Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Enzyme-coated Janus nanoparticles that selectively bind cell receptors as a function of the concentration of glucose†
Gabriele
Rucinskaite
a,
Sebastian A.
Thompson
ab,
Sureyya
Paterson
a and
Roberto
de la Rica
 *ac
*ac
aDepartment of Pure and Applied Chemistry, WestCHEM, University of Strathclyde, Technology and Innovation Centre, 99 George Street, Glasgow, G1 1RD, Scotland, UK. E-mail: roberto.drica@uib.es
bDepartment of Chemistry and Biochemistry, Hunter College – City University of New York, New York 10065, USA
cDepartment of Chemistry, University of the Balearic Islands, Carretera de Valldemossa km 7.5, 07122 Palma de Mallorca, Illes Balears, Spain
First published on 12th April 2017
Abstract
A method is proposed for controlling the number of nanoparticles bound to cell membranes via RGDS peptide–integrin interactions. It consists of propelling nanoparticles bearing the peptides with enzymes (glucose oxidase), which disrupts biomolecular interactions as a function of the concentration of enzyme substrate (glucose).
Biorecognition, that is, the ability of certain biomolecules to specifically bind to a target molecule, is a fundamental phenomenon in biology that underpins the outstanding complexity of the cellular machinery. In vitro, biorecognition reactions can be used to fabricate biosensors for the detection of a wide array of analytes, from disease biomarkers and pathogens, to small molecules and environmental pollutants.1–3 In targeted drug delivery, medicines modified with biorecognition elements preferentially bind to cells overexpressing a particular cell receptor. This increases the number of interactions with the cells, which in turn increases the retention time and cellular uptake.4,5 The selectivity of these biosensing and drug delivery approaches could be enhanced even further if biomolecular interactions could be disrupted as a function of an external stimulus. For example, the signal-to-noise ratio of a biosensor could be greatly improved if low-specificity interactions could be disrupted on demand. In drug delivery, medicines capable of unbinding healthy cells as a function of an external stimulus could reduce unwanted side effects because they would accumulate less in healthy tissue.
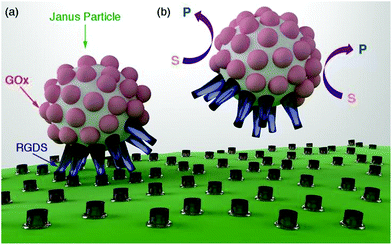
In this manuscript we introduce a new method for disrupting biomolecular interactions as a function of the concentration of glucose. This method consists of attaching biomolecules to Janus particles that are partially covered with enzymes (Fig. 1a). It has been proposed that enzyme-coated Janus nanoparticles can move by means of a diffusiophoretic mechanism.6,7 The velocity of these Janus particles is related to the concentration of the enzyme substrate. It is also well established that biomolecular interactions can be mechanically disrupted, for example, by pulling biomolecules apart with an atomic force microscope or with optical tweezers.8 Here we propose that the mechanical force generated by the motion of enzyme-coated Janus particles can also be used to disrupt biomolecular interactions and unbind the nanoparticles from the cell membrane (Fig. 1b). To prove this concept we prepared nanoparticles partially coated with glucose oxidase (GOx) and a biomolecule that interacts with a cell receptor (RGDS, a peptide that targets integrins,9–12Fig. 1a). GOx increases the diffusion coefficient of the nanoparticles,13,14 which decreases the number of nanoparticles bound to cells as a function of the concentration of the enzyme substrate. The particle design could be easily adapted to respond to the concentration of other metabolites such as urea or hydrogen peroxide by simply changing GOx for urease or catalase, respectively.6,7 This concept is promising for developing a new family of particles that selectively establish interactions with cells as a function of the concentration of different metabolites present in the cell microenvironment.
