Unambiguous identification and discovery of bacterial siderophores by direct injection 21 Tesla Fourier transform ion cyclotron resonance mass spectrometry
Lawrence R.
Walker
,
Malak M.
Tfaily
,
Jared B.
Shaw
,
Nancy J.
Hess
,
Ljiljana
Paša-Tolić
* and
David W.
Koppenaal
*
Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, Richland, Washington, USA. E-mail: Ljiljana.PasaTolic@pnnl.gov; David.koppenaal@pnnl.gov
First published on 1st December 2016
Abstract
Under iron-limiting conditions, bacteria produce low molecular mass Fe(III) binding molecules known as siderophores to sequester the Fe(III), along with other elements, increasing their bioavailability. Siderophores are thought to influence iron cycling and biogeochemistry in both marine and terrestrial ecosystems and hence the need for rapid, confident characterization of these compounds has increased. In this study, the type of siderophores produced by two marine bacterial species, Synechococcus sp. PCC 7002 and Vibrio cyclitrophicus 1F53, were characterized by use of a newly developed 21 T Fourier Transform Ion Cyclotron Resonance Mass Spectrometer (FTICR MS) with direct injection electrospray ionization. This technique allowed for the rapid detection of synechobactins from Synechococcus sp. PCC 7002 as well as amphibactins from Vibrio cyclitrophicus 1F53 based on high mass accuracy and resolution allowing for observation of specific Fe isotopes and isotopic fine structure enabling highly confident identification of these siderophores. When combined with molecular network analysis two new amphibactins were discovered and verified by tandem MS. These results show that high-field FTICR MS is a powerful technique that will greatly improve the ability to rapidly identify and discover metal binding species in the environment.
Significance to metallomicsThis manuscript presents a new approach to the combined metallo-organic characterization of Fe (and other metals) binding siderophores, which serve to increase the bioavailability of these metals in biological and environmental systems. The approach uses a new and unique capability developed at EMSL, Pacific Northwest National Laboratory, namely a 21 Telsa field FTICR MS system, which is capable of providing the ultra-high resolution mass spectrometric detection of these important compounds. Using the new system, we have discovered two new, previously unknown siderophore compounds. |
Introduction
Iron, although abundant in the environment, is a limiting nutrient due to its low solubility. To overcome this problem, microorganisms secrete small (i.e. low molecular mass) molecules known as siderophores, designed to sequester Fe(III) from the environment. These compounds are ecologically important in both terrestrial and marine environments and have received much attention in recent years because of their potential roles and applications in various areas of environmental research (e.g. iron cycling, environmental remediation, and plant growth and health).In terrestrial ecosystems, siderophores play an important role in the extracellular solubilization of iron from minerals thus increasing its availability to microorganisms.1 Siderophores have been shown to improve plant infection and pathogen resistance and to enhance plant growth.2–6 Although the main role of siderophores is to bind Fe(III), they can also play an important role in solubilizing a wide range of other metals (e.g. Pb, Cu, and Zn).7 This has led to the investigation of siderophores in the context of remediation of metal contaminants and pollutants in the environment. For instance, Wang et al. showed that siderophores were able to remove ∼54% of contaminating arsenic from soil.8 Hydroxamate siderophore-like ligands (HO–NH–COOH) were recently reported to be present in the particulate and colloidal organic matter (OM) of soil aggregates. Even though these siderophores constituted only a very minor component of total organic C (<1%), their presence was correlated to Pu concentrations in soil.9
