Increasing H2SO4 contents in disulfates cresting in the new hydrogenium-bis-hydrogendisulfate anion [H(HS2O7)2]−: Li2[S2O7]·H2SO4, Li[HS2O7], and Li[H(HS2O7)2]
Lisa Verena
Schindler
and
Mathias S.
Wickleder
*
Justus-Liebig-University of Gießen, Institute of Inorganic and Analytical Chemistry, Heinrich-Buff-Ring 17, D-35392 Gießen, Germany. E-mail: mathias.wickleder@anorg.chemie.uni-giessen.de; Fax: +49 (641) 99 34109
First published on 25th October 2016
Abstract
The reactions of simple Li salts like sulfates, carbonates, and chlorides with fuming sulfuric acid or neat SO3 led to a couple of new acidic Li disulfates with varying H2SO4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SO3 ratios, namely Li2[S2O7]·H2SO4 (orthorhombic, Pca21, Z = 4, a = 945.67(8) pm, b = 485.77(4) pm, c = 1793.9(1) pm, V = 824.1(1) × 106 pm3, R1 (all data) = 0.138, wR2 (all data) = 0.0380), Li[HS2O7] (monoclinic, Cc, Z = 4, a = 685.87(3) pm, b = 788.49(4) pm, c = 911.48(5) pm, β = 101.618(2)°, V = 482.83(4) × 106 pm3, R1 (all data) = 0.148, wR2 (all data) = 0.0416), and Li[H(HS2O7)2] (monoclinic, P21/n, Z = 2, a = 498.69(2) pm, b = 808.57(3) pm, c = 1297.22(5) pm, β = 94.875(2)°, V = 521.18(3) × 106 pm3, R1 (all data) = 0.157, wR2 (all data) = 0.0481). The stamping structural feature of these salts are moderate and strong hydrogen bonds linking the anions to chains and layers. Especially for Li[H(HS2O7)2] a very strong symmetrical hydrogen bond is found linking two hydrogendisulfate anions via a common hydrogen atom to the new hydrogenium-bis-hydrogendisulfate anion [H(HS2O7)2]−.
SO3 ratios, namely Li2[S2O7]·H2SO4 (orthorhombic, Pca21, Z = 4, a = 945.67(8) pm, b = 485.77(4) pm, c = 1793.9(1) pm, V = 824.1(1) × 106 pm3, R1 (all data) = 0.138, wR2 (all data) = 0.0380), Li[HS2O7] (monoclinic, Cc, Z = 4, a = 685.87(3) pm, b = 788.49(4) pm, c = 911.48(5) pm, β = 101.618(2)°, V = 482.83(4) × 106 pm3, R1 (all data) = 0.148, wR2 (all data) = 0.0416), and Li[H(HS2O7)2] (monoclinic, P21/n, Z = 2, a = 498.69(2) pm, b = 808.57(3) pm, c = 1297.22(5) pm, β = 94.875(2)°, V = 521.18(3) × 106 pm3, R1 (all data) = 0.157, wR2 (all data) = 0.0481). The stamping structural feature of these salts are moderate and strong hydrogen bonds linking the anions to chains and layers. Especially for Li[H(HS2O7)2] a very strong symmetrical hydrogen bond is found linking two hydrogendisulfate anions via a common hydrogen atom to the new hydrogenium-bis-hydrogendisulfate anion [H(HS2O7)2]−.
