence of Fe2O3/Fe3O4 with the same diffraction peak positions and widths before and after the stripping process.
Interestingly, the efficiency of OA-stripping increased with the decrease of size of the anion. Since the smallest anion, F−, is a rather hard base, it should be easily desorbed from the NP surface in protic solvent (protons being a hard acid).19 Hence, in water also at neutral pH the F− layer could be displaced from the NP surface, yielding colloids without any ligand particularly susceptible to the re-grafting of functional ligands with high affinity for the Fe3O4 surface.
In particular, following the successful removal of OA using 15-crown-5/NaF and phase transfer to water to also remove F− ligand, the bare NPs were functionalized through irreversible grafting of nitrodopamide (ND)-bearing ligands, generating a dense shell presenting the desired chemistry (Scheme 2).20 The completely stripped NPs were first collected and added together with nitrodopamide-modified ligand to a DMF![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (3
MeOH (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) mixture; the MeOH was evaporated with rotavapor, NPs precipitated by addition of acetone, collected, repeatedly re-suspended in acetone and centrifuged to remove excess ligand (see ESI† for details).
1) mixture; the MeOH was evaporated with rotavapor, NPs precipitated by addition of acetone, collected, repeatedly re-suspended in acetone and centrifuged to remove excess ligand (see ESI† for details).
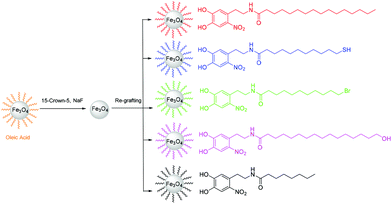 | ||
| Scheme 2 Nitrodopamide-anchored ligands with different lengths and terminal groups re-grafted on NPs stripped by 15-crown-5/NaF. | ||
The TGA result for 11 nm superparamagnetic iron oxide nanoparticles re-grafted with palmitoyl-nitrodopamide (PNDA) using this approach is shown in Fig. 3 (red line). PNDA constitutes an extraordinary 19% total organic content measured by TGA on the NP samples re-grafted and extensively purified of excess ligand (Fig. 3A and Table S4†). This corresponds to 3.1 molecules per nm2 and is higher than achieved after an extensive multi-step protocol for direct ligand exchange (2.7 molecules per nm2) for 3.5 nm iron oxide nanoparticles.21 This grafting density closely corresponds to the theoretical maximum grafting density expected for nitrodopamine on iron oxide.21,22 TEM images after re-grafting with PNDA (Fig. 3B) show no change in size or shape compared to the initial OA-capped NPs. Also, XRD spectra showed no change to the particle cystal structure (Fig. S3†). Iron oxide nanoparticles re-grafted with PNDA were perfectly suspended in THF as expected (Fig. 3B inset). DLS on particles before stripping (OA-capped in toluene) and after re-grafting with PNDA in THF showed the nanoparticle size virtually unchanged (Fig. S4†). This demonstrates that individually modified and colloidally stable nanoparticles were obtained by dense ligand re-grafting. Finally, in the XPS spectrum of stripped NPs re-grafted with PNDA (Fig. 3C), the ratio of C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) O is in good agreement with the stoichiometric ratio of the new ligand and no signals corresponding to Na 1s and F 1s are found. The method of stripping and re-grafting was independent of size in the superparamagnetic nanoparticle range as demonstrated additionally on e.g. cores with a diameter of 4.2 nm (Fig. S5†).
O is in good agreement with the stoichiometric ratio of the new ligand and no signals corresponding to Na 1s and F 1s are found. The method of stripping and re-grafting was independent of size in the superparamagnetic nanoparticle range as demonstrated additionally on e.g. cores with a diameter of 4.2 nm (Fig. S5†).
We also re-grafted other nitrodopamide-anchored ligands with various lengths and terminal groups onto NPs stripped by 15-crown-5/NaF using the same procedure as for PNDA (Scheme 2). The chosen ligand end-functionalities (–SH, –Br, –OH) are useful for further chemical modification via disulfide coupling, atom transfer radical polymerization (ATRP) or esterification, respectively. The functionalized NPs all had a grafting density of ∼3 ligands per nm2 measured by TGA (Table S2†). Thus, optimal re-grafting of NDA-anchored non-polar ligands at the theoretically achievable maximum grafting density onto stripped NPs was consistently achieved independent of chemical functionality.
Conclusions
In summary, we report the first application of crown ethers to completely remove strongly complexed ligands from the surface of NPs at room temperature by simple phase transfer to aqueous suspension using readily available sodium or potassium halide salts. A strong dependence on anion size was observed for displacement of oleic acid. Complete removal of oleic acid from the iron oxide nanoparticle surface was observed only for F−, which is small enough to intercalate between the OA and surface iron ion to displace the ligand from the surface. Furthermore, F− was displaced from the NP surface in water so that stripped NPs could easily be re-grafted with ligands of various length and functional groups thanks to the weak electrostatic interaction of anion and NP surface cation. The theoretical maximum grafting density for nitrodopamide-anchored ligands could thus be achieved regardless of ligand functionality. The presented method should be straightforward to extend to ligand replacement on other types of inorganic nanoparticles synthesized through diffusion-limited, size-focused growth assisted by strongly complexed anionic ligands such as oleic acid.Acknowledgements
We thank Prof. Dieter Baurecht for access to ATR-FTIR. We acknowledge the VIBT Extremophile Center for access to TGA and Tanja Zwölfer for providing iron oxide NPs. Dr Steffen Kurzhals is acknowledged for advice. We thank Martina Schroffenegger and Werner Artner (TU Wien X-ray Center) for assistance with XRD measurements. The research leading to these results received funding from the European Research Council under the European Union's Seventh Framework Program (FP/2007–2013)/ERC Grant Agreement no. 310034.References
- H. Lee, T.-H. Shin, J. Cheon and R. Weissleder, Chem. Rev., 2015, 115, 10690–10724 CrossRef CAS PubMed.