In marine ecosystems, heterotrophic and phototrophic bacteria compete with phytoplankton for Fe by producing a wide range of siderophores to make this essential oceanic nutrient more bioavailable.10 Marine siderophores fall into several structural classes based on binding moieties and hydrophobicity. The three predominant binding moieties are the hydroxamates, catecholates and hydroxy carboxylates. A number of marine siderophores are amphiphilic, containing fatty acid tails; some examples include aquachelins, marinobactins, ochrobactins and synechobactins.11 Marine siderophores are produced by diverse types of oceanic bacteria including proteobacteria and coastal Synechococcus.11–13 Recently, siderophores desferrioxamine E and G have been found to be widely distributed in an Atlantic transect at concentrations of 3–20 pM, accounting for 0.2–4.6% of the total Fe pool.14 Some of the marine organisms that produce siderophores have the ability to degrade petroleum hydrocarbons.15 These siderophores play an indirect role in hydrocarbon degradation by facilitating the iron acquisition needed for the organism to survive and thrive.16
The virulence of some pathogenic bacteria, whether in terrestrial or marine ecosystems, is also related to the organism's ability to uptake iron. In Corynebacterium diphtheria (the causative agent of diphtheria) and virulent mycobacterium species, for example, the virulence factor is regulated by the availability of iron, as is the production of siderophores.17,18 Siderophores can also have an antimicrobial activity. Braun et al. recently discovered two naturally occurring siderophores, albomycin and salmycin, with such activity.19 Additionally, since many siderophores are actively transported across the cell membrane; these compounds have also been investigated for their ability to be a drug delivery system to combat multidrug resistance.20
Methods for characterization of siderophores are available but have limitations. These methods include nuclear magnetic resonance (NMR) spectroscopy, X-ray crystallography, and liquid chromatography-tandem mass spectrometry (LC-MS/MS). While NMR has the ability to give structural information, the sample amount and analysis time required can be too limiting to be practical in many applications. X-ray crystallography has similar drawbacks, including limited sensitivity and throughput, and the need for the sample to be in a purified and crystalline form. LC-MS has the ability to detect siderophores at much lower concentrations than NMR, but throughput and time needed to finish a single run by the conventional LC-MS/MS approach is still limiting. MS data analysis represents an additional challenge due to the complexity of the large data sets. Additionally, siderophore identification by use of these methods is largely limited to those produced by culturable bacterial strains. Abundant but uncultured bacteria are likely to produce additional, unknown and undiscovered siderophores. Studies have also shown that some culturable bacterial and fungal species can produce a variety of siderophores under differing growth conditions, many of these variants are likely also unknown and uncharacterized.12,21,22 Improved methods and approaches are thus needed to identify the range and extent of siderophores produced by all natural bacteria under a variety of environmental conditions.
To address these issues and to better characterize the exo-metabolome of various organisms, a recently developed high magnetic field (21 Tesla, 21 T) Fourier Transform Ion Cyclotron Resonance mass spectrometer (FTICR MS) with direct injection electrospray ionization (ESI) sample introduction was employed. FTICR MS offers the highest resolving power and mass accuracy of any mass spectrometry technique. And, because all key measures of FTICR MS performance improve with increased magnetic field strength, a 21 T FTICR MS now available at EMSL provides an additional level of performance to decipher the chemistry and dynamics of complex molecular systems.23 This approach allows for a highly confident identification of siderophores based on (1) the high mass measurement accuracy yielding very definitive molecular mass assignments and (2) isotopic patterns of both the iron isotopes and other elements present in the molecule. Additionally, as most molecules involved in chelating iron are unique to one or few organisms and as such are not present in publically available databases, molecular network analysis was used as a way to identify candidate new siderophores. Employing direct infusion ESI 21 T FTICR MS, molecular network analysis and an in-house built siderophore database, we characterized several known and unknown siderophores from complex microbial extracts.