Introduction
Sulfuric acid and its salts are important chemicals for both industry and research. The wide array of substances is well investigated and especially the salts containing the hydrogen-free anion SO42− come up with an outstanding structural diversity. The number of compounds containing the protonated anion HSO4− is considerably smaller. The acidic sulfates of the alkaline metals are listed in Table 1 sorted by growing H2SO4 and decreasing H2O content in the first part and growing SO3 and decreasing H2SO4 content in the second part. The stamping feature of all these structures is the formation of strong or moderate hydrogen bonds resulting in a variety of motifs and linking modes. Contemplation of the first part of that table leads to the awareness that Li compounds tend to higher H2SO4 contents in their structures than other alkaline metals. While Na, K, Rb, and Cs bear a couple of structures with a M2[SO4]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2SO4 ratio (M = Na, K, Rb, Cs) greater than one, like M3[H(SO4)2] (M = Na, K, Rb; M2[SO4]
H2SO4 ratio (M = Na, K, Rb, Cs) greater than one, like M3[H(SO4)2] (M = Na, K, Rb; M2[SO4]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2SO4 ratio 1
H2SO4 ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.33)1–3 or M5H3[SO4]4 (M = Rb, Cs; M2[SO4]
0.33)1–3 or M5H3[SO4]4 (M = Rb, Cs; M2[SO4]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2SO4 ratio 1
H2SO4 ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.67),4,5 the acidic Li sulfate with the lowest H2SO4 content, Li[HSO4],6 exhibits a Li2[SO4]
0.67),4,5 the acidic Li sulfate with the lowest H2SO4 content, Li[HSO4],6 exhibits a Li2[SO4]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2SO4 ratio of 1
H2SO4 ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, the two other acidic Li sulfate salts show Li2[SO4]
1, the two other acidic Li sulfate salts show Li2[SO4]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2SO4 ratios smaller than one (1
H2SO4 ratios smaller than one (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 in Li2[HSO4]2·H2SO47 and 1
3 in Li2[HSO4]2·H2SO47 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 in Li[H(HSO4)2]·2H2SO48). Furthermore, Li[H(HSO4)2]·2H2SO4 has the highest H2SO4 content of all acidic alkaline sulfates. The second part of Table 1, the list of acidic alkaline metal sulfates with an excess of SO3, was primarily filled only in the recent past. Just one year ago only two acidic alkaline sulfates with an SO3 excess have been known, namely the double salt K2[S2O7]·K[HSO4]9 and the H2SO4 adduct K2[S2O7]·H2SO4,10 and also these structures have been reported just within the last ten years. With the crystal structure of Rb3[H(HSO4)2][S2O7] we lately presented a Rb salt with an SO3 content comparable to the mentioned K compounds.11 Furthermore, we contributed our comprehensive studies on hydrogenpolysulfates M[HS2O7] (M = K, (NH4), (NO), Rb, Cs)12 and M[HS3O10] (M = Na, K, Rb).13 At the point of publication in each case these compounds were the hydrogensulfates with the highest SO3 content (M2[SO4]
7 in Li[H(HSO4)2]·2H2SO48). Furthermore, Li[H(HSO4)2]·2H2SO4 has the highest H2SO4 content of all acidic alkaline sulfates. The second part of Table 1, the list of acidic alkaline metal sulfates with an excess of SO3, was primarily filled only in the recent past. Just one year ago only two acidic alkaline sulfates with an SO3 excess have been known, namely the double salt K2[S2O7]·K[HSO4]9 and the H2SO4 adduct K2[S2O7]·H2SO4,10 and also these structures have been reported just within the last ten years. With the crystal structure of Rb3[H(HSO4)2][S2O7] we lately presented a Rb salt with an SO3 content comparable to the mentioned K compounds.11 Furthermore, we contributed our comprehensive studies on hydrogenpolysulfates M[HS2O7] (M = K, (NH4), (NO), Rb, Cs)12 and M[HS3O10] (M = Na, K, Rb).13 At the point of publication in each case these compounds were the hydrogensulfates with the highest SO3 content (M2[SO4]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2SO4
H2SO4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SO3: 1
SO3: 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 for M[HS2O7], 1
2 for M[HS2O7], 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 for M[HS3O10]). In the meanwhile we presented an even longer protonated polysulfate chain, namely the unique hydrogenium-bis-tetrasulfate anion [H(S4O13)2]3− in the crystal structure of Li3[H(S4O13)2] with a Li2[SO4]
4 for M[HS3O10]). In the meanwhile we presented an even longer protonated polysulfate chain, namely the unique hydrogenium-bis-tetrasulfate anion [H(S4O13)2]3− in the crystal structure of Li3[H(S4O13)2] with a Li2[SO4]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2SO4
H2SO4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SO3 ratio of 1
SO3 ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.33
0.33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4.14 In the present work we report on a couple of new acidic Li disulfates with varying H2SO4 contents, namely the H2SO4 adduct Li2[S2O7]·H2SO4, the hydrogendisulfate Li[HS2O7], and the unique hydrogenium-bis-hydrogendisulfate Li[H(HS2O7)2] which shows the up to now unknown anion [H(HS2O7)2]− in its structure. Li[H(HS2O7)2] is the alkaline metal sulfate with the highest H2SO4 content known so far.
4.14 In the present work we report on a couple of new acidic Li disulfates with varying H2SO4 contents, namely the H2SO4 adduct Li2[S2O7]·H2SO4, the hydrogendisulfate Li[HS2O7], and the unique hydrogenium-bis-hydrogendisulfate Li[H(HS2O7)2] which shows the up to now unknown anion [H(HS2O7)2]− in its structure. Li[H(HS2O7)2] is the alkaline metal sulfate with the highest H2SO4 content known so far.
