 Open Access Article
Open Access ArticleIron acquisition in fungal pathogens of humans
Gaurav
Bairwa†
a,
Won
Hee Jung†
b and
James W.
Kronstad
 *a
*a
aMichael Smith Laboratories, University of British Columbia, Vancouver, BC V6T 1Z4, Canada. E-mail: kronstad@msl.ubc.ca
bDepartment of Systems Biotechnology, Chung-Ang University, Anseong, 456-756, Republic of Korea
First published on 13th February 2017
Abstract
The devastating infections that fungal pathogens cause in humans are underappreciated relative to viral, bacterial and parasitic diseases. In recent years, the contributions to virulence of reductive iron uptake, siderophore-mediated uptake and heme acquisition have been identified in the best studied and most life-threatening fungal pathogens: Candida albicans, Cryptococcus neoformans and Aspergillus fumigatus. In particular, exciting new work illustrates the importance of iron acquisition from heme and hemoglobin in the virulence of pathogenic yeasts. However, the challenge of establishing how these fungi gain access to hemoglobin in blood and to other sources of heme remains to be fully addressed. Recent studies are also expanding our knowledge of iron uptake in less-well studied fungal pathogens, including dimorphic fungi where new information reveals an integration of iron acquisition with morphogenesis and cell-surface properties for adhesion to host cells. Overall, the accumulating information provides opportunities to exploit iron acquisition for antifungal therapy, and new work highlights the development of specific inhibitors of siderophore biosynthesis and metal chelators for therapeutic use alone or in conjunction with existing antifungal drugs. It is clear that iron-related therapies will need to be customized for specific diseases because the emerging view is that fungal pathogens use different combinations of strategies for iron acquisition in the varied niches of vertebrate hosts.
1. Introduction
Despite being an abundant metal on Earth, the bioavailability of iron in the environment is extremely limited due to its existence as insoluble ferric oxide. Microbial pathogens face an additional challenge to acquire iron and other key metals, such as zinc, manganese and copper, inside vertebrate hosts. The mammalian immune system recognizes the importance of metal ions for the proliferation of pathogens and therefore has evolved elegant mechanisms for controlling the blood, tissue and intracellular levels of metals. For example, the process of host-mediated sequestration of iron has been termed as ‘nutritional immunity’ and its role in the immune response to bacterial pathogens is well established.1 In fact, much is known about the competition for iron between vertebrate hosts and bacterial pathogens or parasites, and an appreciation of this evolutionary host-adaptation process is also starting to emerge for several fungal pathogens of humans.Fungal pathogens cause devastating diseases in humans, but have received far less attention than viral, bacterial and parasitic infections.2–5 This lack of appreciation is surprising given the high risk of fungal infections in people with HIV/AIDS or cancer, or those receiving organ transplants or immunomodulatory therapy. The most common fungal pathogen of humans is Candida albicans and this fungus, along with other Candida species, causes superficial mucosal infections as well as life-threatening systemic disease.6 Additionally, the fungus Cryptococcus neoformans is a devastating agent of meningitis in HIV/AIDS patients with an estimated 1 million cases per year and over 600![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths.5,7 Similarly, Aspergillus fumigatus and other Aspergillus species cause a range of diseases (aspergilloses) that include allergic bronchopulmonary, chronic pulmonary and invasive aspergillosis.8 Given the serious threats posed by fungal pathogens, there is a pressing need for new approaches to treat infections, especially in the face of increasing resistance to an already limited number of antifungal drugs.9 In this context, iron acquisition by fungi during host colonization presents a compelling target for new diagnostic and therapeutic approaches.
000 deaths.5,7 Similarly, Aspergillus fumigatus and other Aspergillus species cause a range of diseases (aspergilloses) that include allergic bronchopulmonary, chronic pulmonary and invasive aspergillosis.8 Given the serious threats posed by fungal pathogens, there is a pressing need for new approaches to treat infections, especially in the face of increasing resistance to an already limited number of antifungal drugs.9 In this context, iron acquisition by fungi during host colonization presents a compelling target for new diagnostic and therapeutic approaches.
In this mini-review, we consider the major systems of iron acquisition in pathogenic fungi including reductive iron uptake, siderophore synthesis and transport, and mechanisms to use heme and hemoglobin, and we highlight recent advances that add depth to our understanding of iron acquisition. With reference to information from the model yeast Saccharomyces cerevisiae, we focus on the specific fungal pathogens C. albicans, Candida glabrata, C. neoformans and A. fumigatus, as illustrated in Fig. 1. In particular, we emphasize the uptake functions for the pathogens that contribute to disease. Additionally, we include new studies for other species of pathogenic fungi where relevant information is available. We also address the iron sources that fungi exploit in vertebrate hosts, and much of this consideration involves exciting new work on fungal exploitation of heme and hemoglobin as abundant sources of iron. The key functions for iron acquisition that we discuss are listed in Table 1. The information on known uptake strategies sets the stage for a consideration of approaches to exploit fungal iron uptake for diagnostic and therapeutic purposes. We refer readers to several recent reviews for additional considerations of iron homeostasis in fungi, including mechanisms of regulation and opportunities for therapy.10–16
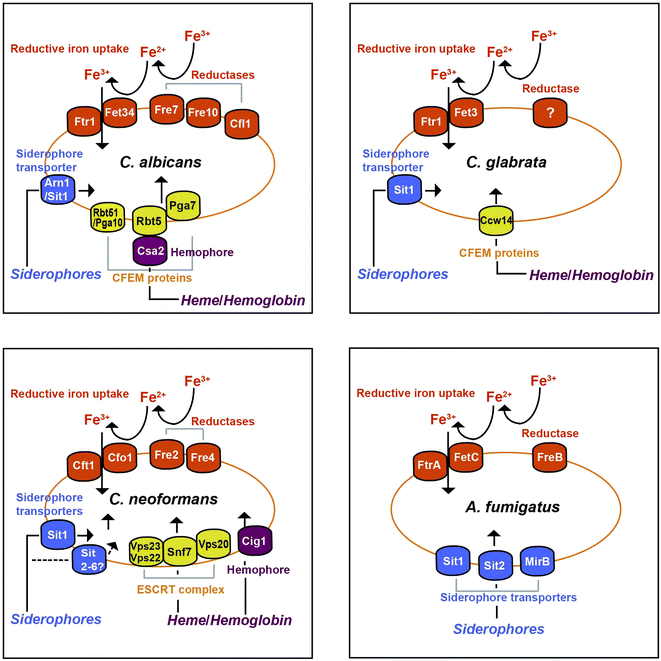 | ||
| Fig. 1 Iron acquisition systems for four of the best-studied fungal pathogens of humans including Aspergillus fumigatus, Candida albicans, Candida glabrata and Cryptococcus neoformans. | ||
| Type of transport system | Species | Protein | Functions | Virulence of mutants | Ref. |
|---|---|---|---|---|---|
| a NA: not applicable. b ND: data not available. c Mutants defective in siderophore biosynthesis are attenuated in mouse models of virulence for A. fumigatus and H. capsulatum.15,16,69 | |||||
| Reductive iron uptake | |||||
| Ferric reductases | |||||
| S. cerevisiae | Fre1 | Ferric iron reduction at the cell surface | NAa | 17 | |
| S. cerevisiae | Fre2 | Ferric iron reduction at the cell surface | NA | 17 | |
| C. albicans | Fre7 | Ferric iron reduction at the cell surface | NDb | 21, 22 and 25 | |
| C. albicans | Fre10 | Ferric iron reduction at the cell surface | ND | 21, 22 and 25 | |
| C. albicans | Cfl1 | Ferric iron reduction at the cell surface | Attenuation in a mouse model of systemic candidiasis | 27 | |
| C. neoformans | Fre2 | Ferric iron reduction at the cell surface | Attenuation in a mouse model of cryptococcosis | 29 | |
| A. fumigatus | FreB | Ferric iron reduction at the cell surface | ND | 41 | |
| Multicopper ferroxidase | |||||
| S. cerevisiae | Fet3 | Multicopper oxidase. Ferrous iron oxidation and high-affinity iron uptake, coupled with Ftr1 | NA | 17 | |
| C. albicans | Fet3 | Putative multicopper oxidase | ND | 32, 38 and 39 | |
| Fet31 | Ferrous iron oxidation and high-affinity iron uptake | Not required | 32 and 38 | ||
| Fet33 | Putative multicopper oxidase | Not required | 39 | ||
| Fet34 | Ferrous iron oxidation and high-affinity iron uptake, coupled with Ftr1 | Attenuation in a mouse model of systemic candidiasis | 38 and 39 | ||
| Fet99 | Putative multicopper oxidase | ND | 32 and 38 | ||
| C. glabrata | Fet3 | Ferrous iron oxidation and high-affinity iron uptake | Reduced kidney colonization | 37 | |
| C. neoformans | Cfo1 | Ferrous iron oxidation and high-affinity iron uptake | Attenuation in a mouse model of cryptococcosis | 44 | |
| C. neoformans | Cfo2 | Putative multicopper oxidase | Not required | 44 | |
| A. fumigatus | fetC | Ferrous iron oxidation and high-affinity iron uptake | ND | 36 | |
| T. marneffei | fetC | Ferrous iron oxidation and high-affinity iron uptake | ND | 49 | |
| Iron permease | |||||
| S. cerevisiae | Ftr1 | High-affinity iron uptake, coupled with Fet3 | NA | 17 | |
| C. albicans | Ftr1 | High-affinity iron uptake, coupled with Fet3 | Avirulent in a mouse model of systemic candidiasis | 23 | |
| C. albicans | Ftr2 | Putative iron permease | Not required | 23 | |
| C. glabrata | Ftr1 | High-affinity iron permease | Reduced kidney colonization | 37 and 46 | |
| C. neoformans | Cft1 | High-affinity iron permease | Attenuation in a mouse model of cryptococcosis | 35 | |
| C. neoformans | Cft2 | Putative iron permease | Not required | 35 | |
| C. neoformans | Cft3 | Putative iron permease | Not required | 43 | |
| A. fumigatus | FtrA | High-affinity iron permease | Not required | 36 | |
| T. marneffei | Ftr1 | High-affinity iron permease | No growth defect in macrophages | 49 | |
| R. oryzae | Ftr1 | High-affinity iron permease | Reduced virulence in a mouse model of mucormycosis | 63 and 64 | |
| Siderophore transportc | |||||
| S. cerevisiae | Arn1 | Ferrichrome and ferrichrome A transport | NA | 17, 24 and 53 | |
| Arn2/Taf1 | Triacetylfusarinine C transport | NA | 17, 24 and 53 | ||