Materials and methods
Materials, reagents and cell cultures
Ferrioxamine B, ferrioxamine E, and pyoverdine samples were purchased from Sigma-Aldrich. Ferrioxamine B came as an iron free form, while ferrioxamine E and pyoverdines came as the iron bound forms. Malonichrome and ferrichrome A were obtained via colleagues and were also iron free. All siderophores were dissolved in deionized water (18.2 MΩ) before being diluted with methanol to working concentration. An iron solution standard (Fluka) was used and diluted as required. Cultures of Synechococcus sp. PCC 7002 and Vibrio cyclitrophicus 1F53 were generously provided by the Repeta group at Woods Hole Oceanographic Institution and were grown in low iron casamino acid based media as described previously.24 Ferrioxamine B-molybdenum complex was formed by adding a three-fold molar excess of molybdenum ICP-MS standard to a solution of ferrioxamine B.Mass spectrometry
Mass spectrometry was performed at the Environmental Molecular Sciences Laboratory (EMSL), a U.S. DOE scientific user facility at Pacific Northwest National Laboratory. High-resolution MS and MS/MS experiments were performed by use of a 21 T FTICR MS that was designed and constructed in-house at the EMSL.23 The spectrometer for the 21 T FTICR-MS consists of a Velos Pro dual linear quadrupole ion trap mass spectrometer (ThermoFisher Scientific, San Jose, CA) front end that is coupled to a custom FTICR mass spectrometer. The Velos Pro provides high sensitivity, efficient ion isolation, tandem mass spectrometry (e.g. collision induced dissociation; CID), and automatic gain control (AGC). Ions are transferred from the Velos Pro to the ICR cell for high resolution mass analysis by RF-only quadrupole ion guides. Three stages of differential pumping yield an ultimate pressure of <10−10 Torr in the analyzer stage. ICR signal was acquire using a harmonized ICR cell that utilizes external shimming to approximate an ideal quadrupolar electric field. Samples were directly infused via ESI at 0.5 μL min−1 by use of 360 μm outer diameter ×50 μm inner diameter etched fused silica emitters. The electrospray voltage was 3.0 kV, and the inlet capillary temperature was 300 °C. CID with a 30 V fragmentation energy (Velos Pro) was employed. Mass spectra generated from the average of 400 transient acquisitions by use of an AGC target of 3 × 106 for the m/z range 240–1200. Typical mass measurement accuracy was below 50 ppb.Siderophore database
A siderophore database was created using available literature sources that provided structural information. The main resource used was a review article by Hider and Kong that contained a vast majority of all known siderophores.26 While this supplemental data was exhaustive in the structural information of known siderophores it contained little information about how the siderophores bind Fe3+ (e.g. what structural/functional group modifications, like hydrogen loss, occur with Fe binding). In our new database, the correct protonation state was determined for each siderophore using available literature and binding motifs based upon known iron binding groups in siderophores (i.e. hydroxamate, catecholate, etc.). We considered both the positive and negative ions, existence of both singly and doubly charged species, and all Fe and apo-Fe species. A list of mass spectral peaks was used as input to determine which siderophores are present based on the detection of the Fe isotopic profiles. Database currently contains 370 unique siderophores, with a total of 743 entries when all isoforms are included. It is important to note that our database currently doesn’t differentiate between isomeric species and thus our technique doesn’t allow for the conclusive molecular structure identification due to the occurrence of highly unresolvable isobaric and isomeric species. A chemical formula calculator developed by the National High Magnetic Field Lab was additionally used in order to determine the chemical formula of previously unobserved siderophores.Network analysis
Network analysis of mass spectral data was performed by use of Cytoscape.27 Data was imported by use of the MetaNetter plugin (http://apps.cytoscape.org/apps/metanetter) and mass accuracy error <1.0 ppm. A sample transformation file that contained a list of 88 common chemical transformations seen in nature was used.28 Network analysis was only performed with use of MS data and not MS/MS data.Results and discussion
Siderophore database creation