M2[SO4]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2SO4 H2SO4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O H2O |
Li | Na | K | Rb | Cs |
|---|---|---|---|---|---|
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.33 0.33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
Na3[H(SO4)2]1 | K3[H(SO4)2]3 | Rb3[H(SO4)2]2 | ||
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.67 0.67![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.2 0.2 |
Cs5H3[SO4]4·0.48H2O15 | ||||
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.67 0.67![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
Rb5H3[SO4]45 | Cs5H3[SO4]44 | |||
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.77 0.77![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.22 0.22 |
K9H7[SO4]8·H2O16 | ||||
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.77 0.77![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
K9H7[SO4]816 | ||||
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
Na[HSO4]·H2O17 | ||||
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
Li[HSO4]6 | Na[HSO4]18 | K[HSO4]19 | Rb[HSO4]20 | Cs[HSO4]21 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
Na[H3O][HSO4]222 | K[H3O][HSO4]223 | |||
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
Li2[HSO4]2·H2SO47 | Na2[HSO4]2·H2SO422 | K[HSO4]·H2SO424 | Rb[HSO4]·H2SO425 | Cs[HSO4]·H2SO425 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
Na[HSO4]·2H2SO422 | ||||
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
Li[H(HSO4)2]·2H2SO48 |
Results and discussion
Li2[S2O7]·H2SO4 crystallizes in the orthorhombic space group Pca21 (no. 29) with four formula units per unit cell. The structure shows the disulfate anion [S2O7]2− adjacent to sulfuric acid H2SO4, a motif that is known from the analogous potassium compound K2[S2O7]·H2SO4.10 The disulfate anion is built up by the vertex connection of two crystallographically distinguishable sulfate tetrahedra, [S1O4] and [S2O4], via the bridging oxygen atom O121 (Fig. 1). The S–O121 bonds are elongated (163.82(6) pm, 162.93(6) pm) compared with the S–O bonds to the terminal oxygen atoms (143.38(7)–145.25(6) pm) which is a common finding for polysulfate anions. The asymmetry of the oxygen bridge is less distinctive than in the according disulfate Li2[S2O7] (161.0(2) pm/166.7(2) pm), the S–O bond lengths to the terminal oxygen atoms are in the same range (143.7(2)–145.2(2) pm).26All atoms of the H2SO4 molecule are located on general positions. The S–O distances to the protonated oxygen atoms O33 and O34 are elongated (153.69(7) pm, 153.89(7) pm) compared with the bonds to the terminal oxygen atoms O31 and O32 (142.61(7) pm, 142.68(7) pm). The sulfuric acid molecules link the disulfate anions via moderate hydrogen bonds (donor–acceptor distance O33–O12: 272.8(1) pm, O34–O22: 269.16(9) pm) to zig-zag shaped chains proceeding along [001] (Fig. 1 and 2), in Fig. 2 a single chain is highlighted in blue. Further details on the hydrogen bonding system are given in Table 2. The hydrogen bonding is significantly weaker than in the analogous potassium salt (donor–acceptor distances: 253.7(1) pm, 259.4(1) pm), furthermore the resulting motif there is a four-membered ring in contrast to the chains in the presented Li salt.10
| Donor atom–hydrogen atom | D–H distance/pm | H–A distance/pm | D–H–A angle/° | D–A distance/pm | Acceptor atom | |
|---|---|---|---|---|---|---|
| Li2[S2O7]·H2SO4 | O33–H33 | 81.4 | 191.9 | 172.81 | 272.8 | O12 |
| O34–H34 | 79.2 | 190.5 | 172.05 | 269.2 | O22 | |
| Li[HS2O7] | O13–H13 | 60.6 | 200.9 | 173.03 | 261.1 | O23 |
| Li[H(HS2O7)2] | O13–H13 | 80.8 | 163.9 | 170.03 | 243.8 | O13 |
| O23–H23 | 87.2 | 175.8 | 164.82 | 261.0 | O22 | |
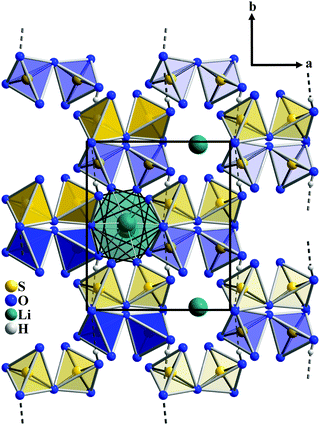
The Li+ cations are located on two crystallographic positions, Li1 and Li2. Both ions are in fivefold coordination of oxygen atoms in the shape of a square pyramid with the distances Li–O ranging from 195.8(2) pm to 218.7(2) pm (Fig. 3). The coordination polyhedra are linked via a common edge to [Li2O8] dimers with a quite short Li–Li distance of 319.3(3) pm; the apices of the pyramids point into opposite directions. A partial occupation of the Li sites that would reduce the Li+–Li+ repulsion within the dimers can be ruled out based on bond valence sum calculations which corroborate the assignment of lithium and hydrogen positions. Primarily oxygen atoms of the disulfate anion take part in the cation coordination, the bridging oxygen atom O121 is not involved. [S2O7]2− acts exclusively as a bidentate ligand. Like the bridging oxygen atom, the protonated oxygen atoms of the molecular sulfuric acid do not take part in the Li+ coordination, a monodentate attack of the cation is realized via each terminal oxygen atom of H2SO4. The [Li2O8] dimers link the [S2O7] units to layers parallel to (110). The layers are stacked along [001] and alternate with layers just containing sulfuric acid as shown in Fig. 2. The same arrangement of layers of the composition M2[S2O7] (M = Li, K) and layers only containing H2SO4 is found in the structure of the analogous potassium salt K2[S2O7]·H2SO4.10 The proceeding of the hydrogen bonded chains in the structure of Li2[S2O7]·H2SO4 is perpendicular to the layers.