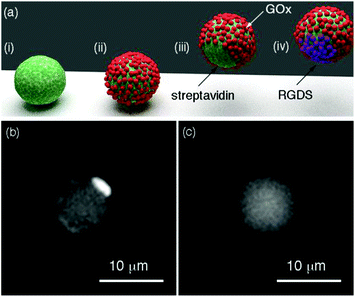
The Janus structure consisting of two hemispheres with different chemical composition (Fig. 1a) enables an asymmetric distribution of the products of the enzymatic reaction around the colloids, which is a crucial factor to trigger the diffusiophoretic mechanism responsible for the motion of the particles.15Fig. 2a depicts the method used here for obtaining Janus particles that are partially covered with enzymes and other biomolecules. Streptavidin-coated magnetic particles are the starting material for preparing Janus particles. An asymmetric distribution of enzymes and biomolecules on the surface of these particles was obtained with a variant of the well-known method of desymmetrization at interfaces.16 It involves holding the streptavidin-coated colloids with a magnet (Fig. 2a(i)) while modifying the surface exposed to the solvent with biotinylated enzymes (Fig. 2a(ii)). After removing the magnet, Janus particles partially covered with enzymes are obtained (Fig. 2a(iii)). The free streptavidin binding sites can now be modified with another biotinylated molecule, for example biotinylated RGDS peptides (Fig. 2a(iv)). The procedure for obtaining Janus particles was tested with micrometric magnetic particles, which enabled detecting the asymmetric coating with optical microscopy. In Fig. 2b when the streptavidin-modified particles were coated with biotinylated fluorescent BSA the Janus structure could be visualized with fluorescence microscopy. In these images the darker areas contain non-fluorescent GOx whereas the bright spots are modified with fluorescent BSA (see also Fig. S2 in ESI†). In Fig. 2c, control colloids that were not modified with fluorescent BSA show a homogenous background fluorescence signal, which demonstrates that the differences in fluorescence intensity observed in Fig. 2b are originated by the asymmetric coating of the particles with GOx and fluorescent BSA. Janus particles based on magnetic iron oxide nanoparticles have been previously obtained with a variety of methods, including seed-and-growth consecutive steps,17 a flame synthetic approach,18 or the deposition of iron oxide on cores with physical vapor deposition.19 The proposed method is particularly advantageous for preparing Janus particles coated with biomolecules because it avoids the utilization of harsh etching,20 or sonication,21 steps that could denature the proteins on the surface of the particles.
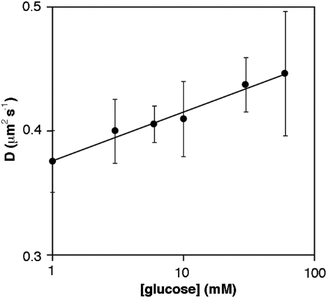
We then studied the motion of the GOx-modified nanoparticles as a function of the concentration of glucose. Janus nanoparticles with a diameter of 220 ± 40 nm were prepared following the method shown in Fig. 2. This size was chosen because it is similar to the size of previously reported enzyme-powered nanomotors.7,14 Nanoparticle motion was studied with dynamic light scattering (DLS), which is a validated technique for studying changes in the diffusion coefficient of enzyme-covered Janus nanoparticles.7 The diffusion coefficient of the nanoparticles in the absence of glucose is 0.36 ± 0.01 μm2 s−1. This value is similar to the diffusion coefficient reported for hollow Janus silica particles decorated with GOx, which have been previously proposed as enzyme-powered nanomotors (0.72 μm2 s−1, diameter 389 nm).7 In Fig. 3 the apparent diffusion coefficient increases with the concentration of glucose in the concentration range between 0 and 60 mM. The apparent diffusion coefficient increased by 23% upon addition of 60 mM glucose (from 0.36 ± 0.01 to 0.44 ± 0.05 μm2 s−1), which is similar to the 24% increase reported for hollow mesoporous nanoparticles in the presence of 75 mM glucose.7D values at 0 and 60 mM glucose are significantly different from each other (p < 0.05). Although the experiments are rather noisy, the general trend observed in Fig. 3 agrees well with the idea that the proposed Janus nanoparticles move in the presence of glucose in a concentration-dependent manner, as previously demonstrated for other GOx-decorated Janus nanoparticles.
