Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Intrinsic functional and architectonic heterogeneity of tumor-targeted protein nanoparticles†
Mireia
Pesarrodona
abc,
Eva
Crosas
d,
Rafael
Cubarsi
e,
Alejandro
Sánchez-Chardi
f,
Paolo
Saccardo
abc,
Ugutz
Unzueta
cg,
Fabian
Rueda
abc,
Laura
Sanchez-García
abc,
Naroa
Serna
abc,
Ramón
Mangues
cg,
Neus
Ferrer-Miralles
abc,
Esther
Vázquez
*abc and
Antonio
Villaverde
 *abc
*abc
aInstitut de Biotecnologia i de Biomedicina, Universitat Autònoma de Barcelona, 08193 Cerdanyola del Vallès, Spain. E-mail: Esther.Vazquez@uab.cat; antoni.villaverde@uab.es
bDepartament de Genètica i de Microbiologia, Universitat Autònoma de Barcelona, 08193 Cerdanyola del Vallès, Spain
cCIBER de Bioingeniería, Biomateriales y Nanomedicina (CIBER-BBN), Spain
dALBA Synchrotron, Carrer de la llum, 2-26, 08290 Cerdanyola del Vallès, Spain
eDepartment of Mathematics, Campus Diagonal Sud, Edifici U, Universitat Politècnica de Catalunya, Carrer de Pau Gargallo, 5, 08028 Barcelona, Spain
fServei de Microscòpia, Universitat Autònoma de Barcelona, Bellaterra 08193 Cerdanyola del Vallès, Barcelona, Spain
gBiomedical Research Institute Sant Pau (IIB-Sant Pau) and Josep Carreras Leukemia Research Institute, Hospital de la Santa Creu i Sant Pau, 08025 Barcelona, Spain
First published on 10th April 2017
Abstract
Self-assembling proteins are gaining attention as building blocks for application-tailored nanoscale materials. This is mostly due to the biocompatibility, biodegradability, and functional versatility of peptide chains. Such a potential for adaptability is particularly high in the case of recombinant proteins, which are produced in living cells and are suitable for genetic engineering. However, how the cell factory itself and the particular protein folding machinery influence the architecture and function of the final material is still poorly explored. In this study we have used diverse analytical approaches, including small-angle X-ray scattering (SAXS) and field emission scanning electron microscopy (FESEM) to determine the fine architecture and geometry of recombinant, tumor-targeted protein nanoparticles of interest as drug carriers, constructed on a GFP-based modular scheme. A set of related oligomers were produced in alternative Escherichia coli strains with variant protein folding networks. This resulted in highly regular populations of morphometric types, ranging from 2.4 to 28 nm and from spherical- to rod-shaped materials. These differential geometric species, whose relative proportions were determined by the features of the producing strain, were found associated with particular fluorescence emission, cell penetrability and receptor specificity profiles. Then, nanoparticles with optimal properties could be analytically identified and further isolated from producing cells for use. The cell's protein folding machinery greatly modulates the final geometry reached by the constructs, which in turn defines the key parameters and biological performance of the material.
Introduction
Controlled self-assembly of synthetic peptides and recombinant proteins is a powerful tool for the generation of functional, micro- and nanostructured materials. Recombinant proteins benefit, over synthetic peptides, from the versatility of biological fabrication. Based on a generic set of genetic engineering procedures, recombinant protein production has largely impacted the biotechnological and biopharmaceutical industries with more than 400 protein drugs approved for human use.1 The identification2,3 and exploitation4,5 of oligomerization domains, the tailored fibrillation of amyloidal protein forms6 and the de novo design of protein–protein interacting patches7,8 offer a wide spectrum of possibilities regarding the generation of supramolecular materials to be used in biological interfaces.9–11 Being not only functional but also biocompatible and biodegradable, protein materials show a still unexplored biomedical potential in both regenerative medicine and conventional or cell-targeted drug delivery.12,13 The natural tendency of GFP to oligomerize14 and the more recent manipulation of GFP assembly15,16 have attracted attention as this beta-sheet rich protein represents a compact, structurally stable building block for the assay of controlled oligomer formation and material characterization.In previous studies, we have developed a protein engineering platform to promote the self-assembly of modular GFP constructs, based on the combination of end-terminal cationic stretches and polyhistidines.17,18 Driven by electrostatic interactions and with a strong involvement of the histidine-rich tail, these peptides promote the formation of stable oligomers of defined average size on the nanoscale irrespective of the amino acid sequence and origin of the core protein placed in between. The resulting nanoparticles, with a toroid-like shape and usually ranging between 12 and 40 nm, are fully stable in vivo.19 Such a stability under physiological conditions, provided by complex forces sustaining protein–protein cross-interactions, enables the material to escape from renal clearance and it also allows a prolonged circulation time in blood.19 When displaying appropriate peptide ligands of cell surface cancer markers CXCR4 or CD44 (T22 and A5G27 respectively) they specifically accumulate in primary tumor and metastasis in colorectal and mammary cancer models, respectively,20,21 being suitable for antitumoral drug delivery. The same platform has been used to construct fluorescent nanoparticles that cross the blood–brain barrier and target the brain.18
Recently,22 we have determined that self-assembled T22-GFP-H6 oligomers elute from Immobilized Metal Affinity Chromatography (IMAC) in two separate fractions with slightly different sizes and CXCR4+ cell penetrability, suggesting alternative solvent exposure of both the His tag and the T22 tumor homing peptide. Also, the bacterial species and strain used for production influence the size and biodistribution of the material upon systemic administration in animal models of CXCR4+ colorectal cancer.23,24 Altogether, these data indicate that bacterial cells can fabricate GFP-based nanoparticles of biomedical interest in alternative conformations. Such a possibility could be relevant to the in vivo use of these materials in a therapeutic context but in general, to the production of self-assembling protein materials with specialized functions. As this suspected architectonic deviation has been so far elusive, we have examined here the functional traits as well as the subunit organization of closely related, GFP-based nanoparticles produced in alternative Escherichia coli strains, specially focusing on intrinsic functional and morphometric variabilities resulting from the biofabrication process. We demonstrate, for the first time, the subtle size- and shape-dependent heterogeneity of protein nanoparticles linked to their functional properties, which determines the performance of the materials as intracellular, cell-targeted vehicles. The identified segregation allows the selection, by their geometry, of specific oligomer populations in which receptor-specificity and cancer cell uptake are dramatically improved.