 | ||
| Fig. 3 Coordination of the Li+ cations in the crystal structure of Li2[S2O7]·H2SO4. The square pyramidal coordination polyhedra share common edges with their apices pointing into opposite directions. | ||
Li[HS2O7] crystallizes in the acentric space group Cc (no. 9) with a flack-x parameter of 0.47(2). A detailed consideration on the choice of the space group is given in the Experimental section. The unit cell contains four formula units, all atoms are located on general positions. The anion is built up by the vertex connection of the protonated sulfate tetrahedra [HS1O4] and the unprotonated sulfate tetrahedra [S2O4] via the bridging oxygen atom O121 (Fig. 4). The S–O bonds to the bridging oxygen atom are elongated, the elongation in the unprotonated tetrahedron is more distinctive (168.2(1) pm vs. 158.2(1) pm) and consequently an asymmetric oxygen bridge is found. These values are in accordance with the values in the structures of comparable hydrogendisulfate salts with low charged counter cations we reported earlier (S1–O121: 159.07(5)–159.80(6) pm, S2–O121: 166.69(6)–168.89(5) pm in the crystal structures of M[HS2O7], M = K, [NH4], [NO], Rb, Cs).12 The S–O bond lengths to the terminal oxygen atoms exhibit values in the range from 141.7(1) pm to 143.1(1) pm, the bond to the oxygen atom O23 which is the acceptor atom in the hydrogen bridge is slightly elongated (145.9(1) pm), the S–OH bond shows a length of 158.2(1) pm.
The hydrogen atom is involved in a hydrogen bond to the acceptor atom O23 linking the anions to chains proceeding along [110] and [−110], respectively (Fig. 5, Table 2). The linkage of [HS2O7]− anions to chains is known from the crystal structure of Cs[HS2O7],12 furthermore the hydrogentrisulfate anions [HS3O10]− in the crystal structures of K[HS3O10] and Rb[HS3O10] are arranged in that way.13 The donor–acceptor distance exhibits a value of 261.1(1) pm which is a moderate hydrogen bond according to the classification of Jeffrey.27 The hydrogen bonding is a little bit weaker than in the other mentioned hydrogendisulfates (D–A distances 252.68(6)–256.88(9) pm).12
The cations are located on a single crystallographic position and coordinated sixfold by oxygen atoms in the distance range from 199.4(3) pm to 244.3(2) pm if distances up to 315 pm are taken into account. The octahedral coordination polyhedron is shown in Fig. 6. The anionic chains cross each other and form channels along [001] providing space for the cations (Fig. 5).
Li[H(HS2O7)2] crystallizes in the monoclinic space group P21/n with two formula units per unit cell. The crystal structure shows the up to now unknown hydrogenium-bis-hydrogendisulfate anion [H(HS2O7)2]− (Fig. 7). The motif, the connection of sulfate tetrahedra via a shared hydrogen atom forming hydrogenium-bis-sulfate anions [H(SO4)2]3−, is known from a couple of acidic sulfates (Table 1),1–3,28,29 the structures of Li[H(HSO4)2](H2SO4)2,8 K3[Pt2(SO4)4H(HSO4)2],30 and Rb3[S2O7][H(HSO4)2]11 bear the even more rarely reported hydrogenium-bis-hydrogensulfate anions [H(HSO4)2]−. Recently we presented the first hydrogenium-bis-polysulfate anion [H(S4O13)2]3− in the crystal structure of Li3[H(S4O13)2] which is longest protonated polysulfate chain ever observed.14 The here described anion [H(HS2O7)2]− is the first hydrogenium-bis-hydrogendisulfate.