| Arn3/Sit1 | Ferrichrome and ferrichrome A transport | NA | 17, 24 and 53 | ||
| Arn4/Enb1 | Enterobactin transport | NA | 17, 24 and 53 | ||
| C. albicans | Arn1/Sit1 | Ferrichrome-type xenosiderophore transport | Required for epithelial invasion | 58–60 | |
| C. glabrata | Sit1 | Ferrichrome transport | Reduced survival within macrophages | 37 and 61 | |
| C. neoformans | Sit1 | Ferrioxamine transport | Not required | 62 | |
| A. fumigatus | Sit1 | Ferrichrome and ferrioxamine B transport | Not required | 67 | |
| Sit2 | Ferrichrome transport | Not required | 67 | ||
| MirB | Triacetylfusarinine C transport | ND | 69 | ||
| H. capsulatum | Mfs1 | Putative siderophore transporter | ND | 70 | |
| Abc1 | Putative siderophore transporter | ND | 70 | ||
| R. oryzae | Fob1, Fob2 | Ferrioxamine binding at the cell surface | Required for virulence in a mouse model of mucormycosis | 63 | |
| Heme assimilation | |||||
| CFEM proteins | |||||
| C. albicans | Rbt5 | Heme uptake, putative GPI-anchored hemoglobin receptor | Required for biofilm formation, not required for virulence | 75 and 78 | |
| C. albicans | Rbt51 | Heme uptake | ND | 75 | |
| C. albicans | Pga7 | Hemoglobin and heme utilization | Attenuation in a mouse model of systemic candidiasis | 80 | |
| C. glabrata | Ccw14 | Unknown | Reduced survival in the liver and the spleen in a mouse model of systemic candidiasis | 37 | |
| P. brasiliensis | Rbt5 | Putative hemoglobin receptor | Impaired survival within macrophages | 84 and 85 | |
| P. lutzii | Rbt5 | Putative hemoglobin receptor | Reduced survival in the spleen | 84 and 85 | |
| ESCRT complex | |||||
| C. neoformans | Vps23 | ESCRT-I complex, heme uptake | Avirulent in a mouse model of cryptococcosis | 88 | |
| C. neoformans | Vps22 | ESCRT-II complex, heme uptake | Avirulent in a mouse model of cryptococcosis | 89 | |
| C. neoformans | Snf7 | ESCRT-III complex, heme uptake | Avirulent in a mouse model of cryptococcosis | 89 | |
| C. neoformans | Vps20 | ESCRT-III complex, heme uptake | ND | 89 | |
| Hemophore | |||||
| C. albicans | Csa2 | Putative hemophore | ND | 77 | |
| C. neoformans | Cig1 | Putative hemophore | Not required, a cig1 and cfo1 double mutant shows attenuated virulence in a mouse model of cryptococcosis | 90 | |
| Haemolysin | |||||
| C. glabrata | Mam3 | Putative haemolysin | Reduced fungal burden in the kidney, liver and brain | 37 | |
2. Reductive iron uptake: connections with virulence and dimorphism
The mechanisms of iron acquisition and homeostasis have been well studied in the model yeast S. cerevisiae, and this information provides a foundation for many of the studies in fungal pathogens.17–19S. cerevisiae primarily employs two different mechanisms for iron uptake that involve reductive and nonreductive systems.17 Reductive iron uptake involves two sequential steps: (1) reduction of ferric iron to soluble ferrous iron by ferric reductases encoded by the FRE genes, FRE1 and FRE2, and; (2) re-oxidation to the ferric form by a multicopper ferroxidase (Fet3) coupled with transport into the cell by a permease (Ftr1). Nonreductive iron uptake involves the use of siderophore-bound iron via specific transporters, although S. cerevisiae does not synthesize its own siderophores.18The fungal pathogens C. albicans, C. neoformans, and A. fumigatus also use a reductive iron uptake system involving conserved orthologs of the cell-surface ferric reductases, ferroxidases and iron permeases identified in S. cerevisiae.19–23 The FRE reductases are integral membrane proteins that require heme, FAD and NADPH for activity.24 At least 17 genes encoding putative ferric reductases have been identified in the C. albicans genome. Of these genes, the FRE10 and FRE7 genes encode highly active cell-surface ferric reductases and deletion of these genes results in decreased ferric iron uptake by the fungus.21,22,25 More recent work on the Cfl1 ferric reductase from C. albicans expands our appreciation of the roles of these enzymes in fungal pathogens. Specifically, transcription of CFL1 is regulated in response to iron and copper, and deletion of the gene results in a remarkably large number of phenotypes.26–28 These phenotypes include increased accumulation of cellular iron, altered cell wall architecture, loss of mitochondrial function, impaired filamentous growth, sensitivity to oxidative stress, and attenuated virulence in mouse model of systemic candidiasis.26–28 Interestingly, a mutant lacking Cfl1 is not defective for iron acquisition, perhaps because of the observed compensatory upregulation of other ferric reductase genes in a cfl1 deletion mutant.27 Cfl1 therefore plays a variety of roles and the recent work illustrates the pleiotropy of iron-related functions with regard to fungal pathogenesis. Transcriptional analysis of several other C. albicans genes encoding ferric reductases indicates that this fungus can regulate the expression of its iron-uptake machinery in different host environments.25 This finding illustrates the need to consider the host niche in determining the relevance of an iron uptake system for disease. In C. neoformans, eight genes encoding putative ferric reductases have been identified (FRE1–7, FRE201).29 Notably, the transcription of FRE2 and FRE4 is regulated by hemin or FeCl3 suggesting a crucial role of these genes in maintaining iron homeostasis during iron-limited conditions. Furthermore, FRE2 is required for robust growth in the presence of heme and transferrin, and for full virulence in a mouse inhalation model of cryptococcosis.29 Similar to the S. cerevisiae FRE genes, the FRE genes in both C. albicans and C. neoformans also show differential transcriptional regulation in the presence of copper.29,30
In S. cerevisiae, the second step of reductive iron uptake involves transport of the reduced ferrous iron via a high-affinity heterodimeric transport complex consisting of the multicopper ferroxidase Fet3 and the permease Ftr1, as mentioned above. Ferrous iron is oxidized to ferric iron by Fet3 and is subsequently transported into the cell by Ftr1. The transport of ferric iron involves a classic metabolite channeling mechanism where amino acid residues in both Fet3 and Ftr1 interact with each other to drive the iron into the cell.31 The corresponding components of the multicopper ferroxidase-iron permease system have been identified and characterized in C. albicans, C. neoformans, A. fumigatus, C. glabrata, and other species (see below).32–37 For example, C. albicans harbors five genes, FET3, FET31, FET33, FET34 and FET99 that are orthologs of the S. cerevisiae FET3 gene.32,38,39 Transcriptional analyses demonstrated that FET3 and FET34 are regulated by iron starvation.22,40 The characterization of FET34 revealed that this gene encodes a functional ferroxidase which localizes to the plasma membrane and contributes to iron accumulation, hyphal development and virulence in a mouse model of systemic candidiasis.39,40 Transcription of the C. albicans ortholog of the S. cerevisiae iron permease FTR1 is also induced upon iron deprivation.33 Importantly, FTR1 in C. albicans, along with the reductase encoded by FRE10, is implicated in iron acquisition from transferrin and ferritin, and Ftr1 is important for virulence in a mouse model of systemic disease.22,23,33
A. fumigatus also possesses the components of the reductive iron uptake system including cell-surface ferric reductases and a ferroxidase (FetC) and iron permease (FtrA).36,41 The ftrA gene of A. fumigatus is highly homologous to C. albicans FTR1, and its expression is induced upon iron depletion. However, the mutant lacking ftrA grew normally in iron-restricted medium and was as virulent as the wild-type strain, thus suggesting that the iron permease is dispensable for virulence in A. fumigatus.36 The fetC gene in A. fumigatus encodes an ortholog of FET3 from S. cerevisiae. Iron-responsive transcriptional regulation of fetC was observed, but the contribution of the gene to virulence has not been determined.36 Notably, a low-affinity iron utilization system, dependent on the putative vacuole iron importer Ccc1, has also been identified in A. fumigatus.42 Ccc1 contributed to the ability of the fungus to grow in the presence of high iron levels when both the high-affinity and siderophore-mediated iron uptake systems were absent.42
Three paralogous genes in C. neoformans (CFT1-3) are orthologs of S. cerevisiae FTR1 and, among them, CFT1 encodes the high-affinity iron permease in the reductive iron uptake system.35,43CFT1 is required for ferric iron uptake and acquisition of iron from holo-transferrin in vitro, and the cft1 mutant is attenuated for virulence in both inhalation and tail vein injection models of cryptococcosis in mice.35 Moreover, fungal colonization in the brain of the infected mice was dramatically reduced for the cft1 mutant implying that transferrin may be an important iron source for C. neoformans during disseminated disease and meningitis.35 The ferroxidase of the reductive uptake system is encoded by CFO1 in C. neoformans, and the cfo1 mutant shows reduced growth in iron-restricted medium.44 Moreover, the cfo1 mutant was unable to utilize holo-transferrin as a sole iron source, while heme and the siderophore feroxamine supported the growth of the mutant (as was found with the cft1 mutant). Deletion of CFO1 also attenuated virulence in a mouse model of cryptococcosis.44 Together, the results with Cft1 and Cfo1 suggest that transferrin may be an important iron source for C. neoformans during infection, and that high-affinity reductive iron uptake is important during cryptococcosis.