Experimental
Protein production
T22-GFP-H6 and A5G27-GFP-H6 are self-assembling modular proteins (ESI Fig. 1†) targeting CXCR4 and CD44, respectively, through amino terminal peptides (T22, RRWCYRKCYKGYCYRKCR and A5G27, RLVSYNGIIFFLK) binding these cell surface receptors.20,21 T22-GFP-H6 was produced in Escherichia coli Origami B [F−ompT hsdSB (rB− mB−) gal dcm lacY1 ahpC (DE3) gor522::Tn10 trxB (KanR, TetR)] (Novagen, Madison, WI, USA), encoded in a pET22b-derived vector, and in the endotoxin-free KPM335 (msbA52, ΔgutQ, ΔkdsD, ΔlpxL, ΔlpxM, ΔpagP, ΔlpxP, ΔeptA, frr181), its parental BW30270 (CGSC#7925–MG1655; F−, rph+, fnr+) and the routine wild type MC4100 (F− [araD139]B/r, Del(argF-lac)169, flhD5301, Δ(fruK-yeiR)725(fruA25), relA1, rpsL150(strR), rbsR22, Del(fimB-fimE)632(::IS1), deoC1) from a pTrc99a-derivative plasmid. A5G27-GFP-H6 and the control GFP-H6 protein with an N-terminal random (non-cationic) peptide were produced in E. coli BL21 DE3 (Novagen, Madison, WI, USA) transformed with pET22b and pET21b plasmids respectively. All encoding gene sequences were optimised for expression according to the E. coli codon usage. Protein production was induced by the addition of 1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) into bacterial cultures that were further cultivated overnight at 20 °C in LB medium with a selective antibiotic (at 16 °C for A5G27-GFP-H6 producing cultures).Protein purification
Proteins were purified as previously described.20 Briefly, cell pellets were resuspended in Wash buffer (10 mM Tris-HCl, 500 mM NaCl, 10 mM Imidazol pH 8.0) in the presence of protease inhibitors. Cell disruption was performed at 1200 psi using a French Press and the lysate was centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 45 min. Protein was purified through the His-tag by Immobilized Metal Affinity Chromatography (IMAC) and protein separation was achieved using an imidazole gradient up to 500 mM. Protein peak fractions were collected, dialysed against carbonate buffer (166 mM NaCO3H, pH 7.4) and centrifuged to remove insoluble aggregates. Protein integrity was analysed by SDS electrophoresis on TGX Stain-Free gels (Bio-Rad, Hercules, CA, USA) and followed by western blotting using an anti-His monoclonal antibody (Santa Cruz Biotechnology, Inc., Heidelberg, Germany). The protein concentration was determined by using an adapted Bradford's assay.25 Proteins were found as nanoparticles upon purification, and no specific assembling protocols were applied. Oligomers are presumably formed already in the producing bacteria.
000g for 45 min. Protein was purified through the His-tag by Immobilized Metal Affinity Chromatography (IMAC) and protein separation was achieved using an imidazole gradient up to 500 mM. Protein peak fractions were collected, dialysed against carbonate buffer (166 mM NaCO3H, pH 7.4) and centrifuged to remove insoluble aggregates. Protein integrity was analysed by SDS electrophoresis on TGX Stain-Free gels (Bio-Rad, Hercules, CA, USA) and followed by western blotting using an anti-His monoclonal antibody (Santa Cruz Biotechnology, Inc., Heidelberg, Germany). The protein concentration was determined by using an adapted Bradford's assay.25 Proteins were found as nanoparticles upon purification, and no specific assembling protocols were applied. Oligomers are presumably formed already in the producing bacteria.
Gel filtration
Protein oligomers were analysed using size-exclusion chromatography. Samples were loaded on a Superdex 200 Increase 10/300 (GE Healthcare, Piscataway, NJ, USA) column pre-equilibrated with carbonate buffer at 0.75 ml min−1. Protein samples intended for small-angle X-ray scattering (SAXS) were prepared by collecting fractions corresponding to each oligomer population and concentrated to 5 mg ml−1, using an Amicon Centrifugal Filters 3 K (Millipore, Cork, Ireland). The supernatant was centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 min to remove possible aggregates and stored at −80 °C. Protein oligomers were re-analysed under the same SEC conditions to assess the structural stability upon separation. The hydrodynamic radius (Rh) was obtained using a protein standard (GE Healthcare, Piscataway, NJ, USA), and the relative amounts of oligomeric forms were calculated by Gaussian deconvolution of the obtained size-exclusion chromatograms using the Peakfit 4.12 software (Systat Software Inc.) and applying a residual method to resolve the overlapped peaks.
000g for 30 min to remove possible aggregates and stored at −80 °C. Protein oligomers were re-analysed under the same SEC conditions to assess the structural stability upon separation. The hydrodynamic radius (Rh) was obtained using a protein standard (GE Healthcare, Piscataway, NJ, USA), and the relative amounts of oligomeric forms were calculated by Gaussian deconvolution of the obtained size-exclusion chromatograms using the Peakfit 4.12 software (Systat Software Inc.) and applying a residual method to resolve the overlapped peaks.
