Methanobactins: from genome to function†
Laura M. K.
Dassama‡
a,
Grace E.
Kenney‡
a and
Amy C.
Rosenzweig
 *ab
*ab
aDepartment of Molecular Biosciences, Northwestern University, Evanston, IL 60208, USA. E-mail: amyr@northwestern.edu
bDepartment of Chemistry, Northwestern University, Evanston, IL 60208, USA
First published on 24th November 2016
Abstract
Methanobactins (Mbns) are ribosomally produced, post-translationally modified peptide (RiPP) natural products that bind copper with high affinity using nitrogen-containing heterocycles and thioamide groups. In some methanotrophic bacteria, Mbns are secreted under conditions of copper starvation and then re-internalized as a copper source for the enzyme particulate methane monooxygenase (pMMO). Genome mining studies have led to the identification and classification of operons encoding the Mbn precursor peptide (MbnA) as well as a number of putative transport, regulatory, and biosynthetic proteins. These Mbn operons are present in non-methanotrophic bacteria as well, suggesting a broader role in and perhaps beyond copper acquisition. Genetic and biochemical studies indicate that specific operon-encoded proteins are involved in Mbn transport and provide insight into copper-responsive gene regulation in methanotrophs. Mbn biosynthesis is not yet understood, but combined analysis of Mbn structures, MbnA sequences, and operon content represents a powerful approach to elucidating the roles of specific biosynthetic enzymes. Future work will likely lead to the discovery of unique pathways for natural product biosynthesis and new mechanisms of microbial metal homeostasis.
1. Introduction
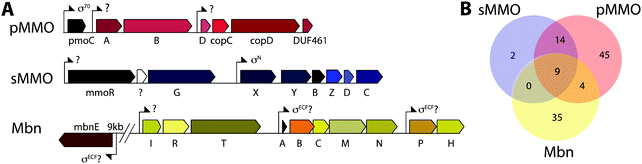
Copper plays an important role in biology, often serving as a cofactor for enzymes that perform difficult chemical transformations. Because of its ability to generate free radicals, bioavailable copper is highly regulated by cells. Thus, organisms that require copper often evolve elaborate strategies for its acquisition and homeostasis.1–3 Among these organisms are methanotrophic bacteria, which use methane as their sole source of carbon and energy.4 Interest in using methanotrophs for methane mitigation and remediation as well as biological gas-to-liquids conversion processes has skyrocketed recently.5,6 To metabolize methane, methanotrophs first convert it to methanol, a reaction that involves cleavage of an aliphatic C–H bond with a 105 kcal mol−1 dissociation energy. This challenging reaction is catalyzed by methane monooxygenase enzymes (MMOs).MMOs are metalloenzymes, and two unrelated forms have been isolated and characterized: a cytoplasmic, iron-dependent enzyme known as soluble methane monooxygenase (sMMO), and a membrane-bound, copper-dependent enzyme called particulate methane monooxygenase (pMMO).6–8 The majority of methanotrophs use pMMO for methane oxidation, but some strains have genes encoding both systems. In this subset of methanotrophs, sMMO and pMMO expression are reciprocally regulated on the basis of copper availability. At very low soluble copper concentrations, sMMO (encoded by the mmoRGXYBZDC operon) is expressed, and when copper is bioavailable, pMMO (encoded by the pmoCAB operon) is expressed. The molecular mechanisms of this intricate regulatory event, termed the “copper switch,” are still largely unknown.9 A key player in methanotroph copper acquisition and the copper switch is methanobactin (Mbn), a ribosomally synthesized and post-translationally modified peptide (RiPP) natural product that binds copper. We abbreviate methanobactin as Mbn rather than Mb to comply with the established naming schema for RiPPs,10 and to avoid confusion with myoglobin, which is typically abbreviated Mb.
Mbn was first isolated from the growth medium of Methylosinus (Ms.) trichosporium OB3b, an obligate methanotroph.11–13 When Ms. trichosporium OB3b is cultured under conditions of copper starvation, it produces and secretes Mbn. Outside of the cell, Mbn scavenges copper from soluble, mineral, and humic sources,14 and can even mediate its removal from borosilicate glass.15 The copper-loaded form (CuMbn) is then taken up by an active transport process16 and can promote the copper switch; soluble copper, which can also promote this regulatory shift, is taken up separately and via a passive process.17 CuMbn from Ms. trichosporium OB3b consists of a peptidic backbone, an “N-terminal” carbonyl group replacing the primary amine of the N-terminal leucine residue, two oxazolone groups with neighboring thioamides, and an intramolecular disulfide bond (Fig. 1A).11 Forms of the Ms. trichosporium OB3b Mbn that lack the C-terminal methionine are sometimes isolated as well.18 The N- and S-ligands of the oxazolones and thioamides chelate a single copper ion in a distorted tetrahedral geometry. Although capable of binding CuII, the copper ion stabilized by Mbn has been identified as CuI.11,19 The reduction of CuII to CuI occurs within 10 minutes, and is mediated by Mbn itself via a mechanism that remains unclear but that does not involve the disulfide bond.19–22 The binding constant of Ms. trichosporium OB3b Mbn for CuI has been measured (Kb ∼ 6 × 1020 M),18 while that for CuII is calculated to be 6.5 × 1011 M.23
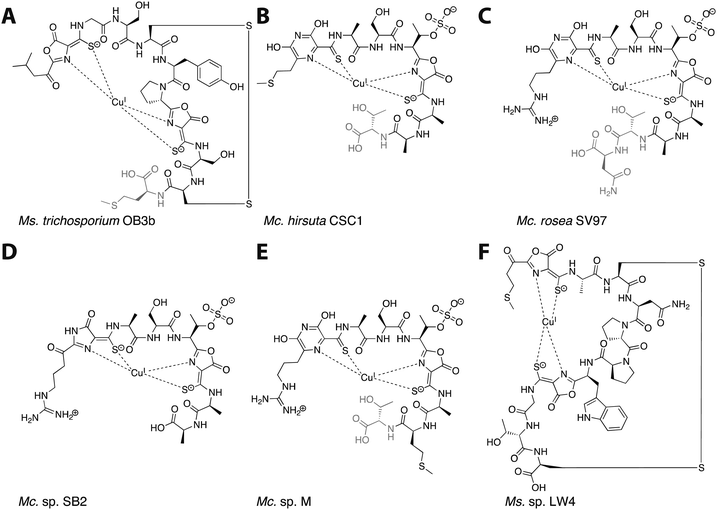 | ||
| Fig. 1 The six characterized Mbns. Light gray C-terminal residues are sometimes lost in compounds isolated from spent medium; based on genomic information, there may be additional residues initially present at the C-terminus of some Mc. Mbns. In all but Mc. sp. M, the five residues following the second heterocycle/thioamide moiety are AATNG (likely AMTNG in Mc. sp. M). (A) Ms. trichosporium OB3b CuMbn, structurally characterized via X-ray crystallography and NMR. (B) Mc. hirsuta CSC1 CuMbn, characterized via X-ray crystallography. (C) Mc. rosea SV97 CuMbn, characterized via X-ray crystallography. (D) Mc. sp. SB2, characterized via NMR. The assignment of the first heterocycle has been called into question.29 (E) Mc. sp. M CuMbn, characterized via X-ray crystallography. (F) Ms. sp. LW4 CuMbn, characterized via NMR. | ||
Although copper is the most physiologically relevant metal ion, Mbn also binds other metal ions, including AuIII, HgII, AgI, UVI, CdII, PbII, MnII, FeII, CoII, and NiII. Only soft metals such as AuIII, HgII, and AgI can bind reductively (with spectral features further supporting a binding mode similar to that of copper),21 and all exhibit binding constants that are approximately five orders of magnitude lower than that of CuII and fifteen orders of magnitude lower than that of CuI.21 Mbn has been used to mediate the synthesis of gold nanoparticles by binding AuIII and reducing it to Au0,24 and optical spectroscopic data suggest that gold has some ability to compete with copper for Mbn binding, thereby limiting copper acquisition via Mbn and affecting sMMO expression.25 Similarly, Mbn binds HgII from HgCl2 (in the absence and to a lesser extent the presence of copper) and in doing so reduces the toxic effects of mercury on the bacteria.26 The ability to bind metals other than copper may be compound-specific; reported metal binding specificity and affinity for Methylocystis (Mc.) sp. SB2 Mbn differs somewhat from that of Ms. trichosporium OB3b Mbn.26 It has been suggested that Mbn plays a role in alleviating mercury toxicity for methanotrophs growing in polluted environments, although no particular correlation is apparent between Mbn-producing species and environments with high levels of heavy metal pollution.27
Following the characterization of Mbn from Ms. trichosporium OB3b, Mbns were isolated from several other methanotroph species. Like Ms. trichosporium OB3b CuMbn, the CuMbns isolated from Mc. rosea SV97,23Mc. sp. M,23Mc. sp. SB2,28Mc. hirsuta CSC1,23 and most recently, Ms. sp. LW4,29 are all peptidic natural products that bind copper with high affinity via two nitrogen-containing heterocycles and two neighboring thioamide groups (Fig. 1B–F). The Mc. Mbns differ from the Ms. Mbns in the presence of a pyrazinedione or an imidazolone (the latter is reported for the Mc. sp. SB2 Mbn,28 though recent data suggest it may in fact also be a pyrazinedione29) in lieu of the N-terminal oxazolone present in the Ms. CuMbns, the absence of a disulfide bond or an “N-terminal” carbonyl group, and the presence of a sulfonated threonine. Like Ms. trichosporium OB3b Mbn, Mbns from the Mc. species are frequently missing one or more C-terminal residues, apparent both in the range of structures obtained and the sequences of the precursor peptides.18,23,30 All four Mc. Mbns are quite similar, differing in at most one or two residues. The backbone of Ms. sp. LW4 Mbn diverges significantly from previously characterized Mbns in both sequence and in the increased distance between the two heterocycle/thioamide moieties, but has several features in common with Ms. trichosporium OB3b Mbn, including the presence of an “N-terminal” carbonyl group, two oxazolones with neighboring thioamides, and an intramolecular disulfide bond.