In the anion the sulfur atoms are located on two crystallographic positions S1 and S2 (Fig. 7). The two sulfate tetrahedra are linked via the bridging oxygen atom O121. The [S2O7] unit is formally protonated twice at the oxygen atoms O13 and O23, but for the hydrogen atom H13 an occupancy of 0.5 results due to the location close to a center of inversion and for reasons of charge compensation. This finding is also reflected in the respective S–O bond lengths. The S–O distance to the fully protonated oxygen atom O23 (152.40(4) pm) is elongated compared with the bond lengths of the terminal S–O bonds (141.99(4)–142.71(4) pm), the length of the S–O bond to the semi protonated oxygen atom O13 is somewhere in between (147.34(4) pm). The S–O bonds to the bridging oxygen atom O121 are yet longer (S1–O121: 165.94(4) pm, S2–O121: 158.51(4) pm). The hydrogen atom H13 links two crystallographically identical hydrogendisulfate units [HS2O7]−via a short (donor–acceptor distance 243.84(8) pm) and accordingly strong hydrogen bond to the new hydrogenium-bis-hydrogendisulfate anion [H(HS2O7)2]−.27Fig. 8 shows a difference-Fourier-map of the hydrogen bond. The electron density is blurred and the two crystallographically equivalent position close to the center of inversion are not clearly distinguishable, a situation that is often observed for symmetrical hydrogen bonds.11,14 In the comparable structures mentioned above only for Rb3[S2O7][H(HSO4)2] the two positions can be visualized in a difference-Fourier-map,11 for Li3[H(S4O13)2] the difference-Fourier-map even shows the location of the hydrogen atom precisely on the center of inversion.14
 | ||
| Fig. 8 Blurred electron density in the difference-Fourier-map of the hydrogen bond O13–H13–O13 in the crystal structure of Li[H(HS2O7)2] (donor–acceptor distance: 243.84(8) pm). | ||
The protonated ends of the hydrogendisulfate anions O23–H23 develop further hydrogen bonds to the oxygen atoms O22 of adjacent hydrogenium-bis-hydrogendisulfate units (donor–acceptor distance 260.99(6) pm, Table 2) connecting the anions to layers stacked along [101] (Fig. 9).
The Li+ cations on a single crystallographic position are coordinated by six oxygen atoms forming an octahedron with Li–O distances in the range from 210.33(4) pm to 211.52(4) pm if distances up to 320 pm are taken into account (Fig. 10). The coordination polyhedra are located between the hydrogen-bonded layers built by the anions and link them to a three dimensional network (Fig. 9).
Conclusion
The reported compounds represent members of the SO3 rich acidic sulfates with molar ratios Li2[SO4]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2SO4
H2SO4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SO3 of 1
SO3 of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Li2[S2O7]·H2SO4), 1
1 (Li2[S2O7]·H2SO4), 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (Li[HS2O7]), and 1
2 (Li[HS2O7]), and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (Li[H(HS2O7)2]). A higher SO3 content in acidic sulfates is found in the recently presented hydrogentrisulfates M[HS3O10] (M = Na, K, Rb)13 (1
4 (Li[H(HS2O7)2]). A higher SO3 content in acidic sulfates is found in the recently presented hydrogentrisulfates M[HS3O10] (M = Na, K, Rb)13 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) and in Li3[H(S4O13)2] (1
4) and in Li3[H(S4O13)2] (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.3
0.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4).14 The highest H2SO4 content in polysulfate salts shows the here presented structure of Li[H(HS2O7)2]. All structures are stamped by the formation of extended hydrogen bonding systems primarily via moderate hydrogen bonds (donor–acceptor distances 260.99(6)–272.8(1) pm) linking the anions to chains and layers. The symmetrical hydrogen bond linking two hydrogendisulfate anions to the new hydrogenium-bis-hydrogendisulfate anion [H(HS2O7)2]− in the structure of Li[H(HS2O7)2] is a short (donor–acceptor distance 243.84(8) pm) and accordingly strong hydrogen bond. The structures give proof of the stabilization of high contents of H2SO4 coexistent with SO3 and thus arouse high expectations on our further investigations on hydrogenpolysulfates and polysulfuric acids.
4).14 The highest H2SO4 content in polysulfate salts shows the here presented structure of Li[H(HS2O7)2]. All structures are stamped by the formation of extended hydrogen bonding systems primarily via moderate hydrogen bonds (donor–acceptor distances 260.99(6)–272.8(1) pm) linking the anions to chains and layers. The symmetrical hydrogen bond linking two hydrogendisulfate anions to the new hydrogenium-bis-hydrogendisulfate anion [H(HS2O7)2]− in the structure of Li[H(HS2O7)2] is a short (donor–acceptor distance 243.84(8) pm) and accordingly strong hydrogen bond. The structures give proof of the stabilization of high contents of H2SO4 coexistent with SO3 and thus arouse high expectations on our further investigations on hydrogenpolysulfates and polysulfuric acids.