Dynamic light scattering (DLS) and fluorescence determination
The volume size distribution of nanoparticles was determined at 1 mg ml−1 in carbonate buffer by dynamic light scattering at 633 nm (Zetasizer Nano ZS, Malvern Instruments Limited, Malvern, UK). Green fluorescence was determined by using a Varian Cary Eclipse fluorescence spectrophotometer (Agilent Technologies, Santa Clara, CA, USA) at a detection wavelength of 510 nm and at 1 mg ml−1 protein concentration, by using an excitation wavelength of 450 nm.Cell lines and protein internalization
The MDA-MB-231 cell line was maintained in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) and 6 mM GlutaMAX (Invitrogen, Carlsbad, CA, USA), whereas HeLa (ATCC-CCL-2) cells were maintained in MEMα (GIBCO, Rockville, MD, USA) supplemented with 10% FBS. Both cell lines were incubated at 37 °C and 5% CO2. To analyse protein internalization, HeLa and MDA-MB-231 cells were cultured on 24-well plates at 3 × 104 cells per well and 8 × 104 cells per well respectively until reaching 70% confluence. The medium was discarded and the cells were washed twice with PBS (Sigma-Aldrich, Steinheim, Germany). Cells were then incubated with OptiPRO™ serum-free medium supplemented with L-glutamine with recombinant proteins dissolved at convenient concentrations for 3 h (T22-GFP-H6) or 24 h (A5G27-GFP-H6). AMD3100, a specific CXCR4 antagonist, was added to the cells at 10 mM, 1 h before protein addition, to assess the specificity of protein internalization. In parallel, to study the protein endosomal escape, 50 mM chloroquine was added 3 h before protein addition. After protein incubation, the medium was removed and cells were washed with PBS. Cells were treated with 1 mg ml−1 trypsin for 15 min to remove protein bound to the cell surface followed by the addition of a complete medium. Then they were centrifuged at 1200 rpm for 15 min and the pellet was resuspended in PBS. Protein internalization was analysed using a FACS-Canto system (Becton Dickinson, Franklin Lakes, NJ, USA) using a 15 mW air-cooled argon ion laser at 488 nm excitation. Experiments were performed in duplicate.SAXS measurements
SAXS profiles were recorded in the Non-Crystalline Diffraction (NCD) beamline at ALBA Synchrotron Light Source (Cerdanyola del Vallès, Barcelona, Spain), by using an imXPAD-S1400 photon-counting detector (imXPAD, La Ciotat, France) placed at 5.9 m from the sample. Multiple frames of 0.5–2 seconds of exposure time were collected at 12.4 keV (λ = 1 Å) without attenuation. Samples were measured in a Teflon cell with a path length of 3 mm and mica windows of 25 μm thickness. SAXS data were processed by using the EMBL-Hamburg ATSAS software package.26 The radius of gyration, Rg, was calculated from the pair-distance distribution function by using GNOM.27 Prior to Rg calculation, the similarity between frames was assessed using the Correlation Map (CorMap) test28 in order to discard frames with radiation damage. The fitting of the scattering profiles after background subtraction to the different form factors was performed by using SasView 3.1.2. The ratio between the radius of gyration Rg determined by SAXS and the hydrodynamic radius Rh determined by SEC, Rg/Rh, was used as an estimator for the geometry of the different nanoparticles.Electron microscopy
The near native state ultrastructure of NPs was assessed by field emission scanning electron microscopy (FESEM) and transmission electron microscopy (TEM). For FESEM, drops of 3 μl of P1 samples (Origami B and KPM335 IMAC fraction 2), P2 (Origami B and KPM335 IMAC fraction 2), P4 (KPM335 IMAC fractions 1 and 2), and P5 (Origami B and KPM335 IMAC fraction 2) were directly deposited on a silicon surface (Ted Pella Inc., Reading, CA, USA) for 1 min, air dried and observed without coating with a FESEM Merlin (Zeiss, Oberkochen, Germany) operating at 2 kV. Images were acquired with a high resolution in-lens secondary electron detector.For TEM, drops of 3 μl from the same 8 samples, at the same concentrations used for FESEM, were deposited for 2 min on 400 mesh carbon coated copper grids, contrasted with 2% uranyl acetate for 2 min, air dried and observed with a transmission electron microscope JeolJEM-1400 (Jeol Ltd, Tokyo, Japan) operating at 80 kV. Images were acquired with a CCD Gatan ES1000 W Erlangshen camera (Gatan, Abingdon, UK).
The lengths and widths of 325 nanoparticles from P1 samples (ASG27, Origami B and KPM335 IMAC fractions 1 and 2), P2 (ASG27, Origami B and KPM335 IMAC fraction 2), P3 (KPM335 IMAC fraction 1), P4 (KPM335 IMAC fractions 1 and 2), and P5 (ASG27, Origami B and KPM335 IMAC fractions 1 and 2) were quantified from TEM images using DigitalMicrograph software (Gatan, Abingdon, UK).
Data analysis
Values are expressed as mean and standard error calculated using Microsoft Excel 2003. Pairwise divergences were detected through Student t-tests using Microsoft Excel 2003. Significant differences were assumed as p < 0.05. Graphical representations were obtained using Sigmaplot 10.0 software.Mathematical methods
Internalization I and receptor-dependent internalization R are variables found depending on the specific fluorescence F and SEC elution volume E. According to Rueda and co-workers,22 the relationship I(F) is given by ln(I) = a + bF. The receptor-dependent specific internalization R(F) is also approximately linear R = p + qF. To interpret the plots of I(E) and R(E) shown in Fig. 5B it was previously necessary to analyze the relationship between the specific fluorescence F and the SEC-elution volume E plotted in Fig. 5A, which presents two main features: first, the symmetry about the maximum specific fluorescence F(e0) (green line); second, the linear and opposite trends of the dots around this symmetry point. To find out the value e0, the data were approximated by a parabola (in grey), by providing the optimal SEC elution volume point at e0 = 14.4 ml. Now it was possible to calculate the least squares approximation for the following relationship (blue lines):| F = m + n|E − e0|. |
Therefore, the compositions of functions I(F(E)) and R(F(E)) necessarily show the same behaviour represented in Fig. 5B. The symmetry point e0 matches the previous value. The regression curve in blue of Fig. 3B was
| ln(I) = a′ + b′|E − e0|; a′ = a + bm; b′ = bn |
| R = p′ + q′|E − e0|; p′ = p + qm; q′ = qn. |
The particular role of the optimal SEC elution volume e0 was also noticeable in terms of other parameters, such as the hydrodynamic diameter and the form.