A siderophore database was created to allow for the identification of known siderophores based on the m/z value and the iron isotopic distribution by use of direct injection ESI FTICR MS. Chelomex, a popular publically available database,29 does an excellent job of identifying siderophores from LC-MS data, but does not allow for interpretation of data collected in direct injection mode or data collected in negative ionization mode. Our custom database was created to take into account the correct protonation state of all known siderophores with iron bound as well as the apo-form of the siderophores. Presumably these siderophores, especially the apo-forms, could exist in multiple protonation states, but multiple protonation states for any siderophore were not previously reported in the literature. Our database also takes into account the possibility of multiple charge states for every siderophore. As shown below, some siderophores were detected as the doubly charged ions and some as the singly charged ions. Hence, multiple charge states need to be taken into account when searching for the 56Fe and 54Fe isotopologues as the m/z difference between these two peaks will be charge state dependent, while the Fe isotopic ratio will be constant. This custom database currently contains 743 entries including all known isoforms of 370 unique siderophores.Characterization of siderophores
Five pure siderophores (ferrioxamine B, ferrioxamine E, ferrichrome A, malonichrome, and pyoverdines) were used to characterize the behavior of siderophores in (positive and negative mode) ESI-MS and to validate the proposed approach. No attempt was made to comprehensively quantify detection limits, but ferrioxamine B was positively identified at 10 ppb in positive ionization mode. This is similar to what has been previously published for FTICR MS analysis of ferrioxamine B.30 For these five pure siderophores and bacterial samples described below, a siderophore was considered positively identified only if both the 56Fe and 54Fe isotopologues were observed, and the isotopic ratio for these peaks was 0.063 ± 0.03.For ferrioxamine B, positive ionization mode resulted in significantly higher signal than negative ionization mode (∼2 orders of magnitude difference), but for ferrioxamine E negative mode ESI resulted in slightly higher signal (<1 order of magnitude). Ferrichrome A and malonichrome ionized more efficiently in positive ionization mode as did the pyoverdines. Ferrichrome A, malonichrome and pyoverdines were predominantly detected as doubly charged ions. Malonichrome showed almost equal intensities for both the singly and double charged ions, while for the ferrioxamines only the singly charged ions were present. For the pyoverdines, the succinic acid and succinamide form were present as the singly and doubly charged ions (pyoverdine 7.12 using the naming convention from Hider et al.).25 For the pyoverdines, the 56Fe peak of the lactamide, α-ketobutyric acid, and α-aminobutyric acid forms were observed at low signal magnitude, and hence the 54Fe peak was not detectable. Data generated for pure siderophores indicated that both positive and negative ionization modes are necessary in order to thoroughly investigate the siderophores present in any sample. Adjusting the pH may result in detection of all siderophores in either positive or negative ionization mode. However, in this study the samples were dissolved in water, before being diluted with methanol and no adjustments to the pH was made. Additional studies and modeling are perhaps necessary to generate a more informed rationale on which ESI parameters to select for comprehensive siderophore characterization.
In the case of ferrioxamine E, additional four siderophores were identified in the sample purified from a microorganism (Table 1). Both the 56Fe and 54Fe peaks corresponding to ferrioxamine G1, ferrioxamine Et2, ferrioxamine Et3, and sabichelin were observed. The ferrioxamine E sample comes from Streptomyces antibioticus and the siderophores positively identified are known to be produced by different species of Streptomyces.31 The 56Fe peak for other ferrioxamines and related siderophores were also detected, but the abundance was too low for the 54Fe peak (∼16× lower abundance) to be observed.
| Siderophore | Formula | Measured m/z | Calculated m/z | Mass error (ppm) | 54Fe/56Fe |
|---|---|---|---|---|---|
| Ferrioxamine E | [C27H44FeN6O9]− | 652.25244 | 652.25246 | −0.033 | 0.052 |
| Ferrioxamine G1 | [C27H46FeN6O10]− | 670.26303 | 670.26303 | 0.002 | 0.056 |
| Sabichelin | [C26H45FeN9O10]− | 699.26418 | 699.26494 | −0.347 | 0.088 |
| Ferrioxamine Et2 | [C29H48FeN6O11]− | 712.27365 | 712.27359 | 0.072 | 0.056 |
| Ferrioxamine Et3 | [C30H50FeN6O12]− | 742.28417 | 742.28416 | 0.009 | 0.063 |
Fig. 1 shows the spectrum of ferrioxamine E. All main isotopologues were detected with mass resolving power of greater than 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. It should be noted that the resolving power, as well as the mass accuracy allows for confident assignment of these peaks as ferrioxamine E without the need for fragmentation or complicated and time consuming LC procedures. A similar Fe isotopic distribution for the other positively identified siderophores from the ferrioxamine E sample was observed. Ferrioxamine Et2, Et3 and G1 showed similar patterns and the low natural abundance (2.119%) 57Fe peak was even visible for these siderophores. Sabichelin did not show enough signal magnitude to allow for the observation of the 57Fe peak, although the 54Fe peak was visible and the isotopic ratio was within the limits set for confident identification of iron bound compounds as discussed previously (54Fe/56Fe = 0.088) (Table 1).