Experimental section
Synthesis of Li2[S2O7]·H2SO4
A 1000 ml three-necked flask was filled with P4O10 (20 g, 97%) and a dropping funnel filled with fuming sulfuric acid (1 ml, 65% SO3; used as received, Merck, Darmstadt, Germany) was connected to a glass ampoule (d = 16 mm; l = 300 mm), which was loaded with Li2CO3 (50 mg, 0.68 mmol, undried, >99%, Chempur, Karlsruhe, Germany) before. Simultaneously to the cooling of the ampoule with liquid nitrogen the fuming sulfuric acid was dropped onto P4O10 within five minutes while heating the three-necked flask with a heatgun. The evaporating SO3 condensed in the ampoule. The ampoule was torch sealed afterwards, placed into a tube furnace and heated up to 80 °C within 12 h. The temperature was maintained for 24 h and the furnace cooled down to room temperature within 120 h afterwards. A small number of colorless and extremely moisture sensitive crystals were obtained. A small excess of SO3 was removed by cooling the end of the ampoule not containing the crystals in liquid nitrogen. A remaining quantity of water in the starting material was sufficient to lead to a H2SO4 adduct.Synthesis of Li[HS2O7] and Li[H(HS2O7)2]
LiCl (300 mg, 7.08 mmol, >99%, Merck, Darmstadt, Germany) was loaded into an ampoule (d = 16 mm; l = 300 mm) for the preparation of Li[HS2O7], whereas Li2SO4 (385 mg, 3.50 mmol, >98%, Chempur, Karlsruhe, Germany) was loaded into a glass tube (d = 16 mm; l = 300 mm) for the preparation of Li[H(HS2O7)2]. Fuming sulfuric acid (1 ml, 65% SO3; used as received, Merck, Darmstadt, Germany) was then added to both ampoules. The tubes were torch-sealed under vacuum, placed in a resistance furnace and heated up to 120 °C within 12 h. The temperature was maintained for 24 h. Upon slow cooling within 120 h colorless and extremely moisture sensitive single crystals were obtained. They were collected by decantation of the mother liquor under inert conditions.Caution! Oleum and SO3 are strong oxidizers which need careful handling. During the reaction and even after cooling down to room temperature the glass tubes might be under pressure. The tubes should be cooled with liquid nitrogen before they are opened.
Structure determination
Single crystals of Li2[S2O7]·H2SO4, Li[HS2O7], and Li[H(HS2O7)2] were selected under protecting oil with the help of a polarization microscope. The crystals were transferred into the cool nitrogen stream (100 K) of a single-crystal diffractometer (BRUKER APEX II) and intensity data were collected.31 The structure solutions assuming the respective space groups were successful applying direct methods (SHELXS).32 Subsequent refinement with SHELXL yielded the complete crystal structures.33 Finally, anisotropic displacement parameters were introduced and a numerical absorption correction was applied to the reflection data. For the structure solution of Li[HS2O7] in the space group Cc a flack-x parameter of 0.47(2) results and correlation between atomic positions is observed (z O12/z O22: 0.634; y O12/y O22: −0.592; x O12/x O22: 0.589; x O21/x O13: 0.559). But the structure solution in the respective centrosymmetric space group C2/c leads to a significant increase of the R values (R1 (Fo > 2σ(Fo)): 0.0566 vs. 0.0144; wR2 (Fo > 2σ(Fo)): 0.1538 vs. 0.0414) and unreasonable bond lengths (S–O bond to a terminal oxygen atom: 148.5(2) pm, S–OH bond to a protonated oxygen atom: 143.1(2) pm). Therefore, the solution in the acentric space group Cc was retained and a twin law was introduced ((−100), (010), (00−1)) leading to a base parameter of 0.56094. All hydrogen atoms in the structures of Li2[S2O7]·H2SO4 and Li[HS2O7] as well as the hydrogen atom H23 in the crystal structure of Li[H(HS2O7)2] are refined without any restraints. For the hydrogen atom H13 in the crystal structure of Li[H(HS2O7)2] an occupancy of 0.5 was defined due to the location close to a center of inversion and for reasons of charge compensation. Table 3 gives details on the data collection and crystallographic data. In Tables 2 and 4 the hydrogen bonding system and important distances are summarized. Atomic positions and further details of the crystal structures can be obtained from the Fachinformationszentrum Karlsruhe, 76344 Eggenstein–Leopoldshafen, Germany, on quoting the deposition number given in Table 3.