Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The molecular mechanism of the ligand exchange reaction of an antibody against a glutathione-coated gold cluster†
Víctor
Rojas-Cervellera
a,
Lluís
Raich
a,
Jaakko
Akola
 bcd and
Carme
Rovira
bcd and
Carme
Rovira
 *ae
*ae
aDepartament de Química Inorgànica i Orgànica (Secció Química Orgànica) & Institut de Química Teòrica i Computacional (IQTCUB), Universitat de Barcelona, Martí i Franquès 1, 08028 Barcelona, Spain. E-mail: c.rovira@ub.edu
bDepartment of Physics, Tampere University of Technology, P.O. Box 692, FI-33101 Tampere, Finland
cCOMP Centre of Excellence, Department of Applied Physics, Aalto University, FI-00076 Aalto, Finland
dDepartment of Physics. Norwegian University of Science and Technology, NO-7491 Trondheim, Norway
eInstitució Catalana de Recerca i Estudis Avançats (ICREA), Passeig Lluís Companys, 23, 08020 Barcelona, Spain
First published on 7th February 2017
Abstract
The labeling of proteins with heavy atom clusters is of paramount importance in biomedical research, but its detailed molecular mechanism remains unknown. Here we uncover it for the particular case of the anti-influenza N9 neuraminidase NC10 antibody against a glutathione-coated gold cluster by means of ab initio QM/MM calculations. We show that the labeling reaction follows an associative double SN2-like reaction mechanism, involving a proton transfer, with low activation barriers only if one of the two distinct peptide/peptidic ligands (the one that occupies the side position) is substituted. Positively charged residues in the vicinity of the incoming thiol result in strong interactions between the antibody and the AuMPC, favoring the ligand exchange reaction for suitable protein mutants. These results pave the way for future investigations aimed at engineering biomolecules to increase their reactivity towards a desired gold atom cluster.
Introduction
Monolayer-protected gold clusters (AuMPCs) are formed by an inner gold cluster that is rendered kinetically metastable by a protecting or stabilizing ligand layer.1,2 For core diameters <2 nm (up to 100 Au atoms), these systems display superatomic electronic shell-closings,3,4 and they are of great interest in biomedicine and nanobiotechnology,5,6 with practical applications in protein labelling, heating,7 and sensing.8 Site-specific biomolecule labelling of AuMPCs is also a promising technique for drug delivery.8 The fact that small enough MPCs can cross the blood–brain barrier without altering its integrity9 opens the door to possible applications for the treatment of cancer and central nervous system diseases.A well-established technique to conjugate molecules to MPCs is based on the place-exchange reaction of Murray and coworkers.10,11 This reaction,10,11 also known as ligand exchange reaction, has been widely used to introduce new functionalities to AuMPCs. During the reaction, one of the ligands of the gold cluster is replaced by a biomolecule bearing a functional group that can bind to the cluster surface (e.g. a thiol group) through a sulfur–gold bond. The gold atoms of the AuMPC that participate in the reaction are those that protrude from the cluster surface, forming the so-called staple-motifs (Fig. 1a).12 Some of these gold atoms are exposed to the solvent,13 being potentially reactive.