A major focus of the last three years has been on mining bacterial genomes to discover genes encoding new Mbn precursor peptides as well as neighboring genes of functional relevance. New bioinformatic, genetic, and biochemical data have provided significant insight into Mbn diversity, its transport and biosynthesis, and its potential role in the methanotroph copper switch mechanism. This minireview summarizes these developments and highlights areas ripe for future investigation.
2. Genome mining for Mbns
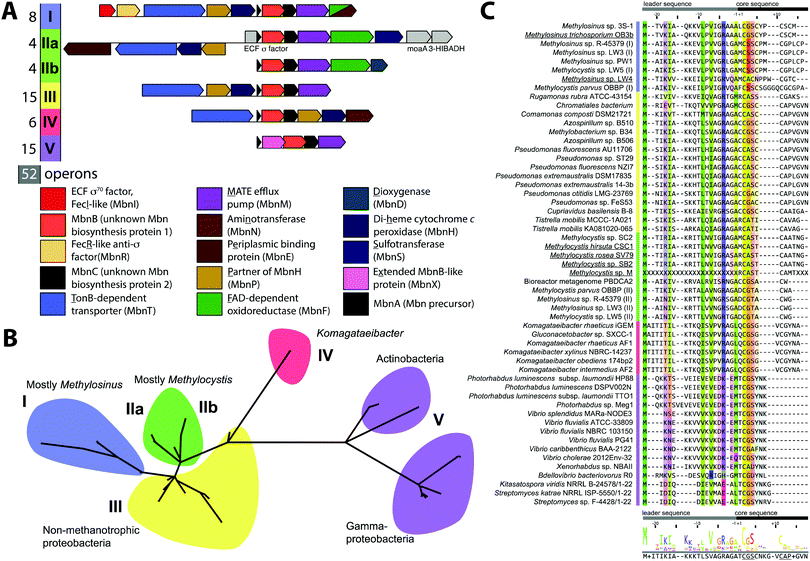
Mbn was initially believed to be the product of non-ribosomal peptide synthesis.31,32 However, sequencing of the Ms. trichosporium OB3b genome33 led to the identification of an open reading frame (ORF) encoding 30 residues, of which the last 11 residues (LCGSCYPCSCM) resemble the peptidic backbone of Mbn.28 The presence of this putative precursor peptide, hereafter referred to as MbnA, and the observations that disruption of its gene abolishes Mbn production34 and that its expression is significantly downregulated by copper35 establish Mbn as a RiPP.10 At the time that the gene encoding MbnA was identified, the only homologue was a 31-residue sequence from Azospirillum sp. B510,36 a diazotrophic endophyte.In 2013, an extensive bioinformatics study that sought to identify MbnA and neighboring genes in newly sequenced bacterial genomes was reported.37 Using a variety of techniques, operons containing the MbnA gene as well as putative biosynthetic and transport genes were detected in 14 species, a number that can now be updated to 52 operons in 48 species (Fig. 2A–C, Table 1 and Fig. S1, ESI†) At the time, only 5 of the species with putative Mbn operons were Type II, α-proteobacterial methanotrophs (like Ms. trichosporium OB3b), and methanotrophic bacteria remain a minority of the species in which Mbn operons have been observed; the presence of mobile genetic elements in the proximity of many Mbn operons suggests that horizontal gene transfer plays a major role in the dissemination of these operons. Time-dependent quantitative real time (qRT)-PCR studies indicate that all genes in the Mbn operon (mbnIRTABCMNPH) of Ms. trichosporium OB3b are downregulated by copper and are coregulated with the sMMO genes (mmoRGXYBZDC), consistent with all the operon-encoded proteins functioning in copper acquisition.35 Furthermore, a potential connection to copper homeostasis is maintained even in non-methanotrophs, in which Mbn operons are frequently found in the proximity of periplasmic and inner member copper homeostasis machinery, including copC, copD, sco1, and DUF461.37
| Gene name | Proposed role | InterProScan ID | TIGRFAM | PFAM |
|---|---|---|---|---|
| mbnA | Precursor peptide | ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[0 with combining low line]](https://www.rsc.org/images/entities/char_0030_0332.gif) ![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) ![[3 with combining low line]](https://www.rsc.org/images/entities/char_0033_0332.gif) ![[9 with combining low line]](https://www.rsc.org/images/entities/char_0039_0332.gif) ![[6 with combining low line]](https://www.rsc.org/images/entities/char_0036_0332.gif) ![[3 with combining low line]](https://www.rsc.org/images/entities/char_0033_0332.gif) |
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[0 with combining low line]](https://www.rsc.org/images/entities/char_0030_0332.gif) ![[4 with combining low line]](https://www.rsc.org/images/entities/char_0034_0332.gif) ![[0 with combining low line]](https://www.rsc.org/images/entities/char_0030_0332.gif) ![[7 with combining low line]](https://www.rsc.org/images/entities/char_0037_0332.gif) ![[1 with combining low line]](https://www.rsc.org/images/entities/char_0031_0332.gif) |
|
| mbnB | Biosynthesis protein I | ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[0 with combining low line]](https://www.rsc.org/images/entities/char_0030_0332.gif) ![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) ![[6 with combining low line]](https://www.rsc.org/images/entities/char_0036_0332.gif) ![[4 with combining low line]](https://www.rsc.org/images/entities/char_0034_0332.gif) ![[3 with combining low line]](https://www.rsc.org/images/entities/char_0033_0332.gif) ![[1 with combining low line]](https://www.rsc.org/images/entities/char_0031_0332.gif) , IPR007801, IPR013022 , IPR007801, IPR013022 |
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[0 with combining low line]](https://www.rsc.org/images/entities/char_0030_0332.gif) ![[4 with combining low line]](https://www.rsc.org/images/entities/char_0034_0332.gif) ![[1 with combining low line]](https://www.rsc.org/images/entities/char_0031_0332.gif) ![[5 with combining low line]](https://www.rsc.org/images/entities/char_0035_0332.gif) ![[9 with combining low line]](https://www.rsc.org/images/entities/char_0039_0332.gif) |
PF05114 |
| mbnC | Biosynthesis protein II | ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[0 with combining low line]](https://www.rsc.org/images/entities/char_0030_0332.gif) ![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) ![[3 with combining low line]](https://www.rsc.org/images/entities/char_0033_0332.gif) ![[9 with combining low line]](https://www.rsc.org/images/entities/char_0039_0332.gif) ![[7 with combining low line]](https://www.rsc.org/images/entities/char_0037_0332.gif) ![[3 with combining low line]](https://www.rsc.org/images/entities/char_0033_0332.gif) |
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[0 with combining low line]](https://www.rsc.org/images/entities/char_0030_0332.gif) ![[4 with combining low line]](https://www.rsc.org/images/entities/char_0034_0332.gif) ![[1 with combining low line]](https://www.rsc.org/images/entities/char_0031_0332.gif) ![[6 with combining low line]](https://www.rsc.org/images/entities/char_0036_0332.gif) ![[0 with combining low line]](https://www.rsc.org/images/entities/char_0030_0332.gif) , TIGR04061 , TIGR04061 |
|
| mbnD | Dioxygenase | IPR003819 | PF02668 | |
| mbnE | Periplasmic binding protein | IPR030678, IPR000914 | PF00496 | |
| mbnF | FAD-dependent monooxygenase | IPR023753, IPR002938 | PF01494 | |
| mbnH | Diheme cytochrome c peroxidase | ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[0 with combining low line]](https://www.rsc.org/images/entities/char_0030_0332.gif) ![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) ![[3 with combining low line]](https://www.rsc.org/images/entities/char_0033_0332.gif) ![[9 with combining low line]](https://www.rsc.org/images/entities/char_0039_0332.gif) ![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) ![[9 with combining low line]](https://www.rsc.org/images/entities/char_0039_0332.gif) , IPR026259 , IPR026259 |
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[0 with combining low line]](https://www.rsc.org/images/entities/char_0030_0332.gif) ![[4 with combining low line]](https://www.rsc.org/images/entities/char_0034_0332.gif) ![[0 with combining low line]](https://www.rsc.org/images/entities/char_0030_0332.gif) ![[3 with combining low line]](https://www.rsc.org/images/entities/char_0033_0332.gif) ![[9 with combining low line]](https://www.rsc.org/images/entities/char_0039_0332.gif) |
PF03150 |
| mbnI | ECF sigma factor | IPR013325, IPR014284, IPR013324 | TIGR02937 |
PF04542,
PF08281 |
| mbnM | MATE exporter | IPR002528 | TIGR00797 | PF01554 |
| mbnN | Aminotransferase | IPR004839 (I), IPR005814 (IV), IPR015424 | TIGR01141 |
PF00155 (I),
PF00202 (IV) |
| mbnP | Unknown | ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[0 with combining low line]](https://www.rsc.org/images/entities/char_0030_0332.gif) ![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) ![[3 with combining low line]](https://www.rsc.org/images/entities/char_0033_0332.gif) ![[9 with combining low line]](https://www.rsc.org/images/entities/char_0039_0332.gif) ![[7 with combining low line]](https://www.rsc.org/images/entities/char_0037_0332.gif) ![[7 with combining low line]](https://www.rsc.org/images/entities/char_0037_0332.gif) |
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[I with combining low line]](https://www.rsc.org/images/entities/char_0049_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) ![[0 with combining low line]](https://www.rsc.org/images/entities/char_0030_0332.gif) ![[4 with combining low line]](https://www.rsc.org/images/entities/char_0034_0332.gif) ![[0 with combining low line]](https://www.rsc.org/images/entities/char_0030_0332.gif) ![[5 with combining low line]](https://www.rsc.org/images/entities/char_0035_0332.gif) ![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) |
|
| mbnR | Anti-sigma factor |
IPR012373, IPR032623,
IPR006860, IPR032508 |
PF04773 | |
| mbnS | Sulfotransferase | IPR000863, IPR027417 | PF00685 | |
| mbnT | TonB-dependent transporter | IPR010105, IPR012910, IPR000531, IPR011662 (I) | TIGR01783 |
PF00593,
PF07715, PF07660 (I) |
| mbnX | Biosynthesis protein III | IPR007801, IPR013022 | PF05114 | |
Mbn operons were initially detected in α-, β-, and γ-proteobacteria, and have since been found in Gram-positive bacteria, indicating that Mbn or Mbn-like molecules are more widespread than originally believed (Fig. S1, ESI†). Interestingly, some well-studied methanotrophic species initially thought/reported to secrete Mbn, including Methylococcus (Mcc.) capsulatus (Bath) and Methylomicrobium album BG8,38 do not contain Mbn operons; no structural characterization of those purported Mbns has been reported and their copper affinities are much lower than that of Ms. trichosporium OB3b CuMbn.38 Thus, if these species make chalkophores (copper-chelating small molecules), they must differ significantly in structure from Mbns. Alternatively, these reported molecules may be metallophores of a different metal specificity with some ability to bind copper, as observed with yersiniabactin,39 or with Mbn and other metal ions.21
Based on phylogenetic analysis of the sequences of MbnA and of the two core operon proteins, MbnB and MbnC (vide infra), as well as the presence of genes related to transport and regulation, Mbn operons were divided into 5 groups, with Ms. trichosporium OB3b assigned to Group I, the Mc. species falling into Group II, and Azospirillum sp. B510 assigned to Group III. This classification scheme remains broadly accurate, with newly identified Gram-positive Mbn operons fitting into the existing Group V based on operon content and core peptide sequence, although increased diversity observed in Group II suggests that it should be subdivided into two distinct subgroups (Fig. 2A–C).40 Some organisms actually possess several Mbn operons (Fig. S1, ESI†), and in all cases to date, the multiple operons belong to different groups. A less granular set of Mbn categorizations has been proposed,9 but phylogenetic analysis and differences in proposed Mbn modifications support the original set of categorizations, described further here.