000. It should be noted that the resolving power, as well as the mass accuracy allows for confident assignment of these peaks as ferrioxamine E without the need for fragmentation or complicated and time consuming LC procedures. A similar Fe isotopic distribution for the other positively identified siderophores from the ferrioxamine E sample was observed. Ferrioxamine Et2, Et3 and G1 showed similar patterns and the low natural abundance (2.119%) 57Fe peak was even visible for these siderophores. Sabichelin did not show enough signal magnitude to allow for the observation of the 57Fe peak, although the 54Fe peak was visible and the isotopic ratio was within the limits set for confident identification of iron bound compounds as discussed previously (54Fe/56Fe = 0.088) (Table 1).
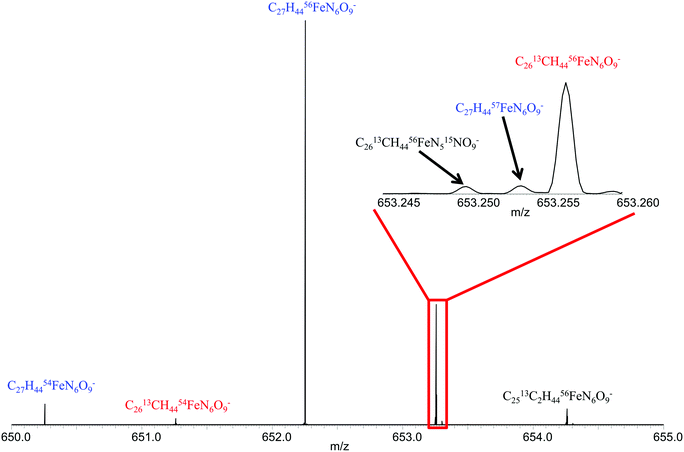 | ||
| Fig. 1 Mass spectrum of ferrioxamine E showing isotopologues detected for singly charged ion. Inset shows isotopic fine structure for m/z = 653.25. | ||
Identification of siderophores in bacterial culture
To further validate this approach, two organisms were investigated for siderophore production, Synechococcus sp. PC7002 and Vibrio cyclitrophicus 1F53.13,24 Earlier studies identified a number of siderophores, called synechobactins, from Synechococcus sp. PC7002 by use of LC-MS/MS in positive ionization mode.13,24 By use of direct injection positive ionization mode ESI FTICR MS in conjunction with the newly created database, no siderophores were identified from the same Synechococcus exo-metabolome. However, synechobactins A, B, C, and C14 were all identified when switched to the negative ionization mode. The 56Fe and corresponding apo-peaks were detected for synechobactin C11 and C13, but there was insufficient signal magnitude to observe the 54Fe peak for these other siderophores (Table 2). Fig. 2 illustrated ability to detect the 56Fe peak, as well as the 54Fe peak, the 57Fe peak, and practically all isotopologues for synechobactin A in direct infusion negative mode ESI FTICR mass spectrum. The ability to detect the isotopic signature of iron as well as other isotopes and isotopologues allows for more confident siderophore identification. These results further confirm that both negative and positive ionization mode are necessary to comprehensively characterize siderophores present in any sample. These results are in good agreement with previously published study of the same six siderophores,24 and confirm that Synechococcus does indeed have a preference for producing siderophores with an even number of carbons in the hydrophobic tail, as those synechobactins appeared to be significantly more abundant.| Siderophore | Formula | Measured m/z | Calculated m/z | Mass error (ppm) | 54Fe/56Fe |