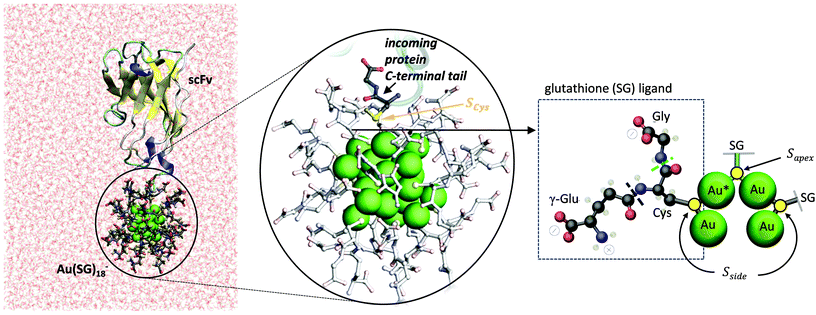
Ligand exchange reactions involving proteins are particularly challenging due to the need to control the site of protein labeling (usually, a solvent-exposed cysteine residue or a cysteine residue introduced by appropriate site-directed mutagenesis) to achieve reasonable reaction yields and avoid perturbation in the protein structure.1 In spite of these limitations, Ackerson and coworkers14 successfully achieved the labeling of proteins with AuMPCs (Fig. 1b). Specifically, AuMPC was conjugated to a single chain fragment (scFv) from a tetrameric protein, the anti-influenza N9 neuraminidase NC10 antibody (the protein C-terminal tail was previously mutated to incorporate an exposed cysteine residue).14 The AuMPC used in experiments contains glutathione (SG, a peptide of Glu-Cys-Gly sequence shown in the right panel of Fig. 2) as ligands and a gold cluster core of uncertain composition.14 Unfortunately, the molecular mechanism of the ligand-exchange reaction involving the NC10 antibody –or any other protein– remains unknown, and this hampers the design of more effective protein mutants and AuMPCs for bioconjugation purposes.
In this work, we uncover the mechanism of the reaction of AuMPC towards the scFv fragment of the NC10 antibody using a combination of molecular dynamics and ab initio QM/MM techniques. Our results provide a microscopic description of the reaction for the first time, allowing predictions regarding the regioselectivity of the reaction (which peptidic ligand will preferentially be substituted) and explaining why certain protein mutants react better than others.
Methods
Model building
The initial coordinates of the protein atoms were taken from the X-ray crystal structure of a complex between the NC10 antibody and the influenza virus neuraminidase (PDB code 1NMB). The heavy chain of the NC10 scFv, which is the one binding to the gold cluster,14 was considered. In silico mutation of the C-terminal tail was carried out to model the reactive protein mutant: scFv—Ala-Lys-Lys-Glu-Cys-Gly (a Au–Cys bond is expected to be formed upon labelling the protein with AuMPC). The three last residues (Glu-Cys-Gly) match the glutathione sequence (Fig. 2, right panel) and will substitute one of the AuMPC ligands during the reaction. Further details can be found in the ESI.†Classical MD simulations
Classical MD simulations using the AMBER software15 (see ESI, pages S2 and S3†) were initially performed to equilibrate the structure of the protein alone. As AuMPC we considered Au25(SG)18−,16,17 a small and stable gold cluster with an 8-electron electronic shell closing.18,19 Au25(SR)18− clusters have been widely used for ligand exchange reactions.20–26 The fact that this cluster includes dimeric staple motifs27 introduces a higher degree of complexity with respect to the reaction involving monomeric staples (Fig. 1), as it is not obvious a priori whether the sulfur atom participating in the reaction (the thiolate being exchanged) will be the apex one or the side one (Fig. 2, right panel) upon bioconjugation to the protein. Classical MD simulations were also performed to check that the protein structure is stable after the ligand exchange reaction (ESI page 11†).QM/MM simulations
The structure of Au25(SG)18− was taken from our previous study on the electronic properties of this AuMPC in the gas-phase and water solution.13Ab initio QM/MM MD simulations, using the Car-Parrinello Molecular Dynamics (CPMD) program,28,29 were used to identify the most solvent-exposed gold atom (hereafter referred as Au*), in the spirit of the work of Heinecke et al.30 (see further details in the ESI†). To build the initial protein⋯AuMPC complex, we manually placed the sulfhydryl group of the protein tail Cys residue in the vicinity (at ≈8 Å) of Au*. The protein⋯Au25(SG)18− complex (Fig. 2, left panel) was equilibrated by classical (120 ns) and QM/MM MD (7 ps) simulations. The following QM-MM partitioning was used: the 25 gold atoms and the cysteine (–C–CH2–S– fragment) of the 18 SG ligands, as well as the nucleophilic cysteine of the protein (Cys118, –C–CH2–SH fragment) (in total 121 atoms) were treated by quantum mechanics, using DFT and the PBE functional.31 Similar methodology has been used in previous works on reaction mechanisms.32–35 The rest of the system (i.e. part of cysteines, glutamates, glycines, sodium cations and water molecules, in total 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 253 atoms) was treated with the classical force-fields. Interestingly, the incoming cysteine residue (the nucleophilic Cys118) approached the Au* atom during the MD equilibration (from 8 Å to 6 Å). The distance was further reduced to 4.6 Å during the QM/MM MD simulation, being well oriented for ligand replacement.