3. Transport of Mbn
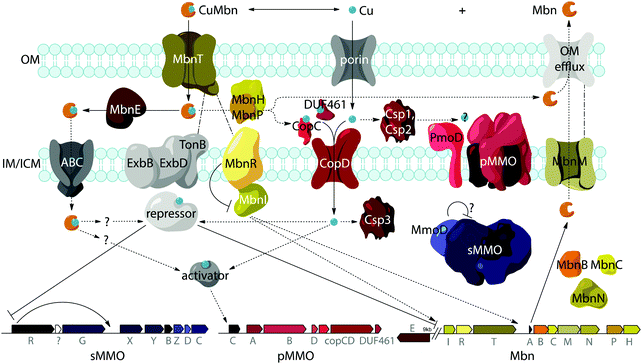
Import by MbnT
Genes encoding TonB-dependent transporters (TBDTs) are often associated with the Mbn operons (MbnTs, Fig. 2A). TBDTs are outer membrane receptors well known for their role in the uptake of siderophores,41,42 iron-chelating small molecules. TBDTs can also import cofactors such as heme, thiamine, and cobalamin, small proteins, natural products like colicins, sugars, and even nickel, copper, and cobalt chelates.43 Thus, the MbnTs are obvious candidates for CuMbn importers, and CuMbn import has been shown previously to occur in an energy-dependent fashion, consistent with the use of a TBDT.16 TBDTs are present in four of the five Mbn operon groups, but the sequences differ significantly. Group I MbnTs (MbnT1s) have an extra ∼100 residue N-terminal extension and are associated with MbnI and MbnR,37 proteins related to E. coli FecI and FecR, a sigma and anti-sigma factor pair that interact with the siderophore TBDT FecA.44–46 MbnT1s also differ from other MbnTs in sequence and domain composition. MbnT2s found in Group II Mbn operons and MbnT3s found in Group III and IV operons form two additional subgroups; sequence similarity network (SSN) analysis indicates that all three are distinct from each other and from any existing characterized TBDT family.40 Notably, genes encoding proteins with known roles in periplasmic copper homeostasis (including sco1, copC, copD, and members of the DUF461 family) are observed near MbnT genes in many Group III and all Group IV genomes, supporting a potential role of these MbnT3s in copper homeostasis. MbnTs are found in a range of species that do not have their own Mbn operons, primarily other methanotrophs and ammonia oxidizers for MbnT1s and MbnT2s, as well as other non-methanotrophic bacteria for MbnT3s, potentially consistent with a scenario in which species with a high copper need but no ability to produce their own Mbns engage in “chalkophore piracy”.40 In preliminary support of this notion, Methylosinus and Methylocystis species have been reported to take up non-native CuMbns.23 MbnTs are not found in any Group V operons (and conserved MbnT1, 2, or 3 genes are not present elsewhere in the genomes), raising the possibility that Group V Mbns are not reimported and may have a function other than copper acquisition.Functional studies of Ms. trichosporium OB3b MbnT1 are complicated by the presence of 45 TBDTs within the genome. qRT-PCR studies reveal significant downregulation of the in-operon MbnT1 in response to copper. Two close homologues of MbnT1, MettrDRAFT_1229 and MettrDRAFT_1241, also exhibit lower transcript levels in the presence of copper, but are downregulated to a lesser extent and with regulatory patterns that differ to some extent from the main Mbn operon.40 It may be that these homologues function in the uptake of non-native Mbns, and thus would also be responsive to copper. The function of the in-operon MbnT1 from Ms. trichosporium OB3b has been further probed experimentally. In an initial study, a gene disruption mutant of MbnT (ΔmbnT) was generated.47 This mutant was found to be capable of Mbn production and secretion, but failed to take up copper provided as CuMbn. Both wildtype (wt) and ΔmbnT cells accumulated copper when supplied with increasing concentrations of soluble copper, consistent with previous work establishing distinct uptake pathways for soluble copper and CuMbn.16 More recently, isotopic labeling experiments on a Ms. trichosporium OB3b ΔmbnT strain generated by a different method also indicate a disruption in CuMbn uptake. In this study, wt and ΔmbnT cells provided with 65CuMbn show clear differences in 63Cu![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 65Cu ratio, indicating that CuMbn uptake is impaired in the mutant.40 Finally, heterologous expression of Ms. trichosporium OB3b MbnT1 confers upon E. coli the ability to take up 65CuMbn.40 These combined data identify MbnT1 as the CuMbn importer.
65Cu ratio, indicating that CuMbn uptake is impaired in the mutant.40 Finally, heterologous expression of Ms. trichosporium OB3b MbnT1 confers upon E. coli the ability to take up 65CuMbn.40 These combined data identify MbnT1 as the CuMbn importer.
The function of MbnT has also been assessed in vitro. Using surface plasmon resonance (SPR), a specific interaction between purified Ms. trichosporium OB3b MbnT1 and Ms. trichosporium OB3b CuMbn has been detected with a measured Kd value of ∼6 μM. The Group II MbnT2 from Mc. rosea SV97 also interacts with Ms. trichosporium OB3b CuMbn, but with an affinity approximately 3-fold less than that measured for Ms. trichosporium OB3b CuMbn binding to its cognate MbnT. The Ms. trichosporium OB3b and Mc. rosea SV97 Mbns contain the same ligands that coordinate the copper (N-heterocycles and thioamide groups), but differ in other structural elements (Fig. 1A and C). Although siderophore TBDTs rarely recognize non-cognate substrates, these observations are reminiscent of the Pseudomonas aeruginosa ATCC 15692 pyoverdine receptor FpvA, which recognizes other pyoverdines by the N-terminal residue and metal coordination elements.48
A potential role for the periplasmic binding protein MbnE
The Group IIa Mbn operons encode a periplasmic binding protein (PBP) (Fig. 2A), MbnE, close homologues of which are found elsewhere in the genomes of other Mbn-producing methanotrophs.40 PBPs are periplasmic in Gram-negative bacteria and membrane-bound (and called solute binding proteins, SBPs) in Gram-positive bacteria, and have attracted attention recently because of their potential use in elucidating new metabolic pathways.49 These proteins often work in concert with ATP-binding cassette (ABC) transporters to import ligands across the inner membrane, although genes encoding ABC transporters are not generally found adjacent to mbnEs and the extent of explicitly cytoplasmic Mbn uptake remains unclear.50 The MbnE sequences belong to SBP Family 5,51 and in a SSN, cluster separately from all characterized PBPs40 except the microcin C7 transporter YejA and the oligopeptide transporters OppA and AppA.52In support of a role for Ms. trichosporium OB3b MbnE in Mbn transport, qRT-PCR data show significant downregulation of mbnE together with the main mbn operon in the presence of copper.35 The closest MbnE homologues in the genome, MettrDRAFT_1622 and MettrDRAFT_3640, are not similarly downregulated.40 In addition, purified MbnE proteins from three species, Ms. trichosporium OB3b (Group I), Mc. hirsuta CSC1 (Group IIa), and Mc. parvus OBBP (Group IIb) were shown by SPR to interact specifically with their cognate CuMbns and exhibit binding affinities of ∼6–12 μM.40 However, unlike MbnT, these MbnEs do not bind non-cognate CuMbns.