Results and discussion
The de novo designed A5G27-GFP-H6 and T22-GFP-H6 nanoparticles show, by dynamic light scattering (DLS), average size peaks of 14 and 12 nm respectively when produced in the conventional E. coli strains BL21 (DE3) and Origami B, respectively19,20 (ESI Fig. 1†). Any intrinsic morphometric heterogeneity, if existing, has been so far unobserved and eclipsed in the analysis of the raw material. However, when T22-GFP-H6 is purified from bacterial cell extracts by IMAC, it is eluted in two separated fractions,22 a fact that indicates alternative solvent exposures of the histidine-rich terminal tail. In this context, and to examine in detail the potential intrinsic heterogeneity in the architecture of these materials, we have screened by SEC the intra-strain size spectrum of T22-GFP-H6 nanoparticles occurring in these two separate IMAC fractions, upon production in different E. coli strains.We included in the analysis the endotoxin-free E. coli strain KPM335, which is particularly interesting not only because of its application in biomedicine as an endotoxin-free cell factory29 but also because of the complex genetic modifications performed to remove endotoxic components from the cell wall. Such a manipulation has resulted in the constitutive up or down regulation of several heat-shock genes involved in the quality control of recombinant proteins.23 As observed (Fig. 1A), the resulting nanoparticles were distributed in numerous peaks, contrasting with the apparent morphometric homogeneity determined by DLS over the pooled material (ESI Fig. 1†). The sizes of nanoparticle populations were highly coincident when comparing bacterial strains, and even when comparing IMAC fractions 1 and 2, indicative of high regularity in the oligomerization states of GFP. In this regard, the differences observed were mostly lying in the relative proportions of these populations. For instance, T22-GFP-H6 oligomers from IMAC fraction 1 accumulated in SEC peaks P3 and P5, while the material present in the IMAC fraction 2 tended to majorly occur in SEC peaks P1 and P2. P5 appeared to correspond to the unassembled protein forms, namely the monomeric or probably dimeric building blocks. Both the proteolytic stability of the modular proteins and the structural stability of the oligomers were fully confirmed by SDS-PAGE on TGX Stain-Free gel (ESI Fig. 3A†) and by a SEC re-analysis of selected SEC fractions (ESI Fig. 3B†) respectively.
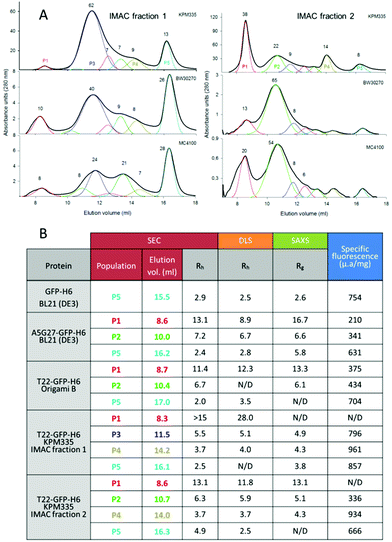 | ||
| Fig. 1 Morphometric variability of T22-GFP-H6. A. SEC chromatograms of T22-GFP-H6 protein produced in KPM335, BW30270 or MC4100 and present in fraction 1 (left, low affinity) or fraction 2 (right, high affinity) of IMAC purification. Black lines indicate the average plot of the elution process. Numbers represent the percentage of the protein amount in each oligomer population (only for nanoparticles represented over 5%), and line colours indicate regularly appearing peaks. Most representative oligomeric populations produced in KPM335 strain are identified as P1–P5 for further evaluation. A SEC calibration curve is depicted in ESI Fig. 2.† B. Summary of the size and fluorescence emission of major populations of GFP variants. Coloured numbers indicate the protein populations, segregated by size and shape in SEC, as indicted in A. | ||
The size (DLS and SAXS) and specific fluorescence (fluorimetry) of the relevant protein populations separated by SEC were determined by independent techniques (Fig. 1B), in an exhaustive analysis that also included A5G27-GFP-H6 from E. coli BL21 and T22-GFP-H6 produced in Origami B (both eluted in a single IMAC fraction). Again, a high coincidence with size data and elution peaks was observed, here also extended to inter protein pairwise comparisons. The unassembled P5 forms of GFP-H6, A5G27-GFP-H6 and T22-GFP-H6 usually ranged between 2 and 3 nm of radius, P4, when observed, around 4 nm, P3 around 5 nm, P2 between 6 and 7 nm and P1 between 11 and 13 nm. The nanoparticle and oligomer sizes were finally determined by TEM imaging of isolated materials (ESI Table 1†), resulting in values again in good agreement with more indirect analytical data (Fig. 1B). The size coincidence when comparing all those analytical approaches confirmed the robustness in the assembling pattern of the GFP oligomers, which appeared to be highly regular despite the nature of the cationic peptide at the amino terminus of the construct. Just a few data among those obtained were out of these ranges. In particular, T22-GFP-H6 nanoparticles from the IMAC elution fraction 1 of KPM335 showed an unusual high comparative size, which is coincident with previous observations,22 indicative of a differential organization of the fully assembled building blocks. Interestingly, all protein fractions were fluorescent, but higher emission values were generally observed in the monomeric forms (P5) when comparing fully assembled nanoparticles and intermediate oligomers (P1 and P2, 50% less fluorescent than the building blocks). The range of fluorescence emission independently confirmed the alternative conformational status of the oligomerized GFP. All these data allowed proposing, in summary, a variable organization of GFP oligomers probably due to alternative protein–protein contact patterns that might be favoured in particular producing strains.