- N. Lee, D. Yoo, D. Ling, M. H. Cho, T. Hyeon and J. Cheon, Chem. Rev., 2015, 115, 10637–10689 CrossRef CAS PubMed.
- N. Koukabi, E. Kolvari, A. Khazaei, M. A. Zolfigol, B. Shirmardi-Shaghasemi and H. R. Khavasi, Chem. Commun., 2011, 47, 9230–9232 RSC.
- D. V. Talapin, J.-S. Lee, M. V. Kovalenko and E. V. Shevchenko, Chem. Rev., 2009, 110, 389–458 CrossRef PubMed.
- J. Park, K. An, Y. Hwang, J.-G. Park, H.-J. Noh, J.-Y. Kim, J.-H. Park, N.-M. Hwang and T. Hyeon, Nat. Mater., 2004, 3, 891–895 CrossRef CAS PubMed.
- S. G. Kwon and T. Hyeon, Small, 2011, 7, 2685–2702 CrossRef CAS PubMed.
- P. R. Brown, D. Kim, R. R. Lunt, N. Zhao, M. G. Bawendi, J. C. Grossman and V. Bulović, ACS Nano, 2014, 8, 5863–5872 CrossRef CAS PubMed.
- J. Tang, K. W. Kemp, S. Hoogland, K. S. Jeong, H. Liu, L. Levina, M. Furukawa, X. Wang, R. Debnath and D. Cha, Nat. Mater., 2011, 10, 765–771 CrossRef CAS PubMed.
- E. L. Rosen, R. Buonsanti, A. Llordes, A. M. Sawvel, D. J. Milliron and B. A. Helms, Angew. Chem., Int. Ed., 2012, 51, 684–689 CrossRef CAS PubMed.
- E. Amstad, M. Textor and E. Reimhult, Nanoscale, 2011, 3, 2819–2843 RSC.
- J. Hühn, C. Carrillo-Carrion, M. G. Soliman, C. Pfeiffer, D. Valdeperez, A. Masood, I. Chakraborty, L. Zhu, M. Gallego and Z. Yue, Chem. Mater., 2016, 29, 399–461 CrossRef.
- M. V. Kovalenko, M. Scheele and D. V. Talapin, Science, 2009, 324, 1417–1420 CrossRef CAS PubMed.
- K. V. Korpany, C. Mottillo, J. Bachelder, S. N. Cross, P. Dong, S. Trudel, T. Friščić and A. S. Blum, Chem. Commun., 2016, 52, 3054–3057 RSC.
- A. Dong, X. Ye, J. Chen, Y. Kang, T. Gordon, J. M. Kikkawa and C. B. Murray, J. Am. Chem. Soc., 2010, 133, 998–1006 CrossRef PubMed.
- E. L. Rosen, R. Buonsanti, A. Llordes, A. M. Sawvel, D. J. Milliron and B. A. Helms, Angew. Chem., Int. Ed., 2012, 51, 684–689 CrossRef CAS PubMed.
- T. Hyeon, S. S. Lee, J. Park, Y. Chung and H. B. Na, J. Am. Chem. Soc., 2001, 123, 12798–12801 CrossRef CAS PubMed.
- A. Verma, O. Uzun, Y. Hu, Y. Hu, H.-S. Han, N. Watson, S. Chen, D. J. Irvine and F. Stellacci, Nat. Mater., 2008, 7, 588–595 CrossRef CAS PubMed.
- L. M. Bronstein, X. Huang, J. Retrum, A. Schmucker, M. Pink, B. D. Stein and B. Dragnea, Chem. Mater., 2007, 19, 3624–3632 CrossRef CAS.
- J. VandeVondele, M. Krack, F. Mohamed, M. Parrinello, T. Chassaing and J. Hutter, Comput. Phys. Commun., 2005, 167, 103–128 CrossRef CAS.
- E. Amstad, T. Gillich, I. Bilecka, M. Textor and E. Reimhult, Nano Lett., 2009, 9, 4042–4048 CrossRef CAS PubMed.
- O. Bixner, A. Lassenberger, D. Baurecht and E. Reimhult, Langmuir, 2015, 31, 9198–9204 CrossRef CAS PubMed.
- E. Amstad, J. Kohlbrecher, E. Müller, T. Schweizer, M. Textor and E. Reimhult, Nano Lett., 2011, 11, 1664–1670 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental and characterization (NMR, TGA, XPS, DLS and XRD). See DOI: 10.1039/c7nr02199b |
| This journal is © The Royal Society of Chemistry 2017 |