More recent studies extended the analysis of reductive iron uptake to another clinically important Candida species, C. glabrata. This pathogen, which is actually more closely related to S. cerevisiae than to C. albicans, has emerged as the second leading cause of candidiasis after C. albicans.45 An ortholog of the FTR1-encoded iron permease was identified in C. glabrata and found to be required for growth in iron-restricted medium.37,46 Additionally, deletion of FTR1 caused reduced colonization of the kidneys of infected mice suggesting that the iron permease is essential for the virulence of C. glabrata.37 The same study also identified the C. glabrata ortholog of the Fet3 ferroxidase of S. cerevisiae. Similar to the ftr1 mutant, the C. glabrata strain lacking the FET3 ortholog also showed reduced colonization of the kidneys of mice, supporting the essentiality of high-affinity reductive iron uptake for virulence.37 Interestingly, plasma membrane-localized Ftr1 moves to the vacuole in a Vps34-dependent manner upon exposure of C. glabrata to excess iron.46VPS34 encodes a phosphoinositide 3-kinase, and the C. glabrata mutant lacking the gene shows impaired iron homeostasis and a significant reduction of organ colonization in mice. The inability of a vps34 mutant to properly traffic Ftr1 as well as other virulence-related phenotypes (e.g., impaired biofilm formation) may contribute to the observed colonization defect.
Reductive iron uptake is also associated with morphogenesis in dimorphic or pleiomorphic fungal pathogens that can switch between yeast-like growth and hyphal or pseudohyphal growth.47–49 The yeast to hyphal morphological transition has been well studied in C. albicans where the ability to switch contributes to virulence.6 Emerging evidence indicates that iron acquisition is intertwined with morphological switching in some dimorphic fungi. For example, the reductive iron uptake system in C. albicans is coordinated with the function of the adhesin-like receptor protein, Als3, to facilitate iron acquisition from ferritin specifically during hyphal but not yeast growth.47 A strong link between the expression of other adhesion proteins (e.g., MP65 and PGA62) and iron was also established in a study of the influence of iron on chromatin remodeling in C. albicans.50 The connection between surface protein expression and iron may be a general feature of pathogenic yeasts because the EPA1 gene encoding the major adhesin in C. glabrata is also regulated by iron.51 Very recently, Pasricha et al. reported a thorough characterization of the connections between cell morphology and both reductive iron uptake and siderophore-mediated uptake in the dimorphic pathogen Talaromyces marneffei.49,52 The pathogenic yeast phase of this fungus proliferates intracellularly in macrophages and causes disease in immunocompromised people, particularly in endemic areas such as Southeast Asia. Initial transcriptional profiling experiments revealed co-incident regulation of iron uptake functions with the temperature-mediated switch from hyphal (25 °C) to yeast (37 °C) growth.52 Subsequent detailed experiments demonstrated that the T. marneffei fetC (ferroxidase) and ftrA (iron permease) genes have higher transcript levels both in response to iron limitation and in yeast cells compared to hyphae. In agreement, a deletion mutant lacking ftrA displayed reduced growth on low iron medium at 37 °C but not at 25 °C.49 These results suggest that an additional iron uptake system may be active at 25 °C or that the yeast cells at 37 °C may have a higher demand for iron. Interestingly, the ftrA mutant did not display a defect in intracellular proliferation in macrophages suggesting that another uptake pathway is used in this context. As discussed below, the siderophore biosynthetic genes in T. marneffei were also characterized, and these genes also show interesting cell-type regulation and contributions to growth.49
3. Siderophore iron uptake: a potential drug target for some but not all fungal pathogens
Along with reductive iron uptake, siderophore-mediated nonreductive iron uptake also contributes to iron acquisition in fungi.14,16S. cerevisiae, C. neoformans and C. albicans do not synthesize their own siderophores, but can use exogenous siderophores (xenosiderophores) produced by other organisms. Early studies in S. cerevisiae again provide a model for uptake mechanisms for xenosiderophore-iron chelates, in this case via Arn/Sit transporters of the major facilitator superfamily.24 These transporters, termed Arn1, Arn2/Taf1, Arn3/Sit1, and Arn4/Enb1, individually show specificity for different bacterial and fungal xenosiderophores such as enterobactin, ferrichrome, ferrichrome A, triacetylfusarine C, and ferrioxmaine B.17,24,53 The siderophore bound Arn transporters are internalized and transported to late-endosomal compartments for vacuolar degradation to release the iron, as revealed by the trafficking of ferrioxamine B and ferrichrome-bound Arn3/Sit1 and Arn1 transporters, respectively.53,54 The trafficking of Sit1 to the vacuolar compartment depends on Rsp5-mediated ubiquitylation and the clathrin adaptor protein Gga2, and is regulated by the presence of the substrate ferrichrome B.55,56 A recent study by Kang et al. added further depth to the connection between intracellular trafficking and iron.57 Specifically, it was found that the transcriptional activator Aft1 in S. cerevisiae, which responds to iron availability and regulates the expression of the FET3, FTR1 and ARN genes, interacts with the Arn3/Sit1 transporter and influences its ubiquitination and vacuole-dependent protein degradation.56,57 The Aft1-Arn3/Sit1 interaction illustrates the potential for additional sensing mechanisms that fine-tune the deployment of iron uptake strategies.As indicated above, some pathogenic yeast exploit xenosiderophores produced by other microbes. C. albicans, for example, employs a Sit1/Arn1 transporter to use xenosiderophores such as ferricrocin, ferrichrysin, ferrirubin, coprogen and triacetylfusarine C.58–60 Given that C. albicans is a human commensal, the fungus may encounter xenosiderophores from bacteria that co-inhabit mucosal or gastrointestinal niches. The Sit1/Arn1 transporter in C. albicans is required for efficient invasion of reconstituted epithelium as a model of the human oral mucosa, but does not contribute to virulence in a mouse model of systemic candidiasis.60 Thus, siderophore-mediated iron uptake may not be important during bloodstream infection by C. albicans.60C. glabrata also possesses a Sit1 siderophore transporter, and Nevitt and Thiele investigated the influence of Sit1 on virulence by monitoring the survival of a sit1 mutant in a mouse macrophage cell line.61 They observed reduced survival of the sit1 mutant within phagolysosomes of the infected macrophages compared to the wild-type strain. However, reduced survival of the mutant was only observed for fungal cells pre-treated with the xenosiderophore ferrichrome, a substrate of Sit1, and no difference was observed between the sit1 mutant and the wild-type strain without ferrichrome pre-treatment. C. neoformans also has a highly conserved transporter, Sit1, for the uptake of ferrioxamine B, and xenosiderophores may be important during proliferation of the fungus in the environment (i.e., in soil, bird excreta and in association with trees).62 However, Sit1 in C. neoformans does not contribute to virulence in a mouse model of cryptococcosis.62 Xenosiderophores are also important in the disease mucormycosis caused by Rhizopus oryzae and other fungi in the order Mucorales.63 Mucormycosis is a threat to patients who are experiencing iron overload as a result of kidney dialysis and are receiving chelation therapy with the bacterial siderophore deferoxamine. The ability of R. oryzae to exploit deferoxamine to grow in the host, and the ability to use the siderophore, depends on cell surface binding proteins (Fob1 and Fob2) as well as the reductive uptake system.63 Consistent with the latter requirement, defects in iron permease activity attenuate virulence in a deferoxamine-treated mouse model of mucormycosis.63,64 In contrast, the reductive uptake system encoded by CFT1 and CFO1 is not required for iron acquisition via the siderophore ferrioxamine B in C. neoformans.33,44