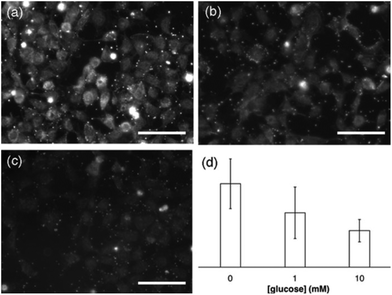
Next we studied the impact of the glucose concentration in the binding of the nanoparticles to integrins present in cell membranes. Integrins are often overexpressed in tumors, and drugs capable of targeting these proteins have been shown to have a greater effect on cancer cells.22 To study the interaction between RGDS and integrins, CHO cells were first fixated with 4%, paraformaldehyde a method that has been previously utilized in similar bioimaging studies.23,24 This procedure allowed us to study the binding of nanoparticles to the plasma membrane independently from cell uptake, since fixated cells do not internalize nanoparticles. Furthermore adding glucose at different concentrations to live cells changes their behavior, cell membrane composition and metabolic profile, making it difficult to study peptide–integrin interactions with confidence.25,26 The proteins on the nanoparticles were modified with fluorescein in order to visualize them easily with fluorescence microscopy. Fig. 4a shows a representative fluorescence microscopy image of the cells after incubation with nanoparticles for 2 h (see also Fig. S3 in ESI†). In this image the cells become fluorescent due to the specific binding of the nanoparticles to the cell membrane, since nanoparticles that were not modified with RGDS did not stain the cell membrane (see Fig. S6 in ESI†). In Fig. 4b and c the same experiment was performed but adding glucose to a final concentration of 1 mM and 10 mM, respectively (see also Fig. S4 and S5 in ESI†). Under these conditions the relative fluorescence intensity around the cells decreases which indicates that less nanoparticles bind to their membranes. In Fig. 4d the average fluorescence intensity was calculated for each glucose concentration. In this figure the fluorescence intensity decreases as the glucose concentration increases. Fluorescence intensity values are significantly different from each other (p < 0.05). This means that less nanoparticles are attached to the cell membrane when the concentration of enzyme substrate increases. These results are in agreement with the hypothesis in Fig. 1 that nanoparticle motion triggered by enzymes disrupts the binding between peptide-coated nanoparticles and cell membranes.
Conclusions
In conclusion, we have demonstrated that it is possible to control the number of GOx-coated Janus nanoparticles bound to cells via peptide–integrin interactions by fine-tuning the concentration of the enzyme substrate glucose. The key factor to control the number of nanoparticles bound the cells is triggering the diffusiophoretic motion of the colloids. Controlling the number of nanoparticles bound to cell membranes as a function of the concentration of metabolites could be useful for designing medicines and bioimaging probes that selectively target cells as a function of biomolecular cues in their microenvironment. For instance, the concentration of glucose in healthy tissue is 10 mM. In the tumor microenvironment this value is 3–10 lower due to the accelerated metabolism of cancer cells.27 The results in Fig. 4 are promising for designing Janus nanoparticles decorated with RGDS and GOx that selectively disrupt RGDS–integrin interactions in healthy tissue, where the concentration of glucose is higher. Drug delivery platforms using these nanoparticles would reduce unwanted side effects in healthy tissue. For example hollow mesoporous Janus nanomotors, which have a high drug-loading capacity,7 could be used to deliver drugs selectively to cancerous tissue. In the context of cancer therapy, the magnetic particles proposed here could be used to kill cancer cells with magnetothermal therapy with reduced side effects.28 Similarly bioimaging nanoprobes based on this design (e.g. MRI probes based on the proposed Janus magnetic nanoparticles)29 could reduce background signals and improve the signal-to-noise ratio when imaging glucose-deficient tumors.Acknowledgements
R. de la Rica acknowledges a Ramón y Cajal contract from MICINN. We are thankful to ScienceConcept3D for Fig. 1 and 2a.Notes and references
- P. D. Howes, R. Chandrawati and M. M. Stevens, Science, 2014, 346, 1247390 CrossRef PubMed.
- R. de la Rica, E. Mendoza, L. M. Lechuga and H. Matsui, Angew. Chem., Int. Ed., 2008, 47, 9752–9755 CrossRef CAS PubMed.
- M. Diaz-gonzalez, M. Gutierrez-capitan, P. Niu, A. Baldi, C. Jimenez-jorquera and C. Fernandez-sanchez, TrAC, Trends Anal. Chem., 2016, 77, 186–202 CrossRef CAS.
- C. L. Modery-pawlowski and A. Sen Gupta, Biomaterials, 2014, 35, 2568–2579 CrossRef CAS PubMed.
- J. M. Stukel, R. C. Li, H. D. Maynard and M. R. Caplan, Biomacromolecules, 2010, 11, 160–167 CrossRef CAS PubMed.
- X. Ma, X. Wang, K. Hahn and S. Sanchez, ACS Nano, 2016, 10, 3597–3605 CrossRef CAS PubMed.
- X. Ma, A. Jannasch, U.-R. Albrecht, K. Hahn, A. Miguel-lópez, E. Schäffer and S. Sánchez, Nano Lett., 2015, 15, 7043–7050 CrossRef PubMed.
- P. Bongrand, Rep. Prog. Phys., 1999, 62, 921–968 CrossRef CAS.
- K. Y. Ahn, H. K. Ko, B. R. Lee, E. J. Lee, J. H. Lee, Y. Byun, I. C. Kwon, K. Kim and J. Lee, Biomaterials, 2014, 35, 6422–6429 CrossRef CAS PubMed.
- T. D. Zaveri, J. S. Lewis, N. V. Dolgova, M. J. Clare-salzler and B. G. Keselowsky, Biomaterials, 2014, 35, 3504–3515 CrossRef CAS PubMed.