Efforts to understand the molecular basis for Mbn binding by MbnEs have thus far yielded a 1.9 Å resolution crystal structure of the Mc. parvus OBBP MbnE in its apo form.40 The structure revealed two major domains, between which is a cavity measuring ∼15 × 30 Å. Docking models generated with the crystal structures of CuMbns from Ms. trichosporium OB3b and Mc. sp. M indicate that this cavity is sufficiently spacious to accommodate both types of CuMbn. Mc. parvus OBBP MbnE is structurally similar to the oligopeptide binding SBP AppA from Bacillus subtilis, which was crystallized with a nonapeptide bound to its cavity,53 and a comparison of the apo MbnE structure with that of substrate-bound AppA suggests structural rearrangements that might occur upon binding of CuMbn.40
Export by MbnM
Most Mbn operons also include a gene encoding a multidrug and toxic compound extrusion (MATE) efflux pump, MbnM.37 MATE proteins are inner membrane exporters that couple H+ or Na+ transport to extrusion of cationic, xenobiotic substrates, including multiple antibiotics.54–56 Export of a natively produced natural product has not been reported for a MATE transporter, but very few of these proteins have been studied, and their conservation even in the divergent Group V Mbn operons is suggestive of a role in Mbn export.Release of copper from Mbn
While aspects of CuMbn uptake have been elucidated, it is not known how copper is released from Mbn once inside the cell. Some siderophores are degraded,57 but it is perhaps more likely that Mbn is recycled, as are similarly complex siderophores such as pyoverdine,58 carboxymycobactin,59 and ferrichrome.60 One way to release the metal ion without Mbn degradation would be oxidation of CuI to CuII,18,23 similar to the alteration of Fe oxidation state for iron release, which is observed in some siderophores.61 The calculated binding affinity of Mbn for CuII is significantly lower than that measured for CuI, and the identity of at least one N-terminal group influences the binding affinity of some Mbns for CuII.23 The high binding affinity is reflective of reduction potentials (Em) that range from 483 mV to 750 mV (against the normal hydrogen electrode). Also notable is the observation that the presence of C-terminal residues in Mbn can impact the Em values.23 Therefore, it is conceivable that oxidation of the CuI could facilitate release23 and transfer to a protein or ligand with greater CuII affinity. Possible transfer candidates include the periplasmic proteins CopC62,63 and DUF461,64 which are coregulated with pMMO in Ms. trichosporium OB3b.35 It is not obvious how CuMbn might be oxidized, but most Mbn operons identified to date include genes encoding a periplasmic diheme cytochrome c peroxidase, MbnH, and its periplasmic partner protein, MbnP (Fig. 2A), which has no domains of known function and is not homologous to any characterized proteins. Both genes are commonly associated with mbnT genes (whether in the context of an Mbn operon or not), potentially supporting functions related to Mbn internalization and/or processing. Some methanotrophs that lack Mbn operons also have copper binding proteins (MopE, CorA),65,66 paired with a diheme cytochrome c peroxidases, but there is no clear relation between these proteins and MbnP (beyond the presence of highly conserved tryptophans and genomic proximity to MauG homologues), and MopE is secreted from the cell, while there is no evidence of this for MbnP.37 Nevertheless, a role for MbnH and MbnP in electron transfer and release of copper from Mbn is conceivable.It is also not known how copper released from Mbn traverses the inner membrane. One possibility is that ABC transporters are involved in CuMbn transport, perhaps with the involvement of MbnE, although no obvious candidates have been identified.35,37 Another potential player has been identified from studies of the mutant strain Ms. trichosporium OB3b PP358, which constitutively produces sMMO and Mbn, even at high concentrations of copper, and does not accumulate intracellular copper.67,68 These phenotypes are supported by recent qRT-PCR data showing that the PP358 mmo and mbn operons are not downregulated in response to copper.35 The genome of PP358 has been sequenced, and contains a frameshift deletion in the copD gene, located adjacent to the pmo operon in α-proteobacterial methanotrophs (Fig. 3A), and also mildly upregulated by copper in wt Ms. trichosporium OB3b.35 CopDs are integral inner membrane proteins postulated to mediate cytoplasmic copper uptake in bacteria.69–72 Unlike most CopD sequences, which include 8 transmembrane helices, Ms. trichosporium OB3b CopD is predicted to have 10 transmembrane helices and a C-terminal periplasmic domain. In PP358, the deletion in copD eliminates the last transmembrane helix and the periplasmic domain. If CopD is a copper importer, whether it acquires copper directly from Mbn-related periplasmic machinery or from the broader pool of periplasmic copper, its dysfunction would affect the cytoplasmic machinery for copper sensing, potentially explaining the disruption of the copper switch. Moreover, copper accumulation in the periplasm could lead to increased expression of efflux systems, which would account for the lower copper content of PP358 cells.68
4. Mbn and the copper switch
The MmoD hypothesis
Mbn itself has been ruled out as the sole mediator of the copper switch because sMMO and pMMO expression in response to copper are similar for wt and ΔmbnA Ms. trichosporium OB3b.34 This is not surprising since most methanotrophs that can produce both sMMO and pMMO do not possess Mbn operons (Fig. 3B), and yet robust evidence for the copper switch in species such as Mcc. capsulatus (Bath) has existed for more than thirty years.73 Instead, Mbn has recently been proposed to work in concert with a protein called MmoD to modulate the copper switch.34 This hypothesis is based on the observation that deletion of almost the entire mmo operon through the first three residues of mmoC (ΔmmoXYBZDC1–3, or the “SMDM mutant”74) alters copper-responsive gene regulation. In particular, MbnA is not downregulated with increasing copper concentrations, and pMMO expression, as monitored by the pmoA gene, is actually upregulated under copper starvation conditions. Based on these findings, the MmoD protein, the only component of the deleted sMMO operon without an assigned functional role, was suggested to play an essential role in the copper switch.34 MmoD is not essential for sMMO-mediated methane oxidation, although it has it been shown to interact with the MMOH complex and to inhibit its activity in vitro.75In the MmoD model, MmoD represses expression of pMMO at low copper concentrations and also upregulates the sMMO and Mbn operons. When more copper is available, MmoD is proposed to bind copper and no longer affect expression of the three operons. Apo and CuMbn are suggested to participate in this model as well, although the mode of regulation (binding DNA, mRNA, or other) has not been elaborated.34 On the basis of BindN sequence analysis of the Ms. trichosporium OB3b derivative strain used in this study (which must be distinct from the type strain based on the low (82%) amino acid sequence identity between its MmoD and that of the type strain), DNA binding regions were suggested,9,34 but the relevant residues are poorly conserved in MmoD sequences from other methanotrophs. In addition, qRT-PCR data indicate the MmoD does not exhibit the same copper-responsive expression as other regulatory genes (vide infra); it is coregulated with the rest of the enzymatic sMMO genes,35 consistent with the previously reported protein–protein interaction and contrary to unpublished data mentioned in a recent review which claim that mmoD is constitutively expressed.9 Moreover, purified MmoD does not bind copper and does not bind to a heparin column, suggesting that it is not a DNA binding protein.35
The MbnIR hypothesis
Another model for regulation involves the MbnI and MbnR proteins encoded by the Group I Mbn operons (Fig. 2A and 4).37 In the related FecIRA system, binding of the ferric citrate substrate to the outer membrane TBDT FecA triggers interaction across the periplasm with the anti-sigma factor FecR via the N-terminal extension, and FecR in turn activates the ECF sigma factor FecI to regulate iron acquisition.44–46 By analogy, CuMbn binding to MbnT could transmit a signal to MbnI via MbnR, regulating some portion of the genes from the mbn operon.37 In support of this model, the genes encoding MbnI and MbnR, along with genes believed to play a role in sMMO regulation (mmoR, mmoG)76 are downregulated within 15 minutes of copper addition to Ms. trichosporium OB3b cells, whereas expression levels for the rest of the Mbn and sMMO operons decrease over a much longer time frame. This finding suggests that each operon has an internal copper-responsive regulator, downregulation of which results in a slower decline in expression for the core operons. Moreover, MbnI binds to a heparin column, consistent with it being an ECF sigma factor.35However, mbnI and mbnR genes are present only in Group I operons or adjacent to genes encoding MbnT1s, suggesting that the other groups of Mbns are regulated differently. It is possible that regulation of Mbn biosynthesis in non-Group I operons occur via a mechanism similar to that of the siderophore pyochelin, which binds directly to a transcriptional regulator after it is taken up into the cytoplasm, and this then regulates subsequent biosynthesis and transport.77–79 A recently revised version of the MmoD hypothesis includes MbnI as a regulator of both the Mbn and sMMO operons, but still includes MmoD as a regulator of Mbn and pMMO expression.9 Given the aforementioned issues with MmoD and the limited presence of MbnIR in Mbn operons, as well as the existence of the copper switch in methanotrophs that lack Mbn operons, it is likely that regulation is significantly more complicated and involves other components. In particular, the regulator that represses mmoRG and mbnIR in response to copper has yet to be identified.
5. Biosynthesis of Mbn
The core operon: genes encoding MbnA, MbnB, and MbnC
MbnAs are short (23–35aa) ribosomally produced peptides. Like most RiPP precursor peptides, they contain an N-terminal leader peptide (not observed in the final natural product) and a C-terminal core peptide that is the site of any post-translational modifications (Fig. 2C). Group II MbnAs are something of an exception, since some appear to be at the C-terminus of an ORF encoding an ECF sigma factor (not closely related to the predicted Mbn regulator MbnI discussed above). It is not known whether this entire ORF is indeed transcribed and translated, as is thought to be the case in operons where a nitrile hydratase/Nif11 sequence replaces a smaller leader peptide for the biosynthesis of certain thiazole/oxazole modified microcins,80 or whether the MbnA sequence is sometimes or always the only transcript. Regardless, MbnA leader peptide sequences exhibit a certain degree of conservation, with positively charged lysine and arginine residues surrounding a hydrophobic patch present prior to the core peptide. Core peptides show comparatively little conservation, except for the cysteines that are the sites of oxazolone/thioamide formation and the two residues following these cysteines. Potential modification targeting motifs can be derived from these sequences: cysteines followed by a small hydrophobic residue (most commonly glycine or alanine) and then a slightly larger, sometimes more hydrophilic residue (most commonly alanine or serine). Cysteines not belonging to a modification motif do not get modified, as observed for the Ms. trichosporium OB3b and Ms. sp. LW4 Mbns, in which cysteines with the wrong neighbors (CYP and CM or CNP and C respectively) instead form an intramolecular disulfide bond.In all genomes with identified Mbn operons, two additional genes are present encoding the proteins MbnB and MbnC, and in Groups I–IV, they immediately follow the gene encoding MbnA (Fig. 2A).37 MbnB proteins contain no domains or motifs with known function, and lack the identifiable leader peptide binding regions observed for some other RiPP families,81 but on the basis of sequence homology and structural prediction, belong to the TIM barrel family.82,83 MbnB proteins are a part of the larger DUF692 family, which is similarly uncharacterized; their closest characterized relatives are xylose isomerases84 and endonuclease IV proteins.85 Although MbnB in Ms. trichosporium OB3b was originally annotated as two ORFs, this appears to have been the result of a nonsense mutation not seen in other Ms. trichosporium OB3b lineages35 or, indeed, other Mbn operons. The gene encoding MbnC is always found adjacent to that encoding MbnB. Like MbnB, MbnC does not contain any domains or motifs for which there are known functions, but unlike MbnB, it does not belong to any identifiable family or superfamily. Because these two genes are present in all complete Mbn operons identified to date, it has been hypothesized that the two proteins are responsible for the biosynthesis of the heterocycles and thioamides observed in all characterized Mbns.37
Whether and how MbnB and MbnC generate the Mbn heterocycles remains a major question. Oxazolones and pyrazinediones are exceedingly uncommon post-translational modifications, and no existing RiPP pathways are known to produce these moieties. However, other heterocycles, including (methyl)oxazoles/thiazoles and (methyl)oxazolines/thiazolines are widespread among RiPP subfamilies, including cyanobactins, microcins, streptolysins, and thiopeptides.86 In these systems, an (N)TP-dependent cyclodehydratase complex is responsible for the initial cyclization of cysteine, serine, or threonine, followed in some cases by oxidation via an FMN-dependent dehydratase.87 It has even been hypothesized that in thioviridamide, a thioamide is produced via homologues of the cyclodehydratase enzymes.88 However, neither MbnB nor MbnC has a recognizable nucleotide-binding motif, and cyclodehydratase homologues are not present in the genomes of many Mbn-producing organisms. A recently proposed mechanism for oxazolone/thioamide biosynthesis involves disulfide formation between a target cysteine in MbnA and a cysteine from a biosynthetic protein,9 but neither MbnB nor MbnC contains a strictly conserved cysteine residue, and it is unclear how MbnB's classification as a DUF692 protein related to xylose isomerases and endonuclease IV proteins fits into this mechanism; similarly, the biochemical relevance of the FAD-dependent monooxygenase MbnF, which is most closely related to RebC, an enzyme involved in rebeccamycin biosynthesis,89 to the proposed pyrazinedione generation pathway is unclear.