|---|---|---|---|---|---|
| NO: 54Fe peak was not observable. | |||||
| Synechobactin C (C8) | [C22H36FeN4O9]− | 556.18370 | 556.18372 | −0.026 | 0.070 |
| Synechobactin B (C10) | [C24H40FeN4O9]− | 584.21502 | 584.21502 | 0.000 | 0.058 |
| Synechobactin C11 | [C25H42FeN4O9]− | 598.23063 | 598.23067 | −0.055 | NO |
| Synechobactin A (C12) | [C26H44FeN4O9]− | 612.24629 | 612.24632 | −0.051 | 0.058 |
| Synechobactin C13 | [C27H46FeN4O9]− | 626.26188 | 626.26197 | −0.144 | NO |
| Synechobactin C14 | [C28H48FeN4O9]− | 640.27760 | 640.27762 | −0.027 | 0.063 |
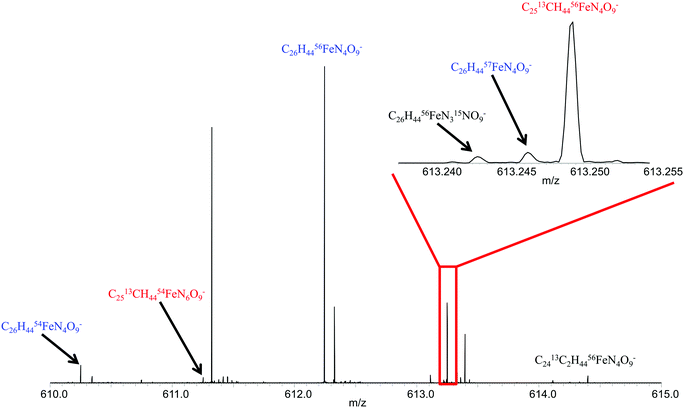
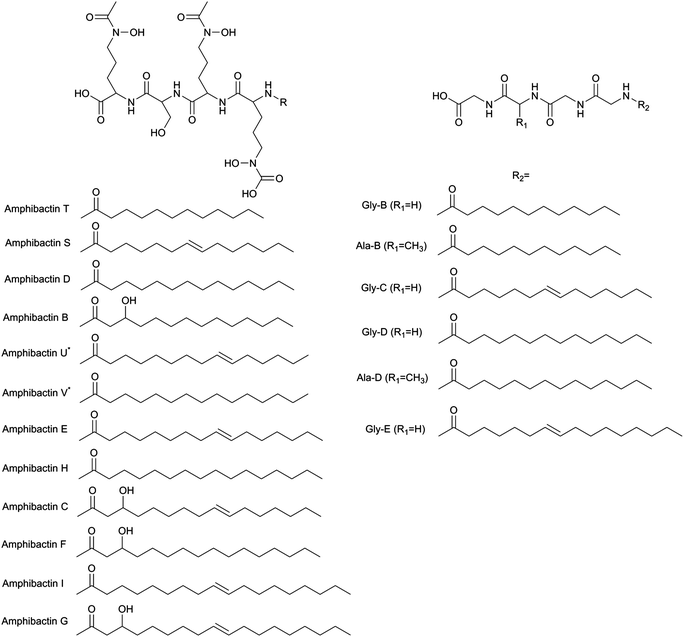
These results led to the investigation of the siderophores produced by a Vibrio species (Vibrio cyclitrophicus 1F53). The siderophore production of other Vibrio species has been reported.15,32 Some Vibrio species have been shown to have the ability to degrade petroleum.33 These earlier studies have shown that some Vibrio species produce siderophores called amphibactins, while other species produce siderophores called moanachelins.32,34 The goal of this current study is to determine if other Vibrio species produce the same siderophores and if so which of the two types would do they produce. The Vibrio cyclitrophicus 1F53 exo-metabolime was investigated by use of the same procedure as described above for the pure siderophores samples and Synechococcus samples. Exo-metabolome samples were directly injected into the 21 T FTICR MS and analyzed in both positive and negative ionization mode. The resultant peaks were imported into the newly created siderophore database to determine which siderophores could be positively identified based on the 56Fe and 54Fe isotopologues. By use of positive ionization mode ESI FTICR MS no known siderophores were identified. However, in negative ionization mode amphibactins B, D, E, and S were successfully identified in the spectrum based on the iron isotopic ratio. The 56Fe peak and corresponding apo-peaks for amphibactins C, F, G, H, I, and T were also observed in the spectrum (Table 3).