253 atoms) was treated with the classical force-fields. Interestingly, the incoming cysteine residue (the nucleophilic Cys118) approached the Au* atom during the MD equilibration (from 8 Å to 6 Å). The distance was further reduced to 4.6 Å during the QM/MM MD simulation, being well oriented for ligand replacement.
To model the ligand exchange reaction, we progressively approached the nucleophilic Cys to the Au* atom, considering as reaction coordinate the difference between the distance of the cysteine sulfur atom and the Au* atom (dSCys–Au*) and the distance between the Au* atom and the sulfur atom of the glutathione leaving group (dAu*–SGSH). A series of constrained QM/MM optimizations were performed varying the reaction coordinate in small steps of 0.2–0.3 Å (see pages S8–S10 of the ESI†) in a sequential way. All the remaining degrees of freedom (including the protein atoms, solvent atoms and the AuMPC) were free to move. Structural optimization was performed by annealing of the ionic velocities in an ab initio molecular dynamics simulation, until the maximum component of the nuclear gradient is lower than 10−4 au per bohr (10−5 au per bohr for the gradient norm).
Results and discussion
As mentioned above, it is unknown whether the preferred glutathione ligand being replaced will be the apex one or the side one (Fig. 1a). Therefore, separate simulations were designed considering two different scenarios, depending on whether the substitution takes place at Sapex or Sside (Fig. 2, right panel).Substitution of the side glutathione
Fig. 3a show the results obtained for ligand exchange at the side glutathione ligand. Consistent with experimental observations of associative mechanisms,10,36 the reaction starts by adsorption of the incoming protein sulfur atom (SCys) on Au*. The adsorbed state exhibits a SCys–Au* distance of 3.54 Å, which is similar to previously reported distances for a ligand exchange reaction in the gas-phase.30 Following adsorption, a two-step reaction was observed. In the first step, the incoming protein sulfur atom approaches Au* from 3.54 Å to 2.41 Å while the leaving SG breaks its bond with Au*, increasing the Au*–Sside distance (from 2.30 Å to 3.57 Å; see Fig. 3a). This leads to an intermediate state in which both the incoming thiol and the leaving AuMPC ligand are covalently bonded to the gold cluster (Fig. 3a and S4†). At this state, the hydrogen atom of the cysteine thiol is hydrogen bonded to Sside, being well prepared for proton transfer to the leaving group. The energy barrier for the first reaction step is 13.5 kcal mol−1 and the energy of the intermediate is 6.5 kcal mol−1 below that of the reactants (Fig. 4). | ||
| Fig. 4 Energy profiles of the ligand exchange reaction of the NC10 antibody towards Au25(SG)18− considering substitution of the side SG (black) or the apex SG (blue). | ||
To complete the reaction, the incoming thiol binds to Au′ and displaces Sside in a second nucleophilic substitution. The abstraction of the cysteine hydrogen atom (Hs in Fig. 3) by the leaving SG group occurs spontaneously during this step, resulting in the SG release. The ligand exchange reaction is completed at this point (i.e. one SG ligand of AuMPC has been replaced by the protein C-terminal tail). The transition state of the second reaction step is 10 kcal mol−1 above the reactants and the overall reaction is exothermic by 13.2 kcal mol−1. Therefore, the reaction is favourable both kinetically and thermodynamically, with the first step being rate-limiting.