Additional biosynthetic genes
Besides MbnB and MbnC, Mbn operons contain a range of other genes encoding proteins with likely biosynthetic functions (Fig. 2A and 3).37,40 A gene encoding an aminotransferase, MbnN, is found in two Group I operons (Ms. trichosporium OB3b, Ms. sp. LW4), and aminotransferases are also present in the four Group IV Mbn operons. It is likely that MbnN catalyzes the insertion of the “N-terminal” carbonyl group in Ms. trichosporium OB3b Mbn, and plays a similar role in other species that have its gene. In either case, and particularly for the hisC-like aminotransferase90 MbnN, the substrate is large and unusual. Group IIa operons contain a gene encoding a sulfotransferase, MbnS, which likely is responsible for the threonine sulfonation observed in the structurally characterized Mc. Mbns;23,28 threonine sulfonation has been observed in some proteins,91 but not previously in natural products. Another putative biosynthetic protein encoded by many, but not all, Group I and II operons is MbnF, which is annotated as a member of the FAD-dependent monooxygenase superfamily.92 Its role in Mbn biosynthesis is unclear, but it could catalyze a hydroxylation reaction as part of the formation of the pyrazinedione present in some Mc. Mbns23 or perform some broader transformation from oxazolone to pyrazinedione.More mysterious are MbnD, a predicted dioxygenase whose gene immediately follows mbnF in Group IIb operons, and MbnX, a DUF692 homologue not belonging to the MbnB subfamily; its gene is found between mbnA and mbnB in both Gram-negative and Gram-positive Group V Mbn operons. One gene is conspicuously absent from Mbn operons: most RiPP families have a protease present within their operons that is responsible for leader peptide cleavage, or in some cases proteolysis of the leader peptide is coupled with export. Mbn operons do not encode the former, and the latter is unlikely due to the need for N-terminal modification of the core peptide in all Mbns characterized to date, which would require cytoplasmic leader peptide loss. Thus, it is unclear whether leader peptide removal is carried out by one of the uncharacterized genes in the Mbn operon or whether it is the result of some other unidentified process. Finally, the diheme cytochrome c peroxidase MbnH and its partner protein MbnP are present in Group I–IV operons; while MbnH has been suggested to play a role in heterocycle biosynthesis,9 homologues of these two proteins are also found in methanotrophs that lack Mbn operons and appear to be most commonly associated with genes encoding MbnTs,37 which may suggest a more general role in copper acquisition (vide supra).
Operon-based Mbn structure prediction
The combination of precursor peptide sequence and the identities of these putative biosynthetic enzymes can be used to predict structures of uncharacterized Mbns (Fig. 5). For example, the Group I operons from Ms. sp. LW4 and Ms. sp. LW3 differ in the presence of MbnN, the putative aminotransferase, in Ms. sp. LW4, and MbnF, the putative FAD-dependent monooxygenase, in Ms. sp. LW3. Although the peptide backbone is different, Ms. sp. LW4 Mbn was predicted to have a similar set of post-translational modifications to Ms. trichosporium OB3b Mbn, whereas the presence of an MbnF gene in the Ms. sp. LW3 operon suggests that the N-terminal oxazolone may undergo further modification, potentially to produce a pyrazinedione. As proof-of-concept, the recently characterized Ms. sp. LW4 structure matches its predicted structure exactly (Fig. 1F and 5A).29 Notably, Ms. sp LW3 also has a Group IIb Mbn operon. Characterization of both its Mbns could help understand what reaction(s) MbnF as well as the dioxygenase MbnD catalyze, and would confirm or refute our current understanding of the modification targeting motif: the second cysteine in Group IIb MbnA core peptides is followed by a tryptophan and a glycine, which do not conform to the motif, and thus is not currently predicted to be subject to coupled oxazolone and thioamide formation. Mbn structure can also be used to predict operons in the absence of genomic information. For example, given the structure of Mc. sp. M Mbn (Fig. 1E) and the core peptide sequences of related species,23 the core peptide is predicted to have the sequence RCASTCAMTNG, and the operon should contain at least mbnABCMFS.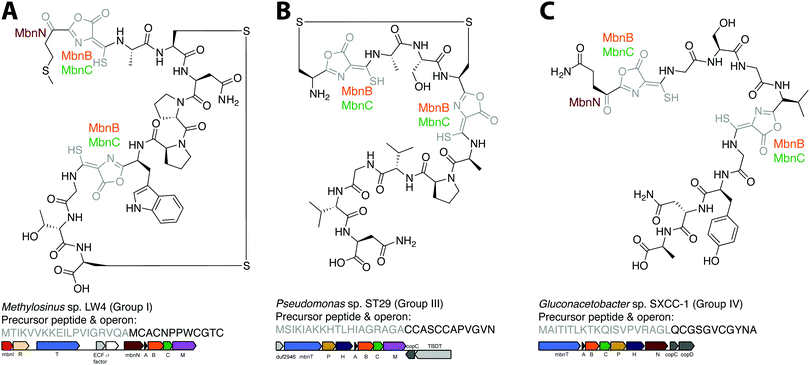 | ||
| Fig. 5 Operon or structure predictions for Mbns from three different groups. (A) Ms. sp. LW4 Mbn. The structure predicted on the basis of the operon content and precursor peptide sequence has recently been confirmed.29 (B) Pseudomonas sp. ST29 Mbn. Structure predicted on the basis of the operon content and precursor peptide sequence. (C) Gluconacetobacter sp. SXCC-1 Mbn. Structure predicted on the basis of the operon content and precursor peptide sequence. | ||
Similar predictions can be made for non-methanotrophic Mbns. Group III Mbn operons encode no predicted biosynthesis-related proteins other than MbnA, MbnB, and MbnC. In addition, like Group I MbnAs, Group III precursor peptides contain four cysteines, of which two belong to typical Mbn modification motifs (Fig. 2C). These Mbns are thus likely to contain two oxazolone/thioamide pairs, with two unaltered cysteine side chains forming an intramolecular disulfide bond; no additional post-translational modifications are predicted in this group (Fig. 5B). Group IV Mbns, generated from MbnA sequences with only two cysteines which are both predicted to be post-translationally modified to form oxazolone/thioamide pairs, should lack the disulfide bond but may be modified at the N-terminus (Fig. 5C), or possibly on the primary amine present in the sidechain of the conserved N-terminal glutamine by the bioA-like aminotransferase93,94 present in the operon. Group V Mbn operons and MbnA sequences are significantly divergent, and the absence of a second cysteine modification motif as well as the presence of a gene encoding the uncharacterized DUF692 protein MbnX between the MbnA and MbnB genes suggests that the resulting Mbns are just as divergent. It may be that these compounds are not copper chelators, but are evolutionarily related natural products that now have a completely different function.
6. Conclusions
A combination of genome mining, bioinformatic, genetic, and biochemical studies has led to major advances in our understanding of Mbn. Specific transporters have been identified and predicted, including MbnT and MbnM, and the first direct protein-Mbn interactions have been reported for MbnT and MbnE. The question of how copper is released from Mbn remains open, as does the possible involvement of Mbn and the operon proteins MbnI and MbnR in the methanotroph copper switch. Predictions based on MbnA precursor peptide sequence and operon content provide a roadmap to understanding Mbn biosynthesis and to characterizing additional Mbns. It is likely that further work in this area will lead to the identification of novel enzymes and biosynthetic mechanisms involved in the production of Mbn's unusual set of post-translational modifications. Finally, the roles that these natural products play in non-methanotrophic bacteria remain completely unexplored. While some methanotrophs likely evolved Mbns to fulfill their unusually high copper requirement, there are no obvious patterns in environment or metabolism identifiable for the non-methanotrophic bacteria with Mbn operons. Thus, whether there is a link between Mbn production and specific ecosystems remains unresolved.The discovery of new Mbns and Mbn-like molecules may have implications far beyond the microbial world. Mbns have been pursued recently as therapeutics for human diseases of copper metabolism,95,96 with a recent study reporting Mbn-mediated reversion of acute liver failure in rats with advanced Wilson disease.97,98 Additionally, Mbn is one of three bacterial-derived natural products that possess thioamides,99 which are known to play a role in antimicrobial activity in at least one natural product.100 A detailed understanding of the Mbn biosynthetic pathway would open up the possibility of generating designer Mbns with desired metal-binding and antimicrobial properties and even cell-specific targeting sequences.
Acknowledgements
This work was supported by NIH grants GM118035 (A. C. R.) and F32GM110934 (L. M. K. D.), a Burroughs Wellcome Fund PDEP award (L. M. K. D.), and AHA grant 14PRE20460104 (G. E. K.).References
- R. A. Festa and D. J. Thiele, Copper: an essential metal in biology, Curr. Biol., 2011, 21, 877–883 CrossRef PubMed.