| Siderophores | Formula | Measured m/z | Calculated m/z | Mass error (ppm) | 54Fe/56Fe |
|---|---|---|---|---|---|
| a New amphibactins detected in this study. NO: 54Fe peak was not observable. | |||||
| Amphibactin B | [C38H65FeN7O14]− | 899.39398 | 899.39444 | −0.512 | 0.066 |
| Amphibactin C | [C40H67FeN7O14]− | 925.14010 | 925.41009 | 0.016 | NO |
| Amphibactin D | [C38H65FeN7O13]− | 883.39968 | 883.39952 | 0.173 | 0.072 |
| Amphibactin E | [C40H67FeN7O13]− | 909.41516 | 909.41517 | −0.010 | 0.058 |
| Amphibactin F | [C40H69FeN7O14]− | 927.42576 | 927.42574 | 0.022 | NO |
| Amphibactin G | [C42H71FeN7O14]− | 953.44149 | 953.44139 | 0.108 | NO |
| Amphibactin H | [C40H69FeN7O13]− | 911.43082 | 911.43082 | −0.000 | NO |
| Amphibactin I | [C42H71FeN7O13]− | 937.44582 | 937.44647 | −0.700 | NO |
| Amphibactin S | [C38H63FeN7O13]− | 881.38371 | 881.38387 | −0.185 | 0.060 |
| Amphibactin T | [C36H61FeN7O13]− | 855.36824 | 855.36822 | 0.023 | NO |
| Amphibactin Ua | [C39H65FeN7O13]− | 895.39959 | 895.39952 | 0.075 | NO |
| Amphibactin Va | [C39H67FeN7O13]− | 897.41517 | 897.41517 | 0.000 | NO |
| Moanachelins Gly-C | [C35H57FeN7O12]− | 823.34266 | 823.34201 | 0.791 | NO |
| Moanachelins Ala-B | [C36H61FeN7O12]− | 839.3767 | 839.37331 | 0.435 | NO |
| Moanachelins Gly-D | [C38H65FeN7O12]− | 867.40478 | 867.40461 | 0.203 | NO |
The 56Fe and apo-peaks for moanachelins Gly-C, Ala-B, and Gly-D were observed in this same spectrum (Table 3). The signal magnitude of the 56Fe peak was much lower than for the majority of the amphibactins, thus precluding the observation of the low abundance 54Fe isotopologue. The relative intensities of the 56Fe and corresponding apo-peaks for each of the moanachelins were roughly the same. Adding a small amount of Fe (1–2 μmoles) to the sample caused a very modest shift in the Fe-bound vs. apo-peak ratios (∼10%).
It is hypothesized that the ability of a single bacterial species to produce a variety of siderophores could be beneficial to the organism.35 Since iron is limiting in the environment, the production of a variety of siderophores should increase the ability of the bacteria to acquire the iron. One potential explanation of the production of closely related siderophores (amphibactins and moanachelins) by this particular species of Vibrio is to keep energy cost for producing the additional array of siderophores at minimum. Also, the similarity of the siderophores would mean the iron could be imported into the cell by the same transporter proteins. The organism would not need to produce multiple, different protein complexes in order to import different siderophores if the siderophores were very closely related in structure. The structural difference between amphibactins and moanachelins, is that the peptide chain of amphibactins always contains N–AC–OH–Orn, serine, N–AC–OH–Orn, N–AC–OH–Orn followed by a hydrophobic tail; whereas, moanachelins replace the serine for either glycine or alanine. What is as yet not understood is if these different siderophores are produced under slightly different environmental conditions. Is there another environmental stress, other than iron depletion, that triggers the production of moanachelins over amphibactins? The variety of carbon chain lengths of the amphibactins might also give these organisms an advantage in oil rich environments. There is also the possibility that some fraction of siderophores will remain tethered to the cell through the hydrophobic tail, similar to how mycobacteria are thought to acquire iron through the use of mycobactins and carboxymycobactins.36 If, however, both types of siderophores are fully secreted into the environment then it could effectively double the chances the bacterial cell will acquire the needed iron for survival.
Identification of new amphibactins
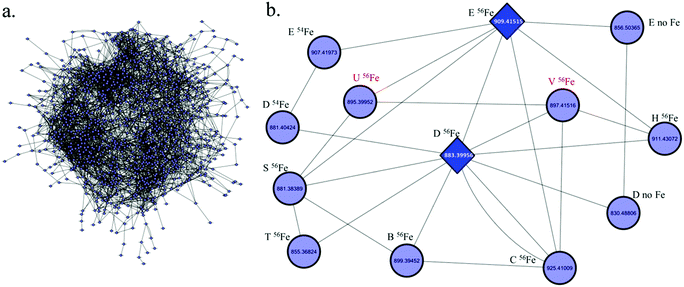