| Chem. formula | Li2[S2O7]·H2SO4 | Li[HS2O7] | Li[H(HS2O7)2] |
|---|---|---|---|
| a R 1 is defined as ∑||Fo| − |Fc||/∑|Fo| for I > 2σ(I). b wR2 is defined as {∑w(Fo2 − Fc2)2/∑w(Fo2)2}1/2. | |||
| Mol. wt/g mol−1 | 288.08 | 184.07 | 362.20 |
| Crystal system | Orthorhombic | Monoclinic | Monoclinic |
| Space group | Pca21 (no. 29) | Cc (no. 9) | P21/n (no. 14) |
| a/pm | 945.67(8) | 685.87(3) | 498.69(2) |
| b/pm | 485.77(4) | 788.49(4) | 808.57(3) |
| c/pm | 1793.9(1) | 911.48(5) | 1297.22(5) |
| β/° | 101.618(2) | 94.875(2) | |
| V/106 pm3 | 824.1(1) | 482.83(4) | 521.18(3) |
| Z | 4 | 4 | 2 |
| T/°C | −173 | −173 | −173 |
| λ/pm | 71.073 | 71.073 | 71.073 |
| D calc/g cm−3 | 2.322 | 2.532 | 2.308 |
| μ/cm−1 | 9.51 | 10.72 | 9.93 |
| Extinction coefficient | 0.0066(9) | 0.010(1) | |
| Measured reflections | 64![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 923 923 |
28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 677 677 |
36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 644 644 |
| Unique reflections | 5186 | 3069 | 3273 |
| With Io > 2σ(I) | 5121 | 3022 | 3147 |
| Number of parameters | 153 | 96 | 97 |
| R 1 (Fo > 2σ(Fo)) | 0.0135 | 0.0144 | 0.0157 |
| wR2b (Fo > 2σ(Fo)) | 0.0377 | 0.0414 | 0.0474 |
| R 1 (all data) | 0.0138 | 0.0148 | 0.0167 |
| wR2b (all data) | 0.0380 | 0.0416 | 0.0481 |
| Goodness of fit | 1.083 | 1.092 | 1.154 |
| Flack-x | 0.165(9) | 0.47(2) | |
| Matrix for twin refinement | (−100), (010), (00−1) | ||
| CSD number | 431426 | 431428 | 431427 |
| Li2[S2O7]·H2SO4 | Li[HS2O7] | Li[H(HS2O7)2] | |
|---|---|---|---|
| S1–O11 | 143.38(7) | 141.7(1) | 142.26(4) |
| S1–O12 | 144.88(6) | 142.3(1) | 142.71(4) |
| S1–O13 | 144.85(7) | 154.0(1) | 147.34(4) |
| S1–O121 | 163.82(6) | 158.2(1) | 165.94(4) |
| S2–O21 | 143.48(7) | 142.7(1) | 141.99(4) |
| S2–O22 | 144.81(6) | 143.1(1) | 142.33(4) |
| S2–O23 | 145.25(6) | 145.9(1) | 152.40(4) |
| S2–O121 | 162.93(6) | 168.2(1) | 158.51(4) |
| S3–O31 | 142.61(7) | ||
| S3–O32 | 142.68(7) | ||
| S3–O33 | 153.69(7) | ||
| S3–O34 | 153.89(7) | ||
| Li1–O11 | 206.1(3) | 210.71(4) (2×) | |
| Li1–O12 | 208.8(3) | 210.33(4) (2×) | |
| Li1–O13 | 218.7(2) | 244.3(2) | |
| Li1–O21 | 195.8(2) | 199.4(3) | 211.52(4) (2×) |
| Li1–O22 | 198.4(2) | 204.7(3) | |
| Li1–O23 | 202.1(2) | 220.1(3) | |
| Li1–O32 | 204.1(2) | ||
| Li2–O11 | 197.8(2) | ||
| Li2–O12 | 199.6(2) | ||
| Li2–O13 | 210.2(2) | ||
| Li2–O23 | 213.3(2) | ||
| Li2–O31 | 197.6(2) | ||
Difference Fourier map
The structure of Li[H(HS2O7)2] was solved and refined without refining the position of the hydrogen atom H13. The remaining electron density was plotted as a difference Fourier map with the software program OLEX2.34Acknowledgements
The authors thank Dr Marc Schmidtmann for the collection of the single crystal data.References
- M. Catti, G. Ferraris and G. Ivaldi, Acta Crystallogr., Sect. B: Struct. Crystallogr. Cryst. Chem., 1979, 35, 525–529 CrossRef.
- S. Fortier, M. E. Fraser and R. D. Heyding, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 1985, 41, 1139–1141 Search PubMed.
- Y. Noda, S. Uchiyama, K. Kafuku, H. Kasatani and H. Terauchi, J. Phys. Soc. Jpn., 1990, 59, 2804–2810 Search PubMed.
- T. Fukami, S. Jin and R. H. Chen, Ionics, 2006, 12, 257–262 CrossRef CAS.