Siderophore-mediated iron uptake has been extensively studied in Aspergillus species including A. nidulans and A. fumigatus, and also in the dimorphic pathogen Histoplasma capsulatum.15,16,36,65,66 Unlike C. albicans and C. neoformans, both the Aspergillus species and H. capsulatum synthesize different hydroxamate-type siderophores including ferricrocin, hydroxyferricrocin, fusarinine C, coprogen B and triacetylfusarinine C.36,65,66 Extensive studies have characterized the genetics and regulation of siderophore production, and defined the role of siderophores in the virulence of A. fumigatus.15,16 In this pathogen, the sidA gene encoding L-ornithine-N5-monooxygenase is essential for siderophore biosynthesis and virulence.36 Putative siderophore transporters have also been identified in both A. fumigatus and A. nidulans.67–69 In a recent study, Park et al. described the roles of highly conserved orthologs of the S. cerevisiae Sit1 and Sit2 transporters in A. fumigatus and demonstrated their participation in the uptake of ferrichrome and ferrioxamine B.67 However, these siderophore transporters did not contribute to the virulence of A. fumigatus in immunosuppressed mice. Another siderophore transporter, MirB, has also been characterized for A. fumigatus, but its contribution to virulence remains to be determined.69 For H. capsulatum, a microarray analysis identified genes with increased expression under iron-poor conditions.70 These genes encoded functions for siderophore synthesis (e.g., SID1 the ortholog of sidA) or transport (MFS1 and ABC1). Importantly, deletion of SID1 in H. capsulatum abolished siderophore biosynthesis as well as growth in bone marrow-derived macrophages and in mice, indicating that siderophore production is essential for the virulence of this fungus.70
As introduced earlier, work in T. marneffei has provided interesting insights into the integration of iron acquisition with virulence-associated dimorphism. With regard to siderophore production, a characterization of the genetics and expression of siderophore biosynthesis revealed a suite of genes orthologous to most of those defined in A. fumigatus for the production of extracellular and intracellular siderophores. Interestingly, two orthologs of the A. fumigatus sidA gene (sidA and sidX) were found, and these genes make distinct contributions to growth in response to iron.49 In particular, the growth of the sidA deletion mutant was not impaired but instead enhanced by iron limitation relative to the wild-type strain at 25 °C or 37 °C.49 In contrast, the sidX deletion mutant grew poorly in low iron conditions at 25 °C or 37 °C, but behaved like wild type upon iron repletion. The sidA and sidX deletion mutants were not impaired for growth in macrophages. Finally, it was observed that the fetC gene for the multicopper ferroxidase was upregulated in the sidA and sidX deletion mutants indicating cross regulation between uptake pathways.49 Taken together, these findings strengthen the view that iron acquisition is integrated with morphogenesis in some fungal pathogens, and they provide insights into the relative contributions of siderophore versus reductive uptake mechanisms for growth in phagocytic cells and for yeast versus hyphal cell types.
4. Heme and hemoglobin are important iron sources for pathogenic yeasts
Approximately 80% of the iron inside a mammalian host is bound to heme in hemoglobin (Hb) and other heme-containing proteins. In addition to hemoglobin, other host iron-containing proteins include lactoferrin (Lf), transferrin (Tf), haptoglobin, haemopexin, lipocalin-1 and lipocalin-2 (Lcn1/Lcn2).71 Additionally, intracellular iron availability to microbes is also limited due to the sequestration of iron in transferrin and ferritin.71,72 Consequently, the ability of fungal pathogens of humans to obtain iron from different host iron-binding proteins to sustain growth and proliferation is a key determinant of the outcome of infection. In this regard, some fungal pathogens target heme and hemoglobin as key iron sources. For example, C. albicans has robust machinery to obtain iron from heme/hemoglobin.73–75 This system is independent of the reductive and siderophore uptake mechanisms and depends on a conserved family of proteins (Rbt5, Rbt51/Pga10, Pga7 and Csa2) which contain the cysteine-rich Common in Fungal Extracellular Membrane (CFEM) domain.74–79 Among these proteins, Rbt5 is a mannosylated plasma membrane-anchored protein that mediates heme uptake.75 The paralog of RBT5, RBT51/PGA10, also confers the ability to use heme as a sole iron source in S. cerevisiae, and this finding provided a fruitful avenue to investigate heme and hemoglobin uptake. Specifically, a large-scale screen of the deletion library in S. cerevisiae expressing C. albicans RBT5 suggested that the Rbt5 protein is the GPI-anchored hemoglobin receptor that facilitates the endocytosis of hemoglobin into the vacuole. The functions identified in the screen implicated the involvement of acidification of the lumen of the late secretory pathway, a type I myosin and the activity of the ESCRT pathway in hemoglobin utilization in C. albicans.76 A recent study also identified a role for another CFEM protein Pga7 in heme uptake in C. albicans. Pga7 was shown to cooperate with Rbt5 within the cell envelope for hemoglobin and heme utilization, and the mutant lacking PGA7 displayed a more significant growth deficiency than the rbt5 mutant in the medium with heme or hemoglobin as a sole iron source.80 Furthermore, the pga7 mutant was attenuated for virulence in a mouse model of systemic infection suggesting that heme iron uptake contributes to C. albicans pathogenesis.80Another CFEM domain protein, Csa2, is required for robust growth on hemoglobin. Csa2 binds hemoglobin and may serve as a hemophore to deliver the protein to other CFEM proteins such as Pga7 and Rbt5.77,80,81 In this context, a recent study presented the structural basis of heme-iron acquisition by Csa2, in C. albicans.81 It was found that the CFEM domain in Csa2 adopts a novel six α-helical basket form with a flat hydrophobic platform on top to allow the attachment of a planer heme molecule.81 A conserved aspartic residue in the CFEM helical basket serves as an axial ligand to confer the specificity of Fe3+ heme binding.81 This aspartic-heme-Fe axial ligand coordination in Csa2 partially resembles the tyrosine-heme-Fe axial ligand coordination of the bacterial hemophore, HasA, as described in the Gram-negative bacteria Serratia marcescens and Pseudomonas aeruginosa.82,83 The work on Csa2 further strengthens the model for heme-iron acquisition in C. albicans that involves removal of heme from hemoglobin by Csa2, transfer to envelope-anchored CFEM proteins Rbt5 and Pga7, and internalization by endocytosis (Fig. 1).80
Other fungi also use heme and hemoglobin as iron sources in various capacities.75 For example, the Candida species C. tropicalis and C. parapsilosis share the ability to use these iron sources with C. albicans, perhaps reflecting the fact that all three species belong to the same clade. However, other Candida species such as C. glabrata and C. krusei are unable to grow with heme and hemoglobin as iron sources. In addition, anti-Rbt51 antiserum reacted with cell lysates of C. tropicalis and C. parapsilosis, but not with lysates of C. glabrata and C. krusei, implying that the first two may possess an Rbt51 ortholog for heme uptake.75 Other heme-related functions have been described in C. glabrata including a putative cell-surface bound CFEM domain-containing protein (Ccw14), a haemolysin-like protein (Mam3), and an intracellular haem oxygenase (Hmx1).37 Both Ccw14 and Mam3 are involved in iron homeostasis and virulence for C. glabrata, although their role in the use of heme or hemoglobin requires further investigation.37 CFEM proteins are important for hemoglobin use by other species including the dimorphic pathogens Paracoccidioides brasiliensis and P. lutzii.84,85 In these fungi, an ortholog of Rbt5 serves as a hemoglobin receptor and loss of the protein results in mutants with impaired survival in macrophages and in the spleens of infected mice. In contrast, deletion of three genes (cfmA-C) encoding CFEM-domain proteins in A. fumigatus did not reveal roles in heme uptake or biofilm formation, although cell wall defects were observed.86 The lack of an involvement of CfmA-C in heme use is interesting given a recent detailed RNA-Seq analysis indicates that A. fumigatus expresses functions for iron homeostasis, secondary metabolism and detoxification to survive in blood, but does not grow.87 This has been termed a “just wait and see” resting stage behavior for the fungus.