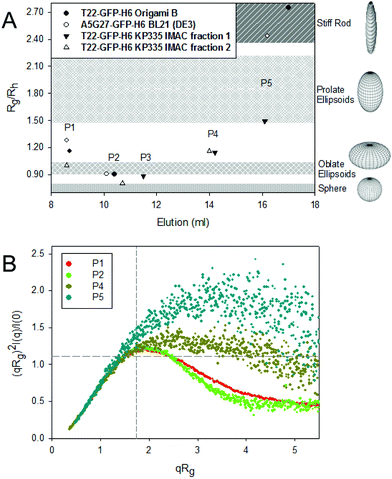
The different architectonic patterns adopted by GFP oligomers would necessarily be connected to distinct morphometries and biophysical properties of the materials, which have been so far generically identified as planar, toroid nanoparticles.19 In this context, fine SAXS analyses revealed a broad range of shapes, from rod forms to spherical forms, depending on the SEC population to which they belong (Fig. 2A). Also, the molecular flexibility of the overhanging peptides from GFP-fusions was also variable (Fig. 2B), revealing a changeable potential of the building block to adopt alternative conformations that might limit, impair or favour specific oligomeric organizations. The best fitting of the SAXS profiles for the materials to different form factors was determined by using SasView, as shown in ESI Fig. 4.† The alternative morphometries in SEC peaks identified by SAXS were fully assessed by high resolution TEM and FESEM imaging (Fig. 3), confirming the nano-architectonic variability of nanoparticles in a fully visual way.
 | ||
| Fig. 3 Morphometry at the ultrastructural level of GFP oligomers. Representative TEM and FESEM images of T22-GFP-H6 oligomers produced in the E. coli strains Origami B and KPM335, classified by their occurrence in SEC peaks. Bar sizes represent 20 nm in general views and 10 nm in detailed images. Shapes within the panels correspond to the expected particle form and their colours to the peak in the plots from Fig. 1. | ||
The protein nanoparticles studied here had been conceived as drug carriers for cancer treatments, which was lately encouraged by their good biodistribution when systemically administered (upon which the material accumulated intracellularly in the tumor and metastatic foci but not in the liver, kidney, spleen or other non-target organs).19,20 Then, how the morphometry and other physical properties of the oligomeric populations might influence receptor-dependent cell penetration is a critical issue that was addressed in a CXCR4+ cell culture model. As observed (Fig. 4A), the unassembled populations of CXCR4-targeted nanoparticles were inefficient in penetrating target cells, although their uptake was clearly over the background values of the non-targeted, parental GFP-H6 (devoid of any receptor ligand). Noteworthily, cell penetrability was progressively gained with the complexity of the oligomers, indicating that the mere presence of a cell ligand is not sufficient to support internalisation. This was fully in agreement with previous data obtained with assembled and disassembled IRPF-based protein nanoparticles30 and with the role of multivalence in the cell binding process and further receptor-mediated internalisation.12 In fact, monovalent building blocks are equally inefficient in reaching their target in vivo.19
Although internalisation is significantly lower for discrete populations compared to larger structures, noticeably, the prevalence of peptide flexibility in P4 and P5 populations shown by the Normalized Kratky Plot (Fig. 2B) could also be involved in a favoured exposure of T22 to the CXCR4 receptor, leading to a major specific internalization. Note also that curves from populations P4 and P5 (Fig. 2B) decay at higher qRg values, which is indicative of domain flexibility. Interestingly, T22-GFP-H6 nanoparticles produced in KPM335 and present in the IMAC fraction 1, were all poorly internalized (in agreement with previous observations using the material pool22), and showed also lower specificity. In these populations, both H6 and T22 tags might be less available for intermolecular interactions, also fitting with the low affinity in IMAC. The surface charge of oligomers from the tested SEC peaks did not appear as a major factor in influencing protein cell interactions as it ranged within a narrow frame from −13.1 mV ± 0.6 to −11.1 mV ± 0.3 (differences not significant, p > 0.07 in a t test; not shown). Therefore, despite the influence that ligand conformation or exposure might have on cell uptake, particle size was found as the major determinant of cell penetrability into target cells, as larger oligomers present enhanced multivalency and internalize more efficiently (compared to P4 and P5) (Fig. 4A and B). Further internalization analysis with A5G27-GFP-H6 protein oligomers in the presence of chloroquine showed an increase in protein lysosomal degradation with a decrease in the oligomer size (Fig. 4B). The endosomal escape of larger oligomers was then more efficient compared to smaller protein assemblies and together with the entrance specificity suggest an unspecific internalization route alternative to a clathrin-mediated endocytosis pathway for P1 populations, which do not lead to late endosome avoiding subsequent protein degradation.31 In this regard, the impact that the nanoparticle geometry (size and shape) has on cell penetrability32 and biodistribution33 has been demonstrated. However, the influence of these parameters may vary depending on bulk material34 and, so far, the effects on protein-based nanoparticles have not been closely examined. In receptor-targeted nanoparticles that are activated with overhanging peptides, uptake might be in addition modulated by the exposure and bioavailability of functional ligands on the particle surface. In our system, these ligands are the tumor homing peptides A5G27 and T22, while the H6 tail has an important role in the endosomal escape upon internalization.35
The variability in the specific GFP fluorescence emission when comparing all the nanoparticles studied here (Fig. 1B) suggested the alternative configurations of the material.22 In this context, and for a global analysis of the potential relationships between the physical and biological properties of the variant oligomers, we performed a numerical screening of dependences between both sets of parameters. Interestingly, highly fluorescent protein particles were regularly collected in the elution SEC peaks around 14 ml (Fig. 5A), formed by a population of nanoparticles that exhibit a medium size. These materials are not particularly efficient as potential drug vehicles, since although the receptor dependent penetrability shows a maximum value here, the cell penetrability (amount of internalized material) is contrarily low (Fig. 5B). Interestingly, GFP oligomers eluted at this stage (around 14 ml in the SEC elution volume, corresponding to the SEC P4), represent a functional and morphometric inflexion in the continuum of forms of the assembled protein. In this regard, the SEC P4 sample represents a virtual frontier between two sets of nanoparticles, regarding the variability of the morphometric traits of the whole protein population (Fig. 5B).