- C. Panithipongwut and S. M. Haile, Solid State Ionics, 2012, 213, 53–57 Search PubMed.
- E. Kemnitz, C. Werner, H. Worzala, S. Trojanov and Y. T. Strutschkov, Z. Anorg. Allg. Chem., 1995, 621, 675–678 Search PubMed.
- C. Werner, E. Kemnitz, S. Troyanov, Y. T. Strutschkov and H. Worzala, Z. Anorg. Allg. Chem., 1995, 621, 1266–1269 Search PubMed.
- C. Werner, S. Trojanov, H. Worzala and E. Kemnitz, Z. Anorg. Allg. Chem., 1996, 622, 337–342 Search PubMed.
- D. Swain and T. N. G. Row, Inorg. Chem., 2008, 47, 8613–8615 Search PubMed.
- A. Weiz, J. Bruns and M. S. Wickleder, Z. Anorg. Allg. Chem., 2014, 640, 27–30 Search PubMed.
- L. V. Schindler and M. S. Wickleder, Z. Naturforsch., 2016 Search PubMed , accepted.
- L. V. Schindler, M. Daub, M. Struckmann, A. Weiz, H. Hillebrecht and M. S. Wickleder, Z. Anorg. Allg. Chem., 2015, 641, 2604–2609 Search PubMed.
- L. V. Schindler, T. Klüner and M. S. Wickleder, Chem. – Eur. J., 2016, 22, 13865–13870 Search PubMed.
- L. V. Schindler, A. Becker and M. S. Wickleder, Chem. – Eur. J., 2016 DOI:10.1002/chem.201603919.
- M. Sakashita, H. Fujihisa, K.-i. Suzuki, S. Hayashi and K. Honda, Solid State Ionics, 2007, 178, 1262–1267 Search PubMed.
- I. Makarova, V. Grebenev, E. Dmitricheva, V. Dolbinina and D. Chernyshov, Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater., 2014, 70, 218–226 Search PubMed.
- G. E. Pringle and T. A. Broadbent, Acta Crystallogr., 1965, 19, 426–432 Search PubMed.
- E. J. Sonneveld and J. W. Visser, Acta Crystallogr., Sect. B: Struct. Crystallogr. Cryst. Chem., 1978, 34, 643–645 Search PubMed.
- L. H. Loopstra and C. H. MacGillavry, Acta Crystallogr., 1958, 11, 349–354 Search PubMed.
- W. Mumme, Acta Crystallogr., Sect. B: Struct. Crystallogr. Cryst. Chem., 1973, 29, 1076–1083 Search PubMed.
- V. Varma, N. Rangavittal and C. N. R. Rao, J. Solid State Chem., 1993, 106, 164–173 Search PubMed.
- S. Trojanov, C. Werner, E. Kemnitz and H. Worzala, Z. Anorg. Allg. Chem., 1995, 621, 1617–1624 Search PubMed.
- E. Kemnitz, C. Werner, S. Trojanov and H. Worzala, Z. Anorg. Allg. Chem., 1994, 620, 1921–1924 Search PubMed.
- E. Kemnitz, C. Werner, H. Worzala and S. Trojanov, Z. Anorg. Allg. Chem., 1995, 621, 1075–1079 Search PubMed.
- C. Werner, S. Trojanov, E. Kemnitz and H. Worzala, Z. Anorg. Allg. Chem., 1996, 622, 380–384 Search PubMed.
- C. Logemann, J. Witt and M. S. Wickleder, Z. Kristallogr. - New Cryst. Struct., 2013, 228, 159–160 Search PubMed.
- G. A. Jeffrey, An Introduction to Hydrogen Bonding, Oxford University Press, 1997 Search PubMed.
- E. Kemnitz, C. Werner and S. Troyanov, Eur. J. Solid State Inorg. Chem., 1996, 33, 563–580 Search PubMed.
- E. Kemnitz, C. Werner and S. Troyanov, Eur. J. Solid State Inorg. Chem., 1996, 33, 581–596 Search PubMed.
- M. Pley and M. S. Wickleder, Z. Anorg. Allg. Chem., 2005, 631, 592–595 Search PubMed.
- BRUKER AXS, Apex2, 2013.
- G. Sheldrick, Acta Crystallogr., Sect. A: Found. Crystallogr., 2008, 64, 112–122 Search PubMed.
- G. Sheldrick, Acta Crystallogr., Sect. C: Struct. Chem., 2015, 71, 3–8 Search PubMed.
- O. V. Dolomanov, L. J. Bourhis, R. J. Gildea, J. A. K. Howard and H. Puschmann, J. Appl. Crystallogr., 2009, 42, 339–341 Search PubMed.
| This journal is © The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2017 |