The use of heme as an iron source has also been described for C. neoformans.88–90 In this fungus, a screen for genes required for growth on heme initially revealed a role for Vps23, a component of the ESCRT-I complex involved in endocytosis or regulation of iron acquisition. Vps23 was also required for virulence in a mouse inhalation model of cryptococcosis.88 An additional study showed that other ESCRT proteins, specifically Vps22 (ESCRT-II) and Snf7/Vps20 (ESCRT-III), also contribute to heme use in C. neoformans.89 Similar to C. albicans, a putative hemophore, Cig1, has also been described in C. neoformans that is involved in heme binding, uptake and virulence.90 The Cig1 protein does not contain a CFEM domain and therefore must interact with heme via a different mechanism than the C. albicans proteins. The reductive iron uptake system can mask the absence of Cig1 during colonization of mice because a cig1 mutant is as virulent as the wild-type strain, and a cig1cfo1 double mutant is more attenuated than a cfo1 mutant. These results contribute to the general view that multiple mechanisms of iron assimilation contribute to host colonization and that additional systems remain to be discovered in C. neoformans.90 One outstanding issue concerns the access of C. neoformans to heme given that hemolytic activity has not been reported. The surface of Cryptococcal cells is unique among fungal pathogens in that the fungus elaborates a large polysaccharide capsule in response to a variety of signals including iron deprivation.91 Interestingly, deletion of the VTC4 gene encoding a polyphosphate polymerase in C. neoformans resulted in impaired ability of the fungus to trigger blood coagulation in vitro. Although virulence was not attenuated for a vtc4 deletion mutant, cells of this strain did accumulate to a higher level than the wild type in the lungs of infected mice. Polyphosphate may therefore function in lung colonization or dissemination of the fungus from the lungs. A connection between the master iron regulator Cir1 in C. neoformans and the expression of functions for phosphate uptake and polyphosphate formation has been established.92 It is therefore tempting to speculate that polyphosphate on the fungal cell surface might contribute to interactions with erythrocytes, and potentially to iron acquisition.
5. Targeting iron acquisition to augment antifungal therapy
The accumulating knowledge on the iron acquisition systems that contribute to fungal virulence promises to inform new approaches to diagnostics and therapy. In general, current strategies to target pathogen iron acquisition include the use of iron chelation therapy, siderophore-drug conjugates, inhibitors of uptake systems or biosynthetic enzymes, vaccines against surface uptake components, and immunological strategies to influence iron availability to intra and extracellular pathogens.93–99 The approach of targeting biosynthetic enzymes for inhibitor development shows considerable promise for pathogens that deploy siderophores during host colonization.97 Success in this area was recently demonstrated in a screen using an assay for the L-ornithine-N5-monooxygenase encoded by sidA in A. fumigatus.100 The crystal structure of the enzyme has been determined and detailed biochemical studies led to the development of a fluorescence polarization binding assay involving a chromophore linked to ADP as a probe for active site binding of small molecules.101,102 A screen of 2320 compounds with the assay yielded celastrol as a specific SidA inhibitor, and follow up work showed that this compound at 50 μM inhibits the growth of A. fumigatus on defined minimal medium without iron and on blood agar.100 As expected, the growth inhibition was rescued by siderophores. Interestingly, celastrol was previously found to have antifungal activity against phytopathogenic fungi.103The use of chelators for treatment of fungal infections also shows potential utility and recent studies illustrate the development of new chelators, the use of these molecules in combination with antifungal drugs and their use in treating topical fungal infections.104–107 In a search for new chelators, Helsel et al. screened a set of metal chelating agents for their ability to inhibit the growth of C. neoformans.105 The tested compounds included general chelators, iron chelators including desferrioxamine B, and extracellular and intracellular molecules that bind copper. Interestingly, chelating compounds that bound iron or copper did not limit fungal growth in the assay conditions, but agents that acted to increase intracellular copper levels were inhibitory. The active molecules were ionophores and, importantly, were fungicidal for C. neoformans. The ability of combinations of iron chelators and antifungal drugs to inhibit the growth of C. neoformans, Cryptococcus gatti and S. cerevisiae has also been tested.107 In particular, the chelators ethylenediamine tetraacetic acid, deferiprone, deferasirox, ciclopirox olamine and lactoferrin reduced the minimal inhibitory concentration of amphotericin B (AmB), a front line antifungal for cryptococcosis. The only combination that showed synergy was lactoferrin with AmB but iron supplementation did not ablate growth inhibition suggesting that another mechanism besides iron chelation may explain the lactoferrin contribution. Finally, recent work examined chelation as a topical treatment for corneal infections caused by A. fumigatus and another opportunistic fungal pathogen Fusarium oxysporum.104 Iron availability as modulated by Fe-dextran and deferrioxamine treatment, and extracellular siderophore production, were shown to influence infection in a murine corneal infection model. Additionally, sequestration with the human siderophore-binding protein lipocalin-1 also reduced fungal growth during infection. Extracellular siderophore biosynthesis by A. fumigatus requires mevalonate as a precursor, and mutants unable to convert mevalonic acid to anhydromevalonyl CoA for incorporation (ΔsidH and ΔsidI) had reduced growth in infected tissue.104 Together, these results led to a demonstration that topical treatment with statins that block HMG-CoA reductase activity for mevalonate production and the chelator deferiprone could protect corneal tissue from infection by A. fumigatus or F. oxysporum.104 Overall, this study provides compelling support for combination treatment approaches, and the results hold particular promise for topical therapy of fungal infections on mucosal surfaces, skin and nails.
6. Conclusions
Our understanding of the mechanisms of iron acquisition systems and their contributions to virulence has expanded in the past three years for the best studied fungal pathogens A. fumigatus, C. albicans, C. glabrata and C. neoformans. Additionally, new insights are emerging for a broader range of pathogens including dimorphic species in the genera Talaromyces and Paracoccidioides. In particular, we gained insights into the roles of candidate ferric reductases at the cell surface and the complexities of the contributions of different enzymes in the face of redundancy. Redundancy is also a challenge in determining the potential contributions of multiple siderophore transporters in fungi that exploit xenosiderophores. Additional information from T. marneffei illustrates the integration of iron acquisition via reductive uptake and siderophores with virulence properties such as the temperature-dependent morphological switch that generates the pathogenic yeast cell type. A major recent accomplishment was the molecular characterization of the relay system of CFEM domain proteins for heme and hemoglobin use in C. albicans. This system will undoubtedly serve as a paradigm for many fungal pathogens, particularly among pathogenic yeasts, but perhaps not for A. fumigatus and other molds. Together, the accumulating information provides the foundation for therapeutic applications and recent studies support the value of targeting siderophore biosynthesis and metal chelation. It will be important to understand the specific strategies of each pathogen so that targeted interventions can be deployed. Many questions remain to be answered, particularly in the context of understanding the relative contributions of uptake systems for specific host iron sources. One challenge will be to fully understand the range of uptake systems when multiple mechanisms support growth in the host, and to develop the experimental models to understand niche-specific access to iron, particularly in the blood.Acknowledgements
We apologize to those authors whose work could not be discussed because of space limitations. Studies of iron acquisition in the Kronstad laboratory are funded by grant 5R01AI053721-13 from the National Institute of Allergy and Infectious Diseases. W. H. J. was supported by a Chung-Ang University research grant in 2016. J. W. K. is a Burroughs Wellcome Fund Scholar in Molecular Pathogenic Mycology.References
- M. I. Hood and E. P. Skaar, Nutritional immunity: transition metals at the pathogen-host interface, Nat. Rev. Microbiol., 2012, 10, 525–537 CrossRef CAS PubMed.
- D. Armstrong-James, G. Meintjes and G. D. Brown, A neglected epidemic: fungal infections in HIV/AIDS, Trends Microbiol., 2014, 22, 120–127 CrossRef CAS PubMed.
- G. D. Brown, D. W. Denning and S. M. Levitz, Tackling human fungal infections, Science, 2012, 336, 647 CrossRef CAS PubMed.
- G. D. Brown, D. W. Denning, N. A. Gow, S. M. Levitz, M. G. Netea and T. C. White, Hidden killers: human fungal infections, Sci. Transl. Med., 2012, 4, 165rv113 Search PubMed.
- S. Vallabhaneni, R. K. Mody, T. Walker and T. Chiller, The Global Burden of Fungal Diseases, Infect. Dis. Clin. North. Am., 2016, 30, 1–11 CrossRef PubMed.
- M. A. Jabra-Rizk, E. F. Kong, C. Tsui, M. H. Nguyen, C. J. Clancy, P. L. Fidel, Jr. and M. Noverr, Candida albicans Pathogenesis: Fitting within the Host-Microbe Damage Response Framework, Infect. Immun., 2016, 84, 2724–2739 CrossRef CAS PubMed.
- B. J. Park, K. A. Wannemuehler, B. J. Marston, N. Govender, P. G. Pappas and T. M. Chiller, Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS, AIDS, 2009, 23, 525–530 CrossRef PubMed.
- S. Krappmann, How to invade a susceptible host: cellular aspects of aspergillosis, Curr. Opin. Microbiol., 2016, 34, 136–146 CrossRef CAS PubMed.
- D. Sanglard, Emerging Threats in Antifungal-Resistant Fungal Pathogens, Frontiers in Medicine, 2016, 3, 11 CrossRef PubMed.
- A. Szebesczyk, E. Olshvang, A. Shanzer, P. L. Carver and E. Gumienna-Kontecka, Harnessing the power of fungal siderophores for the imaging and treatment of human diseases, Coord. Chem. Rev., 2016, 327–328, 84–109 CrossRef CAS.
- A. Brault, T. Mourer and S. Labbe, Molecular basis of the regulation of iron homeostasis in fission and filamentous yeasts, IUBMB Life, 2015, 67, 801–815 CrossRef CAS PubMed.
- J. Choi, W. H. Jung and J. W. Kronstad, The cAMP/protein kinase A signaling pathway in pathogenic basidiomycete fungi: Connections with iron homeostasis, J. Microbiol., 2015, 53, 579–587 CrossRef CAS PubMed.