Substitution of the apex glutathione
The calculations considering ligand exchange at the apex glutathione ligand (Fig. 3b and S5†) also started with absorption of the incoming cysteine, but the adsorbed state shows a longer SCys–Au* distance (3.77 Å) than in the previous case (3.54 Å). In addition, significant structural and energetic differences were found in comparison with the substitution at the side SG ligand. In the first reaction step, the incoming thiol binds to the Au* atom (S–Au* = 2.36 Å; Fig. 3b) and cleavage of the contiguous Au*–Sapex occurs (Au*–Sapex = 3.15 Å; Fig. 3b). The activation barrier for the first reaction step (11.6 kcal mol−1) is similar to the one obtained for the substitution at the side ligand. However, the hydrogen atom of the cysteine thiol at the reaction intermediate is hydrogen bonded to a carbonyl oxygen atom of the leaving SG (1.84 Å), being still far from its proton acceptor (Hs⋯Sapex = 3.98 Å). Moreover, the staple motif is partially open (the Au* atom is away from the other staple gold atoms) to accommodate the incoming thiol (Fig. 3b).As a result, the intermediate is much less stable (6.5 kcal mol−1 above the reactants state) compared to the one obtained for substitution at the side ligand. Afterwards, the cysteine thiol performs a second nucleophilic attack towards Au′′ and the apex SG ligand is released. As in the previous case, abstraction of the cysteine proton occurs simultaneously with the second nucleophilic attack. However, the activation barrier of this step is very large (36.6 kcal mol−1) and the energy of the reaction products is above that of the reactants. Therefore, substitution at the apex SG ligand is unfavorable. Similar conclusions were reached by Fernando & Aikens in a study of the ligand exchange reaction of Au25(SH)18− nanocluster with CH3SH as the incoming thiol ligand.37 Apex substitution (named “central” in the former study) was found less favorable than side (i.e. “terminal”) substitution by ca. 10 kcal mol−1. Considering the different ligands involved in the exchange reaction (CH3SH in Fernando & Aikens and SG in our work, which are much voluminous and charged) and the different environments (protein vs. non-protein), the fact that similar trends are found reflects that the essential features of the reaction are independent of the nature of the thiolate ligands and the presence of the protein.
The different results obtained for the ligand exchange reaction involving the two types of SG ligands (side and apex) can be rationalized in terms of electronic and structural factors at the second reaction step. In particular, they depend on the properties of two atoms involved in the reaction: the hydrogen atom of the sulfhydryl group of the incoming cysteine (Hs) and the gold atom bonded to the leaving SG in the intermediate state (Au′ in the case of side substitution and Au′′ for apex substitution). At the reaction intermediate, the incoming thiol is covalently bonded to Au*, which results in charge transfer from the bonded thiol to the gold cluster.38 As discussed in previous studies,38–41 charge transfer from bound thiols to gold atoms promotes proton transfer. In the case of substitution at the side SG, the Au′ atom is basic at the reaction intermediate, as reflected by its atomic Hirshfeld charge (−0.1e, Fig. S8†), and this facilitates proton transfer. Moreover, Hs is pointing towards Sside, thus being well prepared for proton transfer to the leaving SG group.
In contrast, the atomic charge of Au′′ in the case of substitution at the apex SG is positive (0.3e, Fig. S11†), and no Hs⋯Au interactions can assist the proton transfer to the leaving SG group, resulting in a high energy barrier. We think that this effect is independent of the charge state of the gold cluster, as the additional negative charge (−1) resides in the inner gold core. A second factor that increases the activation energy of the substitution at the apex SG ligand is that the Au* atom needs to recover its position in the dimeric staple and this atomic reorganization has a larger energetic cost compared with substitution at the side SG, which does not involve significant rearrangements of the gold core. Such an atomic rearrangement, previously observed in non-protein environments,30,37 only occurs at the intermediate state, as the Au–Au bond is again formed during the second step of the reaction mechanism. Altogether results show a lower energy barrier for substitution at the side ligand compared with the apex one. Therefore, our calculations predict that the ligand exchange reaction in scFv⋯Au25(SG)18− will preferentially take place at the side SG ligand.