Among the morphometric population spectrum derived from the cell factory, it is possible to identify and separate by SEC the supramolecular organisation of the nanoparticles that display optimal biological properties. In this regard, for this particular system, oligomers from the P2 population (which adopt a spherical shape) exhibit higher cell internalisation and better receptor specificity (Fig. 5B), probably because of a combination of solvent exposure of T22, optimal size and appropriate multivalence of the ligand for cell surface interactions.
In summary, protein nanoparticles generated through the combination of a cationic, cell targeting peptide at the amino terminus of a core GFP and a histidine rich domain at the C-terminus exhibit a spectrum of oligomerization forms previously eclipsed by the pooled analysis of the material (Fig. 1A). The major oligomeric states of the nanoparticles are coincident when comparing proteins constructed with unrelated amino-terminal tags such as A5G27 and T22, and targeted to different cell surface receptors (CD44 and CXCR4, respectively), revealing very regular patterns in the oligomerization process.
These variants exhibit distinguishable biophysical properties including shape, size, fluorescence emission, cellular penetrability and receptor-dependent specificity, some of them critical when considering the applicability of these protein materials as drug carriers in systemic treatments. Interestingly, and considering that high molecular mass of building blocks in protein-only materials are exclusively produced by biological fabrication, the genetic background of the producing cell dramatically influences these abilities. Note for instance the poor penetrability of all nanoparticles produced in KPM335 released in the IMAC fraction 1 contrary to the same material released in fraction 2 (Fig. 4). This is probably due to the impact that altered protein folding machineries have in the conformation of the building block, which might be amplified upon assembly as complex oligomeric species.
At the present stage of understanding of the protein quality control, it is not possible to rationally predict the assembling pattern and functional profile of a protein nanoparticle when produced in a defined bacterial strain. However, since the yield of building block production in bacteria is relatively high (11–23 mg l−1 of culture, before any media and process optimization; not shown), it would be possible to separate, at the downstream stage, particularly convenient material fractions for high performance biological application, and then discard the less efficient. For the materials explored here, proteins eluted in SEC between 8 and 10 ml would appropriately combine high penetrability with high receptor specificity (Fig. 5B), which is not the general case as these parameters tend to evolve in divergent fashions. Also, this set of materials show high particle sizes (Fig. 1B) and geometries ranging from spheres to discoidal (oblate spheres) versions, far from the more elongated, poorly penetrating versions (Fig. 2A and 4A). This is again stressing the regular shaped, high multimerization forms as the most efficient architectonic patterns for receptor-mediated cell penetration.
Conclusions
Compared with other types of nanostructures, protein-only nanoparticles are fully compatible with biological systems, providing a suitable platform for biomedical uses such as drug delivery and imaging probes. Other than chemical composition, size and shape are crucial factors determining the relationships between nanoparticles and increasingly complex biological systems (namely from cells to entire organisms) in critical aspects such as organ specificity and biodistribution,36–40 toxicity,41 and cell uptake and fate.39,42–45 Although previously checked in several types of nanostructures, mainly in crystalline nano- and micromaterials, the present study is the first evaluation of form (geometry) and function (cellular uptake) of protein nanoparticle populations. This has been done using a particular protein oligomerization platform that results in materials highly stable in vivo, which makes possible their applicability as vehicles for cell-targeted delivery of drugs and imaging agents. Although variability in the biofabrication of self-assembling proteins might represent a priori a generic concern for reproducibility in in vivo applications, the high regularity and robustness of the oligomerization patterns instead allows the proper downstream selection of advantageous variants regarding a particular set of applications, such as cell penetrability and specificity in the receptor-dependent uptake of tumor-targeted protein nanoparticles.Acknowledgements
We are indebted to MINECO (BIO2013-41019-P), AGAUR (2014SGR-132) and CIBER de Bioingeniería, Biomateriales y Nanomedicina (project NANOPROTHER) (to AV), Marató de TV3 foundation (TV32013-132031) and AGAUR (2014 PROD 00055) (to RM), Marató de TV3 foundation (to EV) (TV32013-133930) and FIS (to EV and RM) (PI15/00272, PI15/00378 co-founded by FEDER) for funding our research on protein-based therapeutics. This work benefitted from SasView software, originally developed by the DANSE project under NSF award DMR-0520547 (SasView, http://www.sasview.org/). EC was supported by the Spanish Nuclear Safety Council (Consejo de Seguridad Nuclear, CSN). Protein production has been partially performed by the ICTS “NANBIOSIS”, more specifically by the Protein Production Platform of CIBER in Bioengineering, Biomaterials & Nanomedicine (CIBER-BBN)/IBB, at the UAB SepBioES scientific-technical service (http://www.nanbiosis.es/unit/u1-protein-production-platform-ppp/) and DLS measurements have been done at the Biomaterial Processing and Nanostructuring Unit of NANBIOSIS. We are also indebted to Fran Cortés from the Cell Culture and Cytometry Units of the Servei de CultiusCel·lulars, Producciód'AnticossosiCitometria (SCAC), and to the Servei de Microscòpia, both at the UAB. Strain KPM335 was kindly provided by Research Corporation Technologies, Tucson, AZ. AV received an ICREA ACADEMIA award. UU received a Sara Borrell postdoctoral fellowship from ISCIII and LSG was supported by AGAUR (2016FI_B 00034).References
- L. Sanchez-Garcia, L. Martin, R. Mangues, N. Ferrer-Miralles, E. Vazquez and A. Villaverde, Recombinant pharmaceuticals from microbial cells: a 2015 update, Microb. Cell Fact., 2016, 15 Search PubMed.