- S. M. Noble, Candida albicans specializations for iron homeostasis: from commensalism to virulence, Curr. Opin. Microbiol., 2013, 16, 708–715 CrossRef CAS PubMed.
- M. Caza and J. W. Kronstad, Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans, Front. Cell. Infect. Microbiol., 2013, 3, 80 Search PubMed.
- M. Schrettl and H. Haas, Iron homeostasis–Achilles' heel of Aspergillus fumigatus?, Curr. Opin. Microbiol., 2011, 14, 400–405 CrossRef CAS PubMed.
- H. Haas, Fungal siderophore metabolism with a focus on Aspergillus fumigatus, Nat. Prod. Rep., 2014, 31, 1266–1276 RSC.
- C. C. Philpott and O. Protchenko, Response to iron deprivation in Saccharomyces cerevisiae, Eukaryotic Cell, 2008, 7, 20–27 CrossRef CAS PubMed.
- H. Haas, Molecular genetics of fungal siderophore biosynthesis and uptake: the role of siderophores in iron uptake and storage, Appl. Microbiol. Biotechnol., 2003, 62, 316–330 CrossRef CAS PubMed.
- D. J. Kosman, Molecular mechanisms of iron uptake in fungi, Mol. Microbiol., 2003, 47, 1185–1197 CrossRef CAS PubMed.
- J. A. Morrissey, P. H. Williams and A. M. Cashmore, Candida albicans has a cell-associated ferric-reductase activity which is regulated in response to levels of iron and copper, Microbiology, 1996, 142(Pt 3), 485–492 CrossRef CAS PubMed.
- S. A. Knight, E. Lesuisse, R. Stearman, R. D. Klausner and A. Dancis, Reductive iron uptake by Candida albicans: role of copper, iron and the TUP1 regulator, Microbiology, 2002, 148, 29–40 CrossRef CAS PubMed.
- S. A. Knight, G. Vilaire, E. Lesuisse and A. Dancis, Iron acquisition from transferrin by Candida albicans depends on the reductive pathway, Infect. Immun., 2005, 73, 5482–5492 CrossRef CAS PubMed.
- N. Ramanan and Y. Wang, A high-affinity iron permease essential for Candida albicans virulence, Science, 2000, 288, 1062–1064 CrossRef CAS PubMed.
- C. C. Philpott, Iron uptake in fungi: a system for every source, Biochim. Biophys. Acta, 2006, 1763, 636–645 CrossRef CAS PubMed.
- R. E. Jeeves, R. P. Mason, A. Woodacre and A. M. Cashmore, Ferric reductase genes involved in high-affinity iron uptake are differentially regulated in yeast and hyphae of Candida albicans, Yeast, 2011, 28, 629–644 CrossRef CAS PubMed.
- J. E. Hammacott, P. H. Williams and A. M. Cashmore, Candida albicans CFL1 encodes a functional ferric reductase activity that can rescue a Saccharomyces cerevisiae fre1 mutant, Microbiology, 2000, 146(Pt 4), 869–876 CrossRef CAS PubMed.
- N. Xu, K. Qian, Y. Dong, Y. Chen, Q. Yu, B. Zhang, L. Xing and M. Li, Novel role of the Candida albicans ferric reductase gene CFL1 in iron acquisition, oxidative stress tolerance, morphogenesis and virulence, Res. Microbiol., 2014, 165, 252–261 CrossRef CAS PubMed.
- Q. Yu, Y. Dong, N. Xu, K. Qian, Y. Chen, B. Zhang, L. Xing and M. Li, A novel role of the ferric reductase Cfl1 in cell wall integrity, mitochondrial function, and invasion to host cells in Candida albicans, FEMS Yeast Res., 2014, 14, 1037–1047 CrossRef CAS PubMed.
- S. Saikia, D. Oliveira, G. Hu and J. Kronstad, Role of ferric reductases in iron acquisition and virulence in the fungal pathogen Cryptococcus neoformans, Infect. Immun., 2014, 82, 839–850 CrossRef PubMed.
- E. Georgatsou and D. Alexandraki, Regulated expression of the Saccharomyces cerevisiae Fre1p/Fre2p Fe/Cu reductase related genes, Yeast, 1999, 15, 573–584 CrossRef CAS PubMed.
- E. Y. Kwok, S. Severance and D. J. Kosman, Evidence for iron channeling in the Fet3p-Ftr1p high-affinity iron uptake complex in the yeast plasma membrane, Biochemistry, 2006, 45, 6317–6327 CrossRef CAS PubMed.
- R. Eck, S. Hundt, A. Hartl, E. Roemer and W. Kunkel, A multicopper oxidase gene from Candida albicans: cloning, characterization and disruption, Microbiology, 1999, 145(Pt 9), 2415–2422 CrossRef CAS PubMed.
- H. M. Fang and Y. Wang, Characterization of iron-binding motifs in Candida albicans high-affinity iron permease CaFtr1p by site-directed mutagenesis, Biochem. J., 2002, 368, 641–647 CrossRef CAS PubMed.
- E. S. Jacobson, A. P. Goodner and K. J. Nyhus, Ferrous iron uptake in Cryptococcus neoformans, Infect. Immun., 1998, 66, 4169–4175 CAS.
- W. H. Jung, A. Sham, T. Lian, A. Singh, D. J. Kosman and J. W. Kronstad, Iron source preference and regulation of iron uptake in Cryptococcus neoformans, PLoS Pathog., 2008, 4, e45 Search PubMed.
- M. Schrettl, E. Bignell, C. Kragl, C. Joechl, T. Rogers, H. N. Arst, Jr., K. Haynes and H. Haas, Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence, J. Exp. Med., 2004, 200, 1213–1219 CrossRef CAS PubMed.
- V. K. Srivastava, K. J. Suneetha and R. Kaur, A systematic analysis reveals an essential role for high-affinity iron uptake system, haemolysin and CFEM domain-containing protein in iron homoeostasis and virulence in Candida glabrata, Biochem. J., 2014, 463, 103–114 CrossRef CAS PubMed.
- L. Ziegler, A. Terzulli, R. Gaur, R. McCarthy and D. J. Kosman, Functional characterization of the ferroxidase, permease high-affinity iron transport complex from Candida albicans, Mol. Microbiol., 2011, 81, 473–485 CrossRef CAS PubMed.
- X. Cheng, N. Xu, Q. Yu, X. Ding, K. Qian, Q. Zhao, Y. Wang, B. Zhang, L. Xing and M. Li, Novel insight into the expression and function of the multicopper oxidases in Candida albicans, Microbiology, 2013, 159, 1044–1055 CrossRef CAS PubMed.
- C. Y. Lan, G. Rodarte, L. A. Murillo, T. Jones, R. W. Davis, J. Dungan, G. Newport and N. Agabian, Regulatory networks affected by iron availability in Candida albicans, Mol. Microbiol., 2004, 53, 1451–1469 CrossRef CAS PubMed.
- M. Blatzer, U. Binder and H. Haas, The metalloreductase FreB is involved in adaptation of Aspergillus fumigatus to iron starvation, Fungal Genet. Biol., 2011, 48, 1027–1033 CrossRef CAS PubMed.
- F. Gsaller, M. Eisendle, B. E. Lechner, M. Schrettl, H. Lindner, D. Muller, S. Geley and H. Haas, The interplay between vacuolar and siderophore-mediated iron storage in Aspergillus fumigatus, Metallomics, 2012, 4, 1262–1270 RSC.
- K. Han, E. Do and W. H. Jung, A human fungal pathogen Cryptococcus neoformans expresses three distinct iron permease homologs, J. Microbiol. Biotechnol., 2012, 22, 1644–1652 CrossRef CAS PubMed.
- W. H. Jung, G. Hu, W. Kuo and J. W. Kronstad, Role of ferroxidases in iron uptake and virulence of Cryptococcus neoformans, Eukaryotic Cell, 2009, 8, 1511–1520 CrossRef CAS PubMed.
- T. Gabaldon and L. Carrete, The birth of a deadly yeast: tracing the evolutionary emergence of virulence traits in Candida glabrata, FEMS Yeast Res., 2016, 16, fov110 CrossRef PubMed.
- V. Sharma, R. Purushotham and R. Kaur, The Phosphoinositide 3-Kinase Regulates Retrograde Trafficking of the Iron Permease CgFtr1 and Iron Homeostasis in Candida glabrata, J. Biol. Chem., 2016, 291, 24715–24734 CrossRef CAS PubMed.
- R. S. Almeida, S. Brunke, A. Albrecht, S. Thewes, M. Laue, J. E. Edwards, S. G. Filler and B. Hube, the hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin, PLoS Pathog., 2008, 4, e1000217 Search PubMed.
- K. J. Boyce and A. Andrianopoulos, Fungal dimorphism: the switch from hyphae to yeast is a specialized morphogenetic adaptation allowing colonization of a host, FEMS Microbiol. Rev., 2015, 39, 797–811 CrossRef PubMed.