Effect of mutations of the protein C-terminal tail
As a last step in our investigation, we considered how the composition of the C-terminal tail of scFv affects the mechanism and the energy profile of the ligand exchange reaction. Experimentally, it has been found that different mutants of the NC10 antibody react differently with AuMPCs. In particular, the scFv—Ala-Lys-Lys-Glu-Cys-Gly mutant (hereafter named mutant I, the present case) shows quantitative reaction, whereas the scFv—Cys mutant (hereafter named mutant II) does not react at all. The two antibody mutants exhibit a different type of residues in the vicinity of the reactive cysteine. Mutant II displays only neutral residues (either hydrophobic, e.g. leucine and valine, or polar uncharged, e.g. threonine and cysteine), while mutant I exhibits both neutral (Ala and Gly) and charged residues (Lys and Glu). The presence of positively charged groups in the vicinity of the reactive cysteine was suggested to be the main reason of the enhanced reactivity of mutant I towards the negatively charged AuMPC.14To shed light on these variations, we performed additional MD simulations on mutant II scFv⋯Au25(SG)18−. As in the former case, the sulfhydryl group of the reactive cysteine was placed near the Au* atom and the Au25(SG)18− complex was equilibrated by classical and QM/MM MD simulations. The system displayed large oscillations of the SCys–Au* distance compared with mutant I, but the protein tail was still able to get close to AuMPC, up to a distance of 4.8 Å from the Au* atom, i.e. just 0.3 Å longer than the one obtained for the QM/MM optimized structure of mutant I scFv⋯Au25(SG)18− (before adsorption occurs). To test how mutation of the C-terminal tail residues affects the energy barriers, we performed additional QM/MM optimizations of mutant II starting from the stationary states obtained for mutant I, considering substitution at the side SG ligand (Fig. 3a and S4†). The energy barriers for the two reaction steps turned out to be significantly high (31.7 kcal mol−1 and 24.2, respectively, Fig. S15†), explaining why this mutant is inactive towards ligand exchange reaction.14
The reason for the different reactivity of the two mutants can be related to the protein⋯AuMPC interactions formed upon adsorption of the reactive cysteine side chain (Fig. 5). Analysis of the structures obtained along the ligand exchange reaction shows that the two lysine residues of the protein tail of mutant I strongly interact with the carboxylate groups of the AuMPC SG ligands. These interactions, absent in mutant II (the protein tail does not contain charged residues), lead to a tight binding between the antibody and AuMPC. In contrast, mutant II exhibits only much weaker serine–carboxylate interactions, consistent with the large oscillations of the SCys–Au* distance (Fig. S13†). Therefore, we conclude that the higher energy barriers observed for mutant II are due to the weaker adsorption of the reactive cysteine and the looser peptide–AuMPC interactions compared to mutant I. Our results also show that QM/MM techniques can be useful predictive tools to rationalize experiments of AuMPC binding to proteins.
Conclusions
Our combined classical and QM/MM MD study shows that the ligand exchange reaction in the scFv⋯Au25(SG)18− complex consists of two SN2-like reactions, the second one coupled with a proton transfer. However, the molecular mechanism and the identity of the rate-limiting step strongly depend on which SG ligand is being substituted, with the side ligand favored over the apex one. Therefore, our calculations predict that bioconjugation will preferentially take place on the former ligand. Interestingly, a recent X-ray study20 revealed that only the side thiolate bonded to the most solvent-exposed gold atom is substituted when Au25(SC2H4Ph)18 was exposed to a 5-fold molar excess of pBBT for 7 min. This suggests that the mechanism obtained here can be transferred to non-protein environments. Furthermore, the calculations show that the effectiveness of the reaction depends on the presence of positive residues at the protein C-terminal tail that can form attractive intermolecular interactions with the carboxylate groups of the SG ligands, facilitating the adsorption of the protein cysteine on the gold cluster surface.Acknowledgements
This work was supported by the Academy of Finland (Project 284621 to J. A.), Generalitat de Catalunya (AGAUR grant 2014SGR-987 to C. R.) and the Spanish Ministry of Economy and Competitiveness (MINECO. grant CTQ2014-55174-P to C. R.). V. R.-C. and L. R. acknowledge predoctoral fellowships from MINECO (FPU, AP2009-3024) and University of Barcelona (APIF), respectively. We thankfully acknowledge the computer resources and the technical support provided by BSC-CNS (QCM-2011-2-0024) (MareNostrum, Barcelona, Spain) and the CSC – IT Centre for Science (Sisu - Cray XC40, Espoo, Finland).References
- C. J. Ackerson, R. D. Powell and J. F. Hainfeld, Methods Enzymol., 2010, 481, 195–230 CAS.