- S. Tottey, K. J. Waldron, S. J. Firbank, B. Reale, C. Bessant, K. Sato, T. R. Cheek, J. Gray, M. J. Banfield, C. Dennison and N. J. Robinson, Protein-folding location can regulate manganese-binding versus copper- or zinc-binding, Nature, 2008, 455, 1138–1142 CrossRef CAS PubMed.
- L. Macomber and J. A. Imlay, The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 8344–8349 CrossRef CAS PubMed.
- R. S. Hanson and T. E. Hanson, Methanotrophic bacteria, Microbiol. Rev., 1996, 60, 439–471 CAS.
- P. J. Strong, S. Xie and W. P. Clarke, Methane as a resource: can the methanotrophs add value?, Environ. Sci. Technol., 2015, 49, 4001–4018 CrossRef CAS PubMed.
- T. J. Lawton and A. C. Rosenzweig, Methane-oxidizing enzymes: an upstream problem in biological gas-to-liquids conversion, J. Am. Chem. Soc., 2016, 138, 9327–9340 CrossRef PubMed.
- S. Sirajuddin and A. C. Rosenzweig, Enzymatic oxidation of methane, Biochemistry, 2015, 54, 2283–2294 CrossRef CAS PubMed.
- M. H. Sazinsky and S. J. Lippard, Methane monooxygenase: functionalizing methane at iron and copper, Met. Ions Life Sci., 2015, 15, 205–256 Search PubMed.
- A. A. DiSpirito, J. D. Semrau, J. C. Murrell, W. H. Gallagher, C. Dennison and S. Vuilleumier, Methanobactin and the link between copper and bacterial methane oxidation, Microbiol. Mol. Biol. Rev., 2016, 80, 387–409 CrossRef PubMed.
- P. G. Arnison, M. J. Bibb, G. Bierbaum, A. A. Bowers, T. S. Bugni, G. Bulaj, J. A. Camarero, D. J. Campopiano, G. L. Challis, J. Clardy, P. D. Cotter, D. J. Craik, M. Dawson, E. Dittmann, S. Donadio, P. C. Dorrestein, K. D. Entian, M. A. Fischbach, J. S. Garavelli, U. Goransson, C. W. Gruber, D. H. Haft, T. K. Hemscheidt, C. Hertweck, C. Hill, A. R. Horswill, M. Jaspars, W. L. Kelly, J. P. Klinman, O. P. Kuipers, A. J. Link, W. Liu, M. A. Marahiel, D. A. Mitchell, G. N. Moll, B. S. Moore, R. Muller, S. K. Nair, I. F. Nes, G. E. Norris, B. M. Olivera, H. Onaka, M. L. Patchett, J. Piel, M. J. Reaney, S. Rebuffat, R. P. Ross, H. G. Sahl, E. W. Schmidt, M. E. Selsted, K. Severinov, B. Shen, K. Sivonen, L. Smith, T. Stein, R. D. Sussmuth, J. R. Tagg, G. L. Tang, A. W. Truman, J. C. Vederas, C. T. Walsh, J. D. Walton, S. C. Wenzel, J. M. Willey and W. A. van der Donk, Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature, Nat. Prod. Rep., 2013, 30, 108–160 RSC.
- H. J. Kim, D. W. Graham, A. A. DiSpirito, M. A. Alterman, N. Galeva, C. K. Larive, D. Asunskis and P. M. A. Sherwood, Methanobactin, a copper-acquisition compound from methane oxidizing bacteria, Science, 2004, 305, 1612–1615 CrossRef CAS PubMed.
- A. A. DiSpirito, J. A. Zahn, D. W. Graham, H. J. Kim and C. K. Larive, T. S. Derrick, C. D. Cox and A. B. Taylor, Copper-binding compounds from Methylosinus trichosporium OB3b, J. Bacteriol., 1998, 180, 3606–3613 CAS.
- C. M. Téllez, K. P. Gaus, D. W. Graham, R. G. Arnold and R. Z. Guzman, Isolation of copper biochelates from Methylosinus trichosporium OB3b and soluble methane monooxygenase mutants, Appl. Environ. Microbiol., 1998, 64, 1115–1122 Search PubMed.
- M. L. Pesch, M. Hoffmann, I. Christl, S. M. Kraemer and R. Kretzschmar, Competitive ligand exchange between Cu-humic acid complexes and methanobactin, Geobiology, 2013, 11, 44–54 CrossRef CAS PubMed.
- E. Kulczycki and J. A. Roberts, Methanobactin-promoted dissolution of Cu-substituted borosilicate glass, Geobiology, 2007, 5, 251–263 CrossRef CAS.
- R. Balasubramanian, G. E. Kenney and A. C. Rosenzweig, Dual pathways for copper uptake by methanotrophic bacteria, J. Biol. Chem., 2011, 286, 37313–37319 CrossRef CAS PubMed.
- C. W. Knapp, D. A. Fowle, E. Kulczycki, J. A. Roberts and D. W. Graham, Methane monooxygenase gene expression mediated by methanobactin in the presence of mineral copper sources, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 12040–12045 CrossRef CAS PubMed.
- A. El Ghazouani, A. Basle, S. J. Firbank, C. W. Knapp, J. Gray, D. W. Graham and C. Dennison, Copper-binding properties and structures of methanobactins from Methylosinus trichosporium OB3b, Inorg. Chem., 2011, 50, 1378–1391 CrossRef CAS PubMed.
- A. S. Hakemian, C. E. Tinberg, K. C. Kondapalli, J. Telser, B. M. Hoffman, T. L. Stemmler and A. C. Rosenzweig, The copper chelator methanobactin from Methylosinus trichosporium OB3b binds copper(I), J. Am. Chem. Soc., 2005, 127, 17142–17143 CrossRef CAS PubMed.
- D. W. Choi, C. J. Zea, Y. S. Do, J. D. Semrau, W. E. Antholine, M. S. Hargrove, N. L. Pohl, E. S. Boyd, G. G. Geesey, S. C. Hartsel, P. H. Shafe, M. T. McEllistrem, C. J. Kisting, D. Campbell, V. Rao, A. M. de la Mora and A. A. DiSpirito, Spectral, kinetic, and thermodynamic properties of Cu(I) and Cu(II) binding by methanobactin from Methylosinus trichosporium OB3b, Biochemistry, 2006, 45, 1442–1453 CrossRef CAS PubMed.
- D. W. Choi, Y. S. Do, C. J. Zea, M. T. McEllistrem, S.-W. Lee, J. D. Semrau, N. L. Pohl, C. J. Kisting, L. L. Scardino, S. C. Hartsel, E. S. Boyd, G. G. Geesey, T. P. Riedel, P. H. Shafe, K. A. Kranski, J. R. Tritsch, W. E. Antholine and A. A. DiSpirito, Spectral and thermodynamic properties of Ag(I), Au(III), Cd(II), Co(II), Fe(III), Hg(II), Mn(II), Ni(II), Pb(II), U(IV), and Zn(II) binding by methanobactin from Methylosinus trichosporium OB3b, J. Inorg. Biochem., 2006, 100, 2150–2161 CrossRef CAS PubMed.
- M. L. Pesch, I. Christl, M. Hoffmann, S. M. Kraemer and R. Kretzschmar, Copper complexation of methanobactin isolated from Methylosinus trichosporium OB3b: pH-dependent speciation and modeling, J. Inorg. Biochem., 2012, 116, 55–62 CrossRef CAS PubMed.
- A. El Ghazouani, A. Basle, J. Gray, D. W. Graham, S. J. Firbank and C. Dennison, Variations in methanobactin structure influences copper utilization by methane-oxidizing bacteria, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 8400–8404 CrossRef CAS PubMed.
- J.-Y. Xin, K. Lin, T. Wang and C.-G. Xia, Methanobactin-mediated synthesis of gold nanoparticles supported over Al2O3 toward efficient catalyst for glucose oxidation, Int. J. Mol. Sci., 2014, 15, 21603–21620 CrossRef CAS PubMed.
- B. Kalidass, M. F. Ul-Haque, B. S. Baral, A. A. DiSpirito and J. D. Semrau, Competition between metals for binding to methanobactin enables expression of soluble methane monooxygenase in the presence of copper, Appl. Environ. Microbiol., 2015, 81, 1024–1031 CrossRef PubMed.
- B. S. Baral, N. L. Bandow, A. Vorobev, B. C. Freemeier, B. H. Bergman, T. J. Herdendorf, N. Fuentes, L. Ellias, E. Turpin, J. D. Semrau and A. A. DiSpirito, Mercury binding by methanobactin from Methylocystis strain SB2, J. Inorg. Biochem., 2014, 141, 161–169 CrossRef CAS PubMed.
- A. Vorobev, S. Jagadevan, B. S. Baral, A. A. Dispirito, B. C. Freemeier, B. H. Bergman, N. L. Bandow and J. D. Semrau, Detoxification of mercury by methanobactin from Methylosinus trichosporium OB3b, Appl. Environ. Microbiol., 2013, 79, 5918–5926 CrossRef CAS PubMed.
- B. D. Krentz, H. J. Mulheron, J. D. Semrau, A. A. DiSpirito, N. L. Bandow, D. H. Haft, S. Vuilleumier, J. C. Murrell, M. T. McEllistrem, S. C. Hartsel and W. H. Gallagher, A comparison of methanobactins from Methylosinus trichosporium OB3b and Methylocystis strain SB2 predicts methanobactins are synthesized from diverse peptide precursors modified to create a common core for binding and reducing copper ions, Biochemistry, 2010, 49, 10117–10130 CrossRef CAS PubMed.
- G. E. Kenney, A. W. Goering, M. O. Ross, C. J. DeHart, P. M. Thomas, B. M. Hoffman, N. L. Kelleher and A. C. Rosenzweig, Characterization of methanobactin from Methylosinus sp. LW4, J. Am. Chem. Soc., 2016, 138, 11124–11127 CrossRef CAS PubMed.
- N. L. Bandow, W. H. Gallagher, L. Behling, D. W. Choi, J. D. Semrau, S. C. Hartsel, V. S. Gilles and A. A. Dispirito, Isolation of methanobactin from the spent media of methane-oxidizing bacteria, Methods Enzymol., 2011, 495, 259–269 Search PubMed.