- S. Pasricha, L. Schafferer, H. Lindner, K. Joanne Boyce, H. Haas and A. Andrianopoulos, Differentially regulated high-affinity iron assimilation systems support growth of the various cell types in the dimorphic pathogen Talaromyces marneffei, Mol. Microbiol., 2016, 102, 715–737 CrossRef CAS PubMed.
- S. Puri, W. K. Lai, J. M. Rizzo, M. J. Buck and M. Edgerton, Iron-responsive chromatin remodelling and MAPK signalling enhance adhesion in Candida albicans, Mol. Microbiol., 2014, 93, 291–305 CrossRef CAS PubMed.
- V. K. Srivastava, K. J. Suneetha and R. Kaur, The mitogen-activated protein kinase CgHog1 is required for iron homeostasis, adherence and virulence in Candida glabrata, FEBS J., 2015, 282, 2142–2166 CrossRef CAS PubMed.
- S. Pasricha, M. Payne, D. Canovas, L. Pase, N. Ngaosuwankul, S. Beard, A. Oshlack, G. K. Smyth, S. C. Chaiyaroj, K. J. Boyce and A. Andrianopoulos, Cell-type-specific transcriptional profiles of the dimorphic pathogen Penicillium marneffei reflect distinct reproductive, morphological, and environmental demands, G3: Genes, Genomes, Genet., 2013, 3, 1997–2014 Search PubMed.
- C. W. Yun, J. S. Tiedeman, R. E. Moore and C. C. Philpott, Siderophore-iron uptake in Saccharomyces cerevisiae. Identification of ferrichrome and fusarinine transporters, J. Biol. Chem., 2000, 275, 16354–16359 CrossRef CAS PubMed.
- M. Miethke and M. A. Marahiel, Siderophore-based iron acquisition and pathogen control, Microbiol. Mol. Biol. Rev., 2007, 71, 413–451 CrossRef CAS PubMed.
- Z. Erpapazoglou, M. Froissard, I. Nondier, E. Lesuisse, R. Haguenauer-Tsapis and N. Belgareh-Touze, Substrate- and ubiquitin-dependent trafficking of the yeast siderophore transporter Sit1, Traffic, 2008, 9, 1372–1391 CrossRef CAS PubMed.
- M. Y. Jeong, C. M. Kang, J. H. Kim, D. H. Heo, M. Chang, I. J. Baek, H. S. Ro, I. D. Choi, T. H. Kim and C. W. Yun, A novel function of Aft1 in regulating ferrioxamine B uptake: Aft1 modulates Arn3 ubiquitination in Saccharomyces cerevisiae, Biochem. J., 2009, 422, 181–191 CrossRef CAS PubMed.
- C. M. Kang, S. Kang, Y. S. Park and C. W. Yun, Physical interaction between Sit1 and Aft1 upregulates FOB uptake activity by inhibiting protein degradation of Sit1 in Saccharomyces cerevisiae, FEMS Yeast Res., 2015, 15, fov080 CrossRef PubMed.
- M. Holzberg and W. M. Artis, Hydroxamate siderophore production by opportunistic and systemic fungal pathogens, Infect. Immun., 1983, 40, 1134–1139 CAS.
- A. Ismail, G. W. Bedell and D. M. Lupan, Siderophore production by the pathogenic yeast, Candida albicans, Biochem. Biophys. Res. Commun., 1985, 130, 885–891 CrossRef CAS PubMed.
- P. Heymann, M. Gerads, M. Schaller, F. Dromer, G. Winkelmann and J. F. Ernst, The siderophore iron transporter of Candida albicans (Sit1p/Arn1p) mediates uptake of ferrichrome-type siderophores and is required for epithelial invasion, Infect. Immun., 2002, 70, 5246–5255 CrossRef CAS PubMed.
- T. Nevitt and D. J. Thiele, Host iron withholding demands siderophore utilization for Candida glabrata to survive macrophage killing, PLoS Pathog., 2011, 7, e1001322 CAS.
- K. L. Tangen, W. H. Jung, A. P. Sham, T. Lian and J. W. Kronstad, The iron- and cAMP-regulated gene SIT1 influences ferrioxamine B utilization, melanization and cell wall structure in Cryptococcus neoformans, Microbiology, 2007, 153, 29–41 CrossRef CAS PubMed.
- M. Liu, L. Lin, T. Gebremariam, G. Luo, C. D. Skory, S. W. French, T. F. Chou, J. E. Edwards, Jr. and A. S. Ibrahim, Fob1 and Fob2 Proteins Are Virulence Determinants of Rhizopus oryzae via Facilitating Iron Uptake from Ferrioxamine, PLoS Pathog., 2015, 11, e1004842 Search PubMed.
- A. S. Ibrahim, T. Gebremariam, L. Lin, G. Luo, M. I. Husseiny, C. D. Skory, Y. Fu, S. W. French, J. E. Edwards, Jr. and B. Spellberg, The high affinity iron permease is a key virulence factor required for Rhizopus oryzae pathogenesis, Mol. Microbiol., 2010, 77, 587–604 CrossRef CAS PubMed.
- A. H. Hissen, A. N. Wan, M. L. Warwas, L. J. Pinto and M. M. Moore, The Aspergillus fumigatus siderophore biosynthetic gene sidA, encoding L-ornithine N5-oxygenase, is required for virulence, Infect. Immun., 2005, 73, 5493–5503 CrossRef CAS PubMed.
- D. H. Howard, R. Rafie, A. Tiwari and K. F. Faull, Hydroxamate siderophores of Histoplasma capsulatum, Infect. Immun., 2000, 68, 2338–2343 CrossRef CAS PubMed.
- Y. S. Park, J. Y. Kim and C. W. Yun, Identification of ferrichrome- and ferrioxamine B-mediated iron uptake by Aspergillus fumigatus, Biochem. J., 2016, 473, 1203–1213 CrossRef CAS PubMed.
- H. Haas, M. Schoeser, E. Lesuisse, J. F. Ernst, W. Parson, B. Abt, G. Winkelmann and H. Oberegger, Characterization of the Aspergillus nidulans transporters for the siderophores enterobactin and triacetylfusarinine C, Biochem. J., 2003, 371, 505–513 CrossRef CAS PubMed.
- I. Raymond-Bouchard, C. S. Carroll, J. R. Nesbitt, K. A. Henry, L. J. Pinto, M. Moinzadeh, J. K. Scott and M. M. Moore, Structural requirements for the activity of the MirB ferrisiderophore transporter of Aspergillus fumigatus, Eukaryotic Cell, 2012, 11, 1333–1344 CrossRef CAS PubMed.
- L. H. Hwang, J. A. Mayfield, J. Rine and A. Sil, Histoplasma requires SID1, a member of an iron-regulated siderophore gene cluster, for host colonization, PLoS Pathog., 2008, 4, e1000044 Search PubMed.
- M. Nairz, A. Schroll, T. Sonnweber and G. Weiss, The struggle for iron – a metal at the host-pathogen interface, Cell. Microbiol., 2010, 12, 1691–1702 CrossRef CAS PubMed.
- J. E. Cassat and E. P. Skaar, Iron in infection and immunity, Cell Host Microbe, 2013, 13, 509–519 CAS.
- Z. Weissman, R. Shemer and D. Kornitzer, Deletion of the copper transporter CaCCC2 reveals two distinct pathways for iron acquisition in Candida albicans, Mol. Microbiol., 2002, 44, 1551–1560 CrossRef CAS PubMed.
- R. Santos, N. Buisson, S. Knight, A. Dancis, J. M. Camadro and E. Lesuisse, Haemin uptake and use as an iron source by Candida albicans: role of CaHMX1-encoded haem oxygenase, Microbiology, 2003, 149, 579–588 CrossRef CAS PubMed.
- Z. Weissman and D. Kornitzer, A family of Candida cell surface haem-binding proteins involved in haemin and haemoglobin-iron utilization, Mol. Microbiol., 2004, 53, 1209–1220 CrossRef CAS PubMed.
- Z. Weissman, R. Shemer, E. Conibear and D. Kornitzer, An endocytic mechanism for haemoglobin-iron acquisition in Candida albicans, Mol. Microbiol., 2008, 69, 201–217 CrossRef CAS PubMed.
- K. Okamoto-Shibayama, Y. Kikuchi, E. Kokubu, Y. Sato and K. Ishihara, Csa2, a member of the Rbt5 protein family, is involved in the utilization of iron from human hemoglobin during Candida albicans hyphal growth, FEMS Yeast Res., 2014, 14, 674–677 CrossRef CAS PubMed.
- B. R. Braun, W. S. Head, M. X. Wang and A. D. Johnson, Identification and characterization of TUP1-regulated genes in Candida albicans, Genetics, 2000, 156, 31–44 CAS.
- A. Perez, B. Pedros, A. Murgui, M. Casanova, J. L. Lopez-Ribot and J. P. Martinez, Biofilm formation by Candida albicans mutants for genes coding fungal proteins exhibiting the eight-cysteine-containing CFEM domain, FEMS Yeast Res., 2006, 6, 1074–1084 CrossRef CAS PubMed.