- R. Jin, Nanoscale, 2015, 7, 1549–1565 RSC.
- M. Walter, J. Akola, O. Lopez-Acevedo, P. D. Jadzinsky, G. Calero, C. J. Ackerson, R. L. Whetten, H. Gronbeck and H. Hakkinen, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 9157–9162 CrossRef CAS PubMed.
- J. Akola, M. Walter, R. L. Whetten, H. Hakkinen and H. Gronbeck, J. Am. Chem. Soc., 2008, 130, 3756–3757 CrossRef CAS PubMed.
- X. Tan and R. Jin, WIREs Nanomed. Nanobiotechnol., 2013, 5, 569–581 CrossRef CAS PubMed.
- Q. Yuan, Y. Wang, L. Zhao, R. Liu, F. Gao, L. Gao and X. Gao, Nanoscale, 2016, 8, 12095–12104 RSC.
- N. S. Abadeer and C. J. Murphy, J. Phys. Chem. C, 2016, 120, 4691–4716 CAS.
- R. A. Sperling, P. Rivera Gil, F. Zhang, M. Zanella and W. J. Parak, Chem. Soc. Rev., 2008, 37, 1896–1908 RSC.
- S. Guerrero, E. Araya, J. L. Fiedler, J. I. Arias, C. Adura, F. Albericio, E. Giralt, J. L. Arias, M. S. Fernandez and M. J. Kogan, Nanomedicine, 2010, 5, 897–913 CrossRef CAS PubMed.
- M. J. Hostetler, A. C. Templeton and R. W. Murray, Langmuir, 1999, 15, 3782–3789 CrossRef CAS.
- A. C. Templeton, M. J. Hostetler, C. T. Kraft and R. W. Murray, J. Am. Chem. Soc., 1998, 120, 1906–1911 CrossRef CAS.
- P. D. Jadzinsky, G. Calero, C. J. Ackerson, D. A. Bushnell and R. D. Kornberg, Science, 2007, 318, 430–433 CrossRef CAS PubMed.
- V. Rojas-Cervellera, C. Rovira and J. Akola, J. Phys. Chem. Lett., 2015, 6, 3859–3865 CrossRef CAS PubMed.
- C. J. Ackerson, P. D. Jadzinsky, G. J. Jensen and R. D. Kornberg, J. Am. Chem. Soc., 2006, 128, 2635–2640 CrossRef CAS PubMed.
- D. A. Case, T. A. Darden, T. E. Cheatham, C. Simmerling, J. Wang, R. Duke, R. Luo, M. F. Crowley, R. Walker, W. Zhang, K. M. Merz, B. Wang, S. Hayik, A. E. Roitberg, G. Seabra, I. Kolossváry, K. F. Wong, F. Paesani, J. Vanicek, X. Wu, S. Brozell, T. Steinbrecher, H. Gohlke, L. Yang, C. Tan, J. Mongan, V. Hornak, G. Cui, D. H. Mathews, M. G. Seetin, C. Sagui, V. Babin and P. Kollman, AMBER 11, University of California, San Francisco, 2010 Search PubMed.
- M. Zhu, C. M. Aikens, F. J. Hollander, G. C. Schatz and R. Jin, J. Am. Chem. Soc., 2008, 130, 5883–5885 CrossRef CAS PubMed.
- G. Li, H. Abroshan, C. Liu, S. Zhuo, Z. Li, Y. Xie, H. J. Kim, N. L. Rosi and R. Jin, ACS Nano, 2016, 10, 7998–8005 CrossRef CAS PubMed.
- Y. Negishi, N. K. Chaki, Y. Shichibu, R. L. Whetten and T. Tsukuda, J. Am. Chem. Soc., 2007, 129, 11322–11323 CrossRef CAS PubMed.