- L. Marino-Ramirez and J. C. Hu, Isolation and mapping of self-assembling protein domains encoded by the Saccharomyces cerevisiae genome using lambda repressor fusions, Yeast, 2002, 19, 641–650 CrossRef CAS PubMed.
- J. Engel and R. A. Kammerer, What are oligomerization domains good for?, Matrix Biol., 2000, 19, 283–288 CrossRef CAS PubMed.
- H. Gradiar and R. Jerala, Self-assembled bionanostructures: proteins following the lead of DNA nanostructures, J. Nanobiotechnol., 2014, 12 Search PubMed.
- T. A. P. F. Doll, R. Dey and P. Burkhard, Design and optimization of peptide nanoparticles, J. Nanobiotechnol., 2015, 13 Search PubMed.
- D. Li, E. M. Jones, M. R. Sawaya, H. Furukawa, F. Luo and M. Ivanova, et al., Structure-Based Design of Functional Amyloid Materials, J. Am. Chem. Soc., 2014, 136, 18044–18051 CrossRef CAS PubMed.
- J. L. Corchero, E. Vazquez, E. Garcia-Fruitos, N. Ferrer-Miralles and A. Villaverde, Recombinant protein materials for bioengineering and nanomedicine, Nanomedicine, 2014, 9, 2817–2828 CrossRef CAS PubMed.
- N. Ferrer-Miralles, E. Rodriguez-Carmona, J. L. Corchero, E. Garcia-Fruitos, E. Vazquez and A. Villaverde, Engineering protein self-assembling in protein-based nanomedicines for drug delivery and gene therapy, Crit. Rev. Biotechnol., 2015, 35, 209–221 CrossRef CAS PubMed.
- S. Cranford and M. J. Buehler, Materiomics: biological protein materials, from nano to macro, Nanotechnol., Sci. Appl., 2010, 3, 127–148 CAS.
- R. de la Rica and H. Matsui, Applications of peptide and protein-based materials in bionanotechnology, Chem. Soc. Rev., 2010, 39, 3499–3509 RSC.
- M. J. Webber, E. A. Appel, E. W. Meijer and R. Langer, Supramolecular biomaterials, Nat. Mater., 2016, 15, 13–26 CrossRef CAS PubMed.
- U. Unzueta, M. V. Cespedes, E. Vazquez, N. Ferrer-Miralles, R. Mangues and A. Villaverde, Towards protein-based viral mimetics for cancer therapies, Trends Biotechnol., 2015, 33, 253–258 CrossRef CAS PubMed.
- Y. Loo, M. Goktas, A. B. Tekinay, M. O. Guler, C.AE. Hauser and A. Mitraki, Self-Assembled Proteins and Peptides as Scaffolds for Tissue Regeneration, Adv. Healthcare Mater., 2015, 4, 2557–2586 CrossRef CAS PubMed.
- R. K. Jain, P. B. M. Joyce, M. Molinete, P. A. Halban and S. U. Gorr, Oligomerization of green fluorescent protein in the secretory pathway of endocrine cells, Biochem. J., 2001, 360, 645–649 CrossRef CAS PubMed.
- J. C. Y. Tang, S. Rudolph, O. S. Dhande, V. E. Abraira, S. Choi and S. W. Lapan, et al., Cell type-specific manipulation with GFP-dependent Cre recombinase, Nat. Neurosci., 2015, 18, 133441 CrossRef PubMed.
- Y. E. Kim, Y. N. Kim, J. A. Kim, H. M. Kim and Y. Jung, Green fluorescent protein nanopolygons as monodisperse supramolecular assemblies of functional proteins with defined valency, Nat. Commun., 2015, 6 CAS.
- U. Unzueta, N. Ferrer-Miralles, J. Cedano, Z. K. Xu, M. Pesarrodona and P. Saccardo, et al., Non-amyloidogenic peptide tags for the regulatable self-assembling of protein-only nanoparticles, Biomaterials, 2012, 33, 8714–8722 CrossRef CAS PubMed.
- N. Serna, M. V. Cespedes, P. Saccardo, Z. K. Xu, U. Unzueta and P. Alamo, et al., Rational engineering of single-chain polypeptides into protein-only, BBB-targeted nanoparticles, Nanomed. Nanotechnol., 2016, 12, 1241–1251 CrossRef CAS PubMed.
- M. V. Cespedes, U. Unzueta, W. Tatkiewicz, A. Sanchez-Chardi, O. Conchillo-Sole and P. Alamo, et al., In Vivo Architectonic Stability of Fully de Novo Designed Protein-Only Nanoparticles, ACS Nano, 2014, 8, 4166–4176 CrossRef CAS PubMed.
- M. Pesarrodona, N. Ferrer-Miralles, U. Unzueta, P. Gener, W. Tatkiewicz and I. Abasolo, et al., Intracellular targeting of CD44(+) cells with self-assembling, protein only nanoparticles, Int. J. Pharm., 2014, 473, 286–295 CrossRef CAS PubMed.
- U. Unzueta, M. V. Céspedes, N. Ferrer-Miralles, I. Casanova, J. Cedano and J. L. Corchero, et al., Intracellular CXCR4+ cell targeting with T22-empowered protein-only nanoparticles, Int. J. Nanomed., 2012, 7, 4533–4544 CAS.
- F. Rueda, M. V. Cespedes, O. Conchillo-Sole, A. Sanchez-Chardi, J. Seras-Franzoso and R. Cubarsi, et al., Bottom-Up Instructive Quality Control in the Biofabrication of Smart Protein Materials, Adv. Mater., 2015, 27, 7816–7822 CrossRef CAS PubMed.
- F. Rueda, M. V. Cespedes, A. Sanchez-Chardi, J. Seras-Franzoso, M. Pesarrodona and N. Ferrer-Miralles, et al., Structural and functional features of self-assembling protein nanoparticles produced in endotoxin-free Escherichia coli, Microb. Cell Fact., 2016, 15 Search PubMed.