- G. Kuznets, E. Vigonsky, Z. Weissman, D. Lalli, T. Gildor, S. J. Kauffman, P. Turano, J. Becker, O. Lewinson and D. Kornitzer, A relay network of extracellular heme-binding proteins drives C. albicans iron acquisition from hemoglobin, PLoS Pathog., 2014, 10, e1004407 Search PubMed.
- L. Nasser, Z. Weissman, M. Pinsky, H. Amartely, H. Dvir and D. Kornitzer, Structural basis of haem-iron acquisition by fungal pathogens, Nat. Rev. Microbiol., 2016, 1, 16156 CrossRef PubMed.
- P. Arnoux, R. Haser, N. Izadi, A. Lecroisey, M. Delepierre, C. Wandersman and M. Czjzek, The crystal structure of HasA, a hemophore secreted by Serratia marcescens, Nat. Struct. Biol., 1999, 6, 516–520 CrossRef CAS PubMed.
- A. Y. Alontaga, J. C. Rodriguez, E. Schonbrunn, A. Becker, T. Funke, E. T. Yukl, T. Hayashi, J. Stobaugh, P. Moenne-Loccoz and M. Rivera, Structural characterization of the hemophore HasAp from Pseudomonas aeruginosa: NMR spectroscopy reveals protein-protein interactions between Holo-HasAp and hemoglobin, Biochemistry, 2009, 48, 96–109 CrossRef CAS PubMed.
- E. F. Bailao, J. A. Parente, L. L. Pigosso, K. P. de Castro, F. L. Fonseca, M. G. Silva-Bailao, S. N. Bao, A. M. Bailao, M. L. Rodrigues, O. Hernandez, J. G. McEwen and C. M. Soares, Hemoglobin uptake by Paracoccidioides spp. is receptor-mediated, PLoS Neglected Trop. Dis., 2014, 8, e2856 Search PubMed.
- E. F. Bailao, S. Lima Pde, M. G. Silva-Bailao, A. M. Bailao, R. Fernandes Gda, D. J. Kosman and C. M. Soares, Paracoccidioides spp. ferrous and ferric iron assimilation pathways, Front. Microbiol., 2015, 6, 821 Search PubMed.
- Y. Vaknin, Y. Shadkchan, E. Levdansky, M. Morozov, J. Romano and N. Osherov, The three Aspergillus fumigatus CFEM-domain GPI-anchored proteins (CfmA-C) affect cell-wall stability but do not play a role in fungal virulence, Fungal Genet. Biol., 2014, 63, 55–64 CrossRef CAS PubMed.
- H. Irmer, S. Tarazona, C. Sasse, P. Olbermann, J. Loeffler, S. Krappmann, A. Conesa and G. H. Braus, RNAseq analysis of Aspergillus fumigatus in blood reveals a just wait and see resting stage behavior, BMC Genomics, 2015, 16, 640 CrossRef PubMed.
- G. Hu, M. Caza, B. Cadieux, V. Chan, V. Liu and J. Kronstad, Cryptococcus neoformans requires the ESCRT protein Vps23 for iron acquisition from heme, for capsule formation, and for virulence, Infect. Immun., 2013, 81, 292–302 CrossRef CAS PubMed.
- G. Hu, M. Caza, B. Cadieux, E. Bakkeren, E. Do, W. H. Jung and J. W. Kronstad, The endosomal sorting complex required for transport machinery influences haem uptake and capsule elaboration in Cryptococcus neoformans, Mol. Microbiol., 2015, 96, 973–992 CrossRef CAS PubMed.
- B. Cadieux, T. Lian, G. Hu, J. Wang, C. Biondo, G. Teti, V. Liu, M. E. Murphy, A. L. Creagh and J. W. Kronstad, The Mannoprotein Cig1 supports iron acquisition from heme and virulence in the pathogenic fungus Cryptococcus neoformans, J. Infect. Dis., 2013, 207, 1339–1347 CrossRef CAS PubMed.
- J. W. Kronstad, G. Hu and W. H. Jung, An encapsulation of iron homeostasis and virulence in Cryptococcus neoformans, Trends Microbiol., 2013, 21, 457–465 CrossRef CAS PubMed.
- M. Kretschmer, E. Reiner, G. Hu, N. Tam, D. L. Oliveira, M. Caza, J. H. Yeon, J. Kim, C. J. Kastrup, W. H. Jung and J. W. Kronstad, Defects in phosphate acquisition and storage influence virulence of Cryptococcus neoformans, Infect. Immun., 2014, 82, 2697–2712 CrossRef PubMed.
- C. S. Carroll, L. N. Amankwa, L. J. Pinto, J. D. Fuller and M. M. Moore, Detection of a Serum Siderophore by LC-MS/MS as a Potential Biomarker of Invasive Aspergillosis, PLoS One, 2016, 11, e0151260 Search PubMed.
- M. Balhara, R. Chaudhary, S. Ruhil, B. Singh, N. Dahiya, V. S. Parmar, P. K. Jaiwal and A. K. Chhillar, Siderophores; iron scavengers: the novel & promising targets for pathogen specific antifungal therapy, Expert Opin. Ther. Targets, 2016, 20, 1477–1489 CrossRef CAS PubMed.
- H. Haas, M. Petrik and C. Decristoforo, An iron-mimicking, Trojan horse-entering fungi–has the time come for molecular imaging of fungal infections?, PLoS Pathog., 2015, 11, e1004568 Search PubMed.
- B. R. Wilson, A. R. Bogdan, M. Miyazawa, K. Hashimoto and Y. Tsuji, Siderophores in Iron Metabolism: From Mechanism to Therapy Potential, Trends Mol. Med., 2016, 22, 1077–1090 CrossRef CAS PubMed.
- A. L. Lamb, Breaking a pathogen's iron will: Inhibiting siderophore production as an antimicrobial strategy, Biochim. Biophys. Acta, 2015, 1854, 1054–1070 CrossRef CAS PubMed.
- L. J. Pinto and M. M. Moore, Screening method to identify inhibitors of siderophore biosynthesis in the opportunistic fungal pathogen, Aspergillus fumigatus, Lett. Appl. Microbiol., 2009, 49, 8–13 CrossRef CAS PubMed.
- G. Bernier, V. Girijavallabhan, A. Murray, N. Niyaz, P. Ding, M. J. Miller and F. Malouin, Desketoneoenactin-siderophore conjugates for Candida: evidence of iron transport-dependent species selectivity, Antimicrob. Agents Chemother., 2005, 49, 241–248 CrossRef CAS PubMed.
- J. S. Martin Del Campo, N. Vogelaar, K. Tolani, K. Kizjakina, K. Harich and P. Sobrado, Inhibition of the Flavin-Dependent Monooxygenase Siderophore A (SidA) Blocks Siderophore Biosynthesis and Aspergillus fumigatus Growth, ACS Chem. Biol., 2016, 11, 3035–3042 CrossRef CAS PubMed.
- S. Franceschini, M. Fedkenheuer, N. J. Vogelaar, H. H. Robinson, P. Sobrado and A. Mattevi, Structural insight into the mechanism of oxygen activation and substrate selectivity of flavin-dependent N-hydroxylating monooxygenases, Biochemistry, 2012, 51, 7043–7045 CrossRef CAS PubMed.
- J. Qi, K. Kizjakina, R. Robinson, K. Tolani and P. Sobrado, A fluorescence polarization binding assay to identify inhibitors of flavin-dependent monooxygenases, Anal. Biochem., 2012, 425, 80–87 CrossRef CAS PubMed.
- D. Q. Luo, H. Wang, X. Tian, H. J. Shao and J. K. Liu, Antifungal properties of pristimerin and celastrol isolated from Celastrus hypoleucus, Pest Manage. Sci., 2005, 61, 85–90 CrossRef CAS PubMed.
- S. M. Leal, Jr., S. Roy, C. Vareechon, S. Carrion, H. Clark, M. S. Lopez-Berges, A. Di Pietro, M. Schrettl, N. Beckmann, B. Redl, H. Haas and E. Pearlman, Targeting iron acquisition blocks infection with the fungal pathogens Aspergillus fumigatus and Fusarium oxysporum, PLoS Pathog., 2013, 9, e1003436 Search PubMed.
- M. E. Helsel, E. J. White, S. Z. Razvi, B. Alies and K. J. Franz, Chemical and functional properties of metal chelators that mobilize copper to elicit fungal killing of Cryptococcus neoformans, Metallomics, 2017, 9, 69–81 RSC.
- H. Nazik, J. C. Penner, J. A. Ferreira, J. A. Haagensen, K. Cohen, A. M. Spormann, M. Martinez, V. Chen, J. L. Hsu, K. V. Clemons and D. A. Stevens, Effects of Iron Chelators on the Formation and Development of Aspergillus fumigatus Biofilm, Antimicrob. Agents Chemother., 2015, 59, 6514–6520 CrossRef CAS PubMed.
- Y. W. Lai, L. T. Campbell, M. R. Wilkins, C. N. Pang, S. Chen and D. A. Carter, Synergy and antagonism between iron chelators and antifungal drugs in Cryptococcus, Int. J. Antimicrob. Agents, 2016, 48, 388–394 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally and should be considered co-first authors. |
| This journal is © The Royal Society of Chemistry 2017 |