- N. Van Steerteghem, S. Van Cleuvenbergen, S. Deckers, C. Kumara, A. Dass, H. Hakkinen, K. Clays, T. Verbiest and S. Knoppe, Nanoscale, 2016, 8, 12123–12127 RSC.
- T. W. Ni, M. A. Tofanelli, B. D. Phillips and C. J. Ackerson, Inorg. Chem., 2014, 53, 6500–6502 CrossRef CAS PubMed.
- S. Si, C. Gautier, J. Boudon, R. Taras, S. Gladiali and T. Bürgi, J. Phys. Chem. C, 2009, 113, 12966–12969 CAS.
- J. F. Parker, K. A. Kacprzak, O. Lopez-Acevedo, H. Häkkinen and R. W. Murray, J. Phys. Chem. C, 2010, 114, 8276–8281 CAS.
- C. A. Fields-Zinna, J. F. Parker and R. W. Murray, J. Am. Chem. Soc., 2010, 132, 17193–17198 CrossRef CAS PubMed.
- V. R. Jupally, R. Kota, E. V. Dornshuld, D. L. Mattern, G. S. Tschumper, D.-E. Jiang and A. Dass, J. Am. Chem. Soc., 2011, 133, 20258–20266 CrossRef CAS PubMed.
- S. Knoppe and T. Burgi, Phys. Chem. Chem. Phys., 2013, 15, 15816–15820 RSC.
- T. M. Carducci, R. E. Blackwell and R. W. Murray, J. Phys. Chem. Lett., 2015, 6, 1299–1302 CrossRef CAS PubMed.
- M. W. Heaven, A. Dass, P. S. White, K. M. Holt and R. W. Murray, J. Am. Chem. Soc., 2008, 130, 3754–3755 CrossRef CAS PubMed.
- CPMD program, Copyright IBM Corp. 1990–2015, Copyright MPI für Festkörperforschung, Stuttgart 1997–2001. http://www.cpmd.org.
- A. Laio, J. VandeVondele and U. Rothlisberger, J. Chem. Phys., 2002, 116, 6941–6947 CrossRef CAS.
- C. L. Heinecke, T. W. Ni, S. Malola, V. Mäkinen, O. A. Wong, H. Häkkinen and C. J. Ackerson, J. Am. Chem. Soc., 2012, 134, 13316–13322 CrossRef CAS PubMed.
- J. P. Perdew, K. Burke and M. Ernzerhof, Phys. Rev. Lett., 1996, 77, 3865–3868 CrossRef CAS PubMed.
- J. Iglesias-Fernandez, L. Raich, A. Ardèvol and C. Rovira, Chem. Sci., 2015, 6, 1167–1177 RSC.
- A. Ardevol, J. Iglesias-Fernandez, V. Rojas-Cervellera and C. Rovira, Biochem. Soc. Trans., 2016, 44, 51–60 CrossRef CAS PubMed.
- A. Ardèvol and C. Rovira, J. Am. Chem. Soc., 2015, 137, 7528–7547 CrossRef PubMed.
- M. Boero, K. Terakura and M. Tateno, J. Am. Chem. Soc., 2002, 124, 8949–8957 CrossRef CAS PubMed.
- Y. Song and R. W. Murray, J. Am. Chem. Soc., 2002, 124, 7096–7102 CrossRef CAS PubMed.
- A. Fernando and C. M. Aikens, J. Phys. Chem. C, 2015, 119, 20179–20187 CAS.
- V. Rojas-Cervellera, E. Giralt and C. Rovira, Inorg. Chem., 2012, 51, 11422–11429 CrossRef CAS PubMed.
- M. Hasan, D. Bethell and M. Brust, J. Am. Chem. Soc., 2002, 124, 1132–1133 CrossRef CAS PubMed.
- H. Gronbeck, A. Curioni and W. Andreoni, J. Am. Chem. Soc., 2000, 122, 3839–3842 CrossRef.
- W. Andreoni, A. Curioni and H. Grönbeck, Int. J. Quantum Chem., 2000, 80, 598 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Further computational details and data analysis. See DOI: 10.1039/c6nr08498b |
| This journal is © The Royal Society of Chemistry 2017 |