- R. H. Harrison, J. A. M. Steele, R. Chapman, A. J. Gormley, L. W. Chow, M. M. Mahat, L. Podhorska, R. G. Palgrave, D. J. Payne, S. P. Hettiaratchy, I. E. Dunlop and M. M. Stevens, Adv. Funct. Mater., 2015, 25, 5748–5757 CrossRef CAS PubMed.
- N. J. Walters and E. Gentleman, Acta Biomater., 2015, 11, 3–16 CrossRef CAS PubMed.
- L. K. E. A. Abdelmohsen, M. Nijemeisland, G. M. Pawar, G. J. A. Janssen, R. J. M. Nolte, J. C. M. Van Hest and D. A. Wilson, ACS Nano, 2016, 10, 2652–2660 CrossRef CAS PubMed.
- P. Schattling, B. Thingholm and B. Städler, Chem. Mater., 2015, 27, 7412–7418 CrossRef CAS.
- S. Sanchez, L. Soler and J. Katuri, Angew. Chem., Int. Ed., 2015, 54, 1414–1444 CrossRef CAS PubMed.
- J. Hu, S. Zhou, Y. Sun, X. Fang and L. Wu, Chem. Soc. Rev., 2012, 41, 4356 RSC.
- J. Reguera, D. Jiménez de Aberasturi, N. Winckelmans, J. Langer, S. Bals and L. M. Liz-marzán, Faraday Discuss., 2016, 191, 47–59 RSC.
- N. Zhao and M. Gao, Adv. Mater., 2009, 21, 184–187 CrossRef CAS.
- B. Ren, A. Ruditskiy, J. H. Song and I. Kretzschmar, Langmuir, 2012, 28, 1149–1156 CrossRef CAS PubMed.
- Z. Li, E. Cheng, W. Huang, T. Zhang, Z. Yang, D. Liu and Z. Tang, J. Am. Chem. Soc., 2011, 133, 15284–15287 CrossRef CAS PubMed.
- J. Li, V. V. Singh, S. Sattayasamitsathit, J. Orozco, K. Kaufmann, R. Dong, W. Gao, B. Jurado-sanchez, Y. Fedorak and J. Wang, ACS Nano, 2014, 8, 11118–11125 CrossRef CAS PubMed.
- D. Chen, B. Li, S. Cai, P. Wang, S. Peng, Y. Sheng, Y. He, Y. Gu and H. Chen, Biomaterials, 2016, 100, 1–16 CrossRef CAS PubMed.
- K. Hu, H. Wang, G. Tang, T. Huang, X. Tang, X. Liang, S. Yao and D. Nie, J. Nucl. Med., 2015, 56, 1278–1284 CrossRef CAS PubMed.
- H.-B. Bae, J.-M. Tadie, S. Jiang, D. W. Park, C. P. Bell, L. C. Thompson, C. B. Peterson, V. J. Thannickal, E. Abraham and J. W. Zmijewski, J. Immunol., 2013, 190, 2273–2281 CrossRef CAS PubMed.
- N. A. Graham, M. Tahmasian, B. Kohli, E. Komisopoulou, M. Zhu, I. Vivanco, M. A. Teitell, H. Wu, A. Ribas, R. S. Lo, I. K. Mellinghoff, P. S. Mischel and T. G. Graeber, Mol. Syst. Biol., 2012, 8, 1–16 CrossRef PubMed.
- M. Golpour, H. Akhavan Niaki, H. R. Khorasani, A. Hajian, R. Mehrasa and A. Mostafazadeh, Int. J. Mol. Cell. Med., 2014, 3, 74–80 Search PubMed.
- K. Birsoy, R. Possemato, F. K. Lorbeer, E. C. Bayraktar, P. Thiru, B. Yucel, T. Wang, W. W. Chen, C. B. Clish and D. M. Sabatini, Nature, 2014, 508, 108–112 CrossRef CAS PubMed.
- J. H. Fang, Y. T. Lee, W. H. Chiang and S. H. Hu, Small, 2015, 11, 2417–2428 CrossRef CAS PubMed.
- X. Ji, R. Shao, A. M. Elliott, R. Jason Stafford, E. Esparza-coss, J. A. Bankson, G. Liang, Z. P. Luo, K. Park, J. T. Markert and C. Li, J. Phys. Chem. C, 2007, 111, 6245–6251 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental section and Fig. S1–S5. See DOI: 10.1039/c7nr00298j |
| This journal is © The Royal Society of Chemistry 2017 |