- O. Cano-Garrido, M. V. Cespedes, U. Unzueta, P. Saccardo, M. Roldan and A. Sanchez-Chardi, et al., CXCR4(+)-targeted protein nanoparticles produced in the food-grade bacterium Lactococcus lactis, Nanomedicine, 2016, 11, 2387–2398 CrossRef CAS PubMed.
- M. Pesarrodona, Y. Fernandez, L. Foradada, A. Sanchez-Chardi, O. Conchillo-Sole and U. Unzueta, et al., Conformational and functional variants of CD44-targeted protein nanoparticles bio-produced in bacteria, Biofabrication, 2016, 8 Search PubMed.
- M. V. Petoukhov, D. Franke, A. V. Shkumatov, G. Tria, A. G. Kikhney and M. Gajda, et al., New developments in the ATSAS program package for small-angle scattering data analysis, J. Appl. Crystallogr., 2012, 45, 342–350 CAS.
- D. I. Svergun, Determination of the Regularization Parameter in Indirect-Transform Methods Using Perceptual Criteria, J. Appl. Crystallogr., 1992, 25, 495–503 CrossRef.
- D. Franke, C. M. Jeffries and D. I. Svergun, Correlation Map, a goodness-of-fit test for one-dimensional X-ray scattering spectra, Nat. Methods, 2015, 12, 419–422 CrossRef CAS PubMed.
- U. Mamat, K. Wilke, D. Bramhill, A. B. Schromm, B. Lindner and T. A. Kohl, et al., Detoxifying Escherichia coli for endotoxin-free production of recombinant proteins, Microb. Cell Fact., 2015, 14 Search PubMed.
- Z. K. Xu, U. Unzueta, M. Roldan, R. Mangues, A. Sanchez-Chardi and N. Ferrer-Miralles, et al., Formulating tumor-homing peptides as regular nanoparticles enhances receptor-mediated cell penetrability, Mater. Lett., 2015, 154, 140–143 CrossRef CAS.
- L. K. Medina-Kauwe, J. Xie and S. Hamm-Alvarez, Intracellular trafficking of nonviral vectors, Gene Ther., 2005, 12, 1734–1751 CrossRef CAS PubMed.
- W. Jiang, B. Y. S. Kim, J. T. Rutka and W. C. W. Chan, Nanoparticle-mediated cellular response is size-dependent, Nat. Nanotechnol., 2008, 3, 145–150 CrossRef CAS PubMed.
- E. Blanco, H. Shen and M. Ferrari, Principles of nanoparticle design for overcoming biological barriers to drug delivery, Nat. Biotechnol., 2015, 33, 941–951 CrossRef CAS PubMed.
- K. Murugan, Y. E. Choonara, P. Kumar, D. Bijukumar, L. C. du Toit and V. Pillay, Parameters and characteristics governing cellular internalization and trans-barrier trafficking of nanostructures, Int. J. Nanomed., 2015, 10, 2191–2206 CAS.
- N. Ferrer-Miralles, J. L. Corchero, P. Kumar, J. A. Cedano, K. C. Gupta and A. Villaverde, et al., Biological activities of histidine-rich peptides; merging biotechnology and nanomedicine, Microb. Cell Fact., 2011, 10 Search PubMed.
- P. Decuzzi, B. Godin, T. Tanaka, S. Y. Lee, C. Chiappini and X. Liu, et al., Size and shape effects in the biodistribution of intravascularly injected particles, J. Controlled Release, 2010, 141, 320–327 CrossRef CAS PubMed.
- S. Hirn, M. Semmler-Behnke, C. Schleh, A. Wenk, J. Lipka and M. Schaffler, et al., Particle size-dependent and surface charge-dependent biodistribution of gold nanoparticles after intravenous administration, Eur. J. Pharm. Biopharm., 2011, 77, 407–416 CrossRef CAS PubMed.
- O. J. Osborne, S. J. Lin, C. H. Chang, Z. X. Ji, X. C. Yu and X. Wang, et al., Organ-Specific and Size-Dependent Ag Nanoparticle Toxicity in Gills and Intestines of Adult Zebrafish, ACS Nano, 2015, 9, 9573–9584 CrossRef CAS PubMed.
- R. Toy, P. M. Peiris, K. B. Ghaghada and E. Karathanasis, Shaping cancer nanomedicine: the effect of particle shape on the in vivo journey of nanoparticles, Nanomedicine, 2014, 9, 121–134 CrossRef CAS PubMed.
- J. F. Tan, S. Shah, A. Thomas, H. D. Ou-Yang and Y. L. Liu, The influence of size, shape and vessel geometry on nanoparticle distribution, Microfluid. Nanofluid., 2013, 14, 77–87 CrossRef CAS PubMed.
- K. W. Powers, P. L. Carpinone and K. N. Siebein, Characterization of nanomaterials for toxicological studies, Methods Mol. Biol., 2012, 926, 13–32 CAS.
- A. Albanese, P. S. Tang and W. C. W. Chan, The Effect of Nanoparticle Size, Shape, and Surface Chemistry on Biological Systems, Annu. Rev. Biomed. Eng., 2012, 14, 1–16 CrossRef CAS PubMed.
- B. D. Chithrani, A. A. Ghazani and W. C. W. Chan, Determining the size and shape dependence of gold nanoparticle uptake into mammalian cells, Nano Lett., 2006, 6, 662–668 CrossRef CAS PubMed.
- H. Herd, N. Daum, A. T. Jones, H. Huwer, H. Ghandehari and C. M. Lehr, Nanoparticle Geometry and Surface Orientation Influence Mode of Cellular Uptake, ACS Nano, 2013, 7, 1961–1973 CrossRef CAS PubMed.
- L. Shang, K. Nienhaus and G. U. Nienhaus, Engineered nanoparticles interacting with cells: size matters, J. Nanobiotechnol., 2014, 12 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6nr09182b |
| This journal is © The Royal Society of Chemistry 2017 |