- R. Balasubramanian and A. C. Rosenzweig, Copper methanobactin: a molecule whose time has come, Curr. Opin. Chem. Biol., 2008, 12, 245–249 CrossRef CAS PubMed.
- H. J. Kim, N. Galeva, C. K. Larive, M. Alterman and D. W. Graham, Purification and physical-chemical properties of methanobactin: a chalkophore from Methylosinus trichosporium OB3b, Biochemistry, 2005, 44, 5140–5148 CrossRef CAS PubMed.
- L. Y. Stein, S. Yoon, J. D. Semrau, A. A. DiSpirito, A. Crombie, J. C. Murrell, S. Vuilleumier, M. G. Kalyuzhnaya, H. J. Op den Camp, F. Bringel, D. Bruce, J. F. Cheng, A. Copeland, L. Goodwin, S. Han, L. Hauser, M. S. Jetten, A. Lajus, M. L. Land, A. Lapidus, S. Lucas, C. Medigue, S. Pitluck, T. Woyke, A. Zeytun and M. G. Klotz, Genome sequence of the obligate methanotroph Methylosinus trichosporium strain OB3b, J. Bacteriol., 2010, 192, 6497–6498 CrossRef CAS PubMed.
- J. D. Semrau, S. Jagadevan, A. A. DiSpirito, A. Khalifa, J. Scanlan, B. H. Bergman, B. C. Freemeier, B. S. Baral, N. L. Bandow, A. Vorobev, D. H. Haft, S. Vuilleumier and J. C. Murrell, Methanobactin and MmoD work in concert to act as the ‘copper-switch’ in methanotrophs, Environ. Microbiol., 2013, 15, 3077–3086 CAS.
- G. E. Kenney, M. Sadek and A. C. Rosenzweig, Copper-responsive gene expression in the methanotroph Methylosinus trichosporium OB3b, Metallomics, 2016, 8, 931–940 RSC.
- G. E. Kenney and A. C. Rosenzweig, Chemistry and biology of the copper chelator methanobactin, ACS Chem. Biol., 2012, 7, 260–268 CrossRef CAS PubMed.
- G. E. Kenney and A. C. Rosenzweig, Genome mining for methanobactins, BMC Biol., 2013, 11, 17–35 CrossRef CAS PubMed.
- D. W. Choi, N. L. Bandow, M. T. McEllistrem, J. D. Semrau, W. E. Antholine, S. C. Hartsel, W. Gallagher, C. J. Zea, N. L. Pohl, J. A. Zahn and A. A. DiSpirito, Spectral and thermodynamic properties of methanobactin from gamma-proteobacterial methane oxidizing bacteria: A case for copper competition on a molecular level, J. Inorg. Biochem., 2010, 104, 1240–1247 CrossRef CAS PubMed.
- K. S. Chaturvedi, C. S. Hung, J. R. Crowley, A. E. Stapleton and J. P. Henderson, The siderophore yersiniabactin binds copper to protect pathogens during infection, Nat. Chem. Biol., 2012, 8, 731–736 CrossRef CAS PubMed.
- L. M. K. Dassama, G. E. Kenney, S. Y. Ro, E. L. Zielazinski and A. C. Rosenzweig, Methanobactin transport machinery, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 13027–13032 CrossRef CAS PubMed.
- N. Noinaj, M. Guillier, T. J. Barnard and S. K. Buchanan, TonB-dependent transporters: regulation, structure, and function, Annu. Rev. Microbiol., 2010, 64, 43–60 CrossRef CAS PubMed.
- I. J. Schalk, W. W. Yue and S. K. Buchanan, Recognition of iron-free siderophores by TonB-dependent iron transporters, Mol. Microbiol., 2004, 54, 14–22 CrossRef CAS PubMed.
- K. Schauer, D. A. Rodionov and H. de Reuse, New substrates for TonB-dependent transport: do we only see the ‘tip of the iceberg’?, Trends Biochem. Sci., 2008, 33, 330–338 CrossRef CAS PubMed.
- V. Braun, S. Mahren and M. Ogierman, Regulation of the Fecl-type ECF sigma factor by transmembrane signalling, Curr. Opin. Microbiol., 2003, 6, 173–180 CrossRef CAS PubMed.
- R. Koebnik, TonB-dependent trans-envelope signalling: the exception or the rule?, Trends Microbiol., 2005, 13, 343–347 CrossRef CAS PubMed.
- V. Braun and F. Endriss, Energy-coupled outer membrane transport proteins and regulatory proteins, BioMetals, 2007, 20, 219–231 CrossRef CAS PubMed.
- W. Gu, M. Farhan Ul Haque, B. S. Baral, E. A. Turpin, N. L. Bandow, E. Kremmer, A. Flatley, H. Zischka, A. A. DiSpirito and J. D. Semrau, A TonB-dependent transporter is responsible for methanobactin uptake by Methylosinus trichosporium OB3b, Appl. Environ. Microbiol., 2016, 82, 1917–1923 CrossRef CAS PubMed.
- J. Greenwald, M. Nader, H. Celia, C. Gruffaz, V. Geoffroy, J. M. Meyer, I. J. Schalk and F. Pattus, FpvA bound to non-cognate pyoverdines: molecular basis of siderophore recognition by an iron transporter, Mol. Microbiol., 2009, 72, 1246–1259 CrossRef CAS PubMed.
- M. W. Vetting, N. Al-Obaidi, S. Zhao, B. San Francisco, J. Kim, D. J. Wichelecki, J. T. Bouvier, J. O. Solbiati, H. Vu, X. Zhang, D. A. Rodionov, J. D. Love, B. S. Hillerich, R. D. Seidel, R. J. Quinn, A. L. Osterman, J. E. Cronan, M. P. Jacobson, J. A. Gerlt and S. C. Almo, Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes, Biochemistry, 2015, 54, 909–931 CrossRef CAS PubMed.
- K. J. Linton, Structure and function of ABC transporters, Physiology, 2007, 22, 122–130 CrossRef CAS PubMed.
- R. P. A. Berntsson, S. H. J. Smits, L. Schmitt, D.-J. Slotboom and B. Poolman, A structural classification of substrate-binding proteins, FEBS Lett., 2010, 584, 2606–2617 CrossRef CAS PubMed.
- K. Severinov and S. K. Nair, Microcin C: biosynthesis and mechanisms of bacterial resistance, Future Microbiol., 2012, 7, 281–289 CrossRef CAS PubMed.
- V. M. Levdikov, E. V. Blagova, J. A. Brannigan, L. Wright, A. A. Vagin and A. J. Wilkinson, The structure of the oligopeptide-binding protein, AppA, from Bacillus subtilis in complex with a nonapeptide, J. Mol. Biol., 2005, 345, 879–892 CrossRef CAS PubMed.
- H. Omote, M. Hiasa, T. Matsumoto, M. Otsuka and Y. Moriyama, The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations, Trends Pharmacol. Sci., 2006, 27, 587–593 CrossRef CAS PubMed.
- T. Kuroda and T. Tsuchiya, Multidrug efflux transporters in the MATE family, Biochim. Biophys. Acta, 2009, 1794, 763–768 CrossRef CAS PubMed.
- X. A. He, P. Szewczyk, A. Karyakin, M. Evin, W. X. Hong, Q. H. Zhang and G. Chang, Structure of a cation-bound multidrug and toxic compound extrusion transporter, Nature, 2010, 467, 991–994 CrossRef CAS PubMed.
- I. J. Schalk and L. Guillon, Fate of ferrisiderophores after import across bacterial outer membranes: different iron release strategies are observed in the cytoplasm or periplasm depending on the siderophore pathways, Amino Acids, 2013, 44, 1267–1277 CrossRef CAS PubMed.
- F. Imperi, F. Tiburzi and P. Visca, Molecular basis of pyoverdine siderophore recycling in Pseudomonas aeruginosa, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 20440–20445 CrossRef CAS PubMed.
- C. M. Jones, R. M. Wells, A. V. Madduri, M. B. Renfrow, C. Ratledge, D. B. Moody and M. Niederweis, Self-poisoning of Mycobacterium tuberculosis by interrupting siderophore recycling, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 1945–1950 CrossRef CAS PubMed.
- M. Hannauer, Y. Barda, G. L. Mislin, A. Shanzer and I. J. Schalk, The ferrichrome uptake pathway in Pseudomonas aeruginosa involves an iron release mechanism with acylation of the siderophore and recycling of the modified desferrichrome, J. Bacteriol., 2010, 192, 1212–1220 CrossRef CAS PubMed.
- M. Miethke, J. Hou and M. A. Marahiel, The siderophore-interacting protein YqjH acts as a ferric reductase in different iron assimilation pathways of Escherichia coli, Biochemistry, 2011, 50, 10951–10964 CrossRef CAS PubMed.
- C. J. Wijekoon, T. R. Young, A. G. Wedd and Z. Xiao, CopC protein from Pseudomonas fluorescens SBW25 features a conserved novel high-affinity Cu(II) binding site, Inorg. Chem., 2015, 54, 2950–2959 CrossRef CAS PubMed.
- T. J. Lawton, G. E. Kenney, J. D. Hurley and A. C. Rosenzweig, The CopC Family: structural and bioinformatic insights into a diverse group of periplasmic copper binding proteins, Biochemistry, 2016, 55, 2278–2290 CrossRef CAS PubMed.
- F. E. Jen, K. Y. Djoko, S. J. Bent, C. J. Day, A. G. McEwan and M. P. Jennings, A genetic screen reveals a periplasmic copper chaperone required for nitrite reductase activity in pathogenic Neisseria, FASEB J., 2015, 29, 3828–3838 CrossRef CAS PubMed.
- T. Ve, K. Mathisen, R. Helland, O. A. Karlsen, A. Fjellbirkeland, A. K. Røhr, K. K. Andersson, R. B. Pedersen, J. R. Lillehaug and H. B. Jensen, The Methylococcus capsulatus (Bath) secreted protein, MopE*, binds both reduced and oxidized copper, PLoS One, 2012, 7, e43146 CAS.
- K. A. Johnson, T. Ve, O. Larsen, R. B. Pedersen, J. R. Lillehaug, H. B. Jensen, R. Helland and O. A. Karlsen, CorA Is a copper repressible surface-associated copper(I)-binding protein produced in Methylomicrobium album BG8, PLoS One, 2014, 9, e87750 Search PubMed.
- P. A. Phelps, S. K. Agarwal, G. E. Speitel and G. Georgiou, Methylosinus trichosporium OB3b mutants having constitutive expression of soluble methane monooxygenase in the presence of high levels of copper, Appl. Environ. Microbiol., 1992, 58, 3701–3708 CAS.
- M. W. Fitch, D. W. Graham, R. G. Arnold, S. K. Agarwal, P. Phelps, G. E. Speitel, Jr. and G. Georgiou, Phenotypic characterization of copper-resistant mutants of Methylosinus trichosporium OB3b, Appl. Environ. Microbiol., 1993, 59, 2771–2776 CAS.
- S. Chillappagari, M. Miethke, H. Trip, O. P. Kuipers and M. A. Marahiel, Copper acquisition is mediated by YcnJ and regulated by YcnK and CsoR in Bacillus subtilis, J. Bacteriol., 2009, 191, 2362–2370 CrossRef CAS PubMed.
- F. Serventi, Z. A. Youard, V. Murset, S. Huwiler, D. Buhler, M. Richter, R. Luchsinger, H. M. Fischer, R. Brogioli, M. Niederer and H. Hennecke, Copper starvation-inducible protein for cytochrome oxidase biogenesis in Bradyrhizobium japonicum, J. Biol. Chem., 2012, 287, 38812–38823 CrossRef CAS PubMed.
- J. S. Cha and D. A. Cooksey, Copper hypersensitivity and uptake in Pseudomonas syringae containing cloned components of the copper resistance operon, Appl. Environ. Microbiol., 1993, 59, 1671–1674 CAS.
- X. X. Zhang and P. B. Rainey, Regulation of copper homeostasis in Pseudomonas fluorescens SBW25, Environ. Microbiol., 2008, 10, 3284–3294 CrossRef CAS PubMed.
- S. H. Stanley, S. D. Prior, D. J. Leak and H. Dalton, Copper stress underlies the fundamental change in intracellular location of methane monooxygenase in methane-oxidizing organisms: studies in batch and continuous cultures, Biotechnol. Lett., 1983, 5, 487–492 CrossRef CAS.
- E. Borodina, T. Nichol, M. G. Dumont, T. J. Smith and J. C. Murrell, Mutagenesis of the “leucine gate” to explore the basis of catalytic versatility in soluble methane monooxygenase, Appl. Environ. Microbiol., 2007, 73, 6460–6467 CrossRef CAS PubMed.
- M. Merkx and S. J. Lippard, Why OrfY? Characterization of MMOD, a long overlooked component of the soluble methane monooxygenase from Methylococcus capsulatus (Bath), J. Biol. Chem., 2002, 277, 5858–5865 CrossRef CAS PubMed.
- R. Csáki, L. Bodrossy, J. Klem, J. C. Murrell and K. L. Kovács, Genes involved in the copper-dependent regulation of soluble methane monooxygenase of Methylococcus capsulatus (Bath): cloning, sequencing and mutational analysis, Microbiology, 2003, 149, 1785–1795 CrossRef PubMed.
- L. Michel, A. Bachelard and C. Reimmann, Ferripyochelin uptake genes are involved in pyochelin-mediated signalling in Pseudomonas aeruginosa, Microbiology, 2007, 153, 1508–1518 CrossRef CAS PubMed.
- D. E. Heinrichs and K. Poole, PchR, a regulator of ferripyochelin receptor gene (fptA) expression in Pseudomonas aeruginosa, functions both as an activator and as a repressor, J. Bacteriol., 1996, 178, 2586–2592 CrossRef CAS PubMed.
- L. Michel, N. Gonzalez, S. Jagdeep, T. Nguyen-Ngoc and C. Reimmann, PchR-box recognition by the AraC-type regulator PchR of Pseudomonas aeruginosa requires the siderophore pyochelin as an effector, Mol. Microbiol., 2005, 58, 495–509 CrossRef CAS PubMed.
- D. H. Haft, M. K. Basu and D. A. Mitchell, Expansion of ribosomally produced natural products: a nitrile hydratase-and Nif11-related precursor family, BMC Biol., 2010, 8, 70 CrossRef PubMed.
- B. J. Burkhart, G. A. Hudson, K. L. Dunbar and D. A. Mitchell, A prevalent peptide-binding domain guides ribosomal natural product biosynthesis, Nat. Chem. Biol., 2015, 11, 564–570 CrossRef CAS PubMed.
- R. R. Copley and P. Bork, Homology among (betaalpha)8 barrels: implications for the evolution of metabolic pathways, J. Mol. Biol., 2000, 303, 627–640 CrossRef CAS PubMed.
- N. Nagano, C. A. Orengo and J. M. Thornton, One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions, J. Mol. Biol., 2002, 321, 741–765 CrossRef CAS PubMed.
- T. D. Fenn, D. Ringe and G. A. Petsko, Xylose isomerase in substrate and inhibitor michaelis states: atomic resolution studies of a metal-mediated hydride shift, Biochemistry, 2004, 43, 6464–6474 CrossRef CAS PubMed.
- I. Ivanov, J. A. Tainer and J. A. McCammon, Unraveling the three-metal-ion catalytic mechanism of the DNA repair enzyme endonuclease IV, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 1465–1470 CrossRef CAS PubMed.
- M. A. Ortega and W. A. van der Donk, New insights into the biosynthetic logic of ribosomally synthesized and post-translationally modified peptide natural products, Cell Chem. Biol., 2016, 23, 31–44 CrossRef CAS PubMed.
- C. L. Cox, J. R. Doroghazi and D. A. Mitchell, The genomic landscape of ribosomal peptides containing thiazole and oxazole heterocycles, BMC Genomics, 2015, 16, 16 CrossRef PubMed.
- M. Izawa, T. Kawasaki and Y. Hayakawa, Cloning and heterologous expression of the thioviridamide biosynthetic gene cluster from Streptomyces olivoviridis, Appl. Environ. Microbiol., 2013, 79, 7110–7113 CrossRef CAS PubMed.
- H. Onaka, S. Taniguchi, Y. Igarashi and T. Furumai, Characterization of the biosynthetic gene cluster of rebeccamycin from Lechevalieria aerocolonigenes ATCC 39243, Biosci., Biotechnol., Biochem., 2003, 67, 127–138 CrossRef CAS PubMed.
- V. Grisolia, M. S. Carlomagno, A. G. Nappo and C. B. Bruni, Cloning, structure, and expression of the Escherichia coli K-12 hisC gene, J. Bacteriol., 1985, 164, 1317–1323 CAS.
- K. F. Medzihradszky, Z. Darula, E. Perlson, M. Fainzilber, R. J. Chalkley, H. Ball, D. Greenbaum, M. Bogyo, D. R. Tyson, R. A. Bradshaw and A. L. Burlingame, O-sulfonation of serine and threonine: mass spectrometric detection and characterization of a new posttranslational modification in diverse proteins throughout the eukaryotes, Mol. Cell. Proteomics, 2004, 3, 429–440 CAS.
- I. R. Phillips and E. A. Shephard, Flavin-containing monooxygenases: mutations, disease and drug response, Trends Pharmacol. Sci., 2008, 29, 294–301 CrossRef CAS PubMed.
- S. W. Van Arsdell, J. B. Perkins, R. R. Yocum, L. Luan, C. L. Howitt, N. P. Chatterjee and J. G. Pero, Removing a bottleneck in the Bacillus subtilis biotin pathway: bioA utilizes lysine rather than S-adenosylmethionine as the amino donor in the KAPA-to-DAPA reaction, Biotechnol. Bioeng., 2005, 91, 75–83 CrossRef CAS PubMed.
- G. L. Stoner and M. A. Eisenberg, Purification and properties of 7,8-diaminopelargonic acid aminotransferase, J. Biol. Chem., 1975, 250, 4029–4036 CAS.
- K. H. Summer, J. Lichtmannegger, N. Bandow, D. W. Choi, A. A. DiSpirito and B. Michalke, The biogenic methanobactin is an effective chelator for copper in a rat model for Wilson disease, J. Trace Elem. Med. Biol., 2011, 25, 36–41 CAS.
- H. Zischka, J. Lichtmannegger, S. Schmitt, N. Jägemann, S. Schulz, D. Wartini, L. Jennen, C. Rust, N. Larochette, L. Galluzzi, V. Chajes, N. Bandow, V. S. Gilles, A. A. DiSpirito, I. Esposito, M. Goettlicher, K. H. Summer and G. Kroemer, Liver mitochondrial membrane crosslinking and destruction in a rat model of Wilson disease, J. Clin. Invest., 2011, 121, 1508–1518 Search PubMed.
- J. Lichtmannegger, C. Leitzinger, R. Wimmer, S. Schmitt, S. Schulz, Y. Kabiri, C. Eberhagen, T. Rieder, D. Janik, F. Neff, B. K. Straub, P. Schirmacher, A. A. DiSpirito, N. Bandow, B. S. Baral, A. Flatley, E. Kremmer, G. Denk, F. P. Reiter, S. Hohenester, F. Eckardt-Schupp, N. A. Dencher, J. Adamski, V. Sauer, C. Niemietz, H. H. J. Schmidt, U. Merle, D. N. Gotthardt, G. Kroemer, K. H. Weiss and H. Zischka, Methanobactin reverses acute liver failure in a rat model of Wilson disease, J. Clin. Invest., 2016, 126, 2721–2735 Search PubMed.
- S. G. Kaler, Microbial peptide de-coppers mitochondria: implications for Wilson disease, J. Clin. Invest., 2016, 126, 2412–2414 CrossRef PubMed.
- S. Banala and R. D. Süssmuth, Thioamides in nature: in search of secondary metabolites in anaerobic microorganisms, ChemBioChem, 2010, 11, 1335–1337 CrossRef CAS PubMed.
- F. Kloss, A. I. Chiriac and C. Hertweck, Mapping the modular closthioamide architecture reveals crucial motifs of polythioamide antibiotics, Chemistry, 2014, 17, 15451–15458 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6mt00208k |
| ‡ These authors contributed equally to this manuscript. |
| This journal is © The Royal Society of Chemistry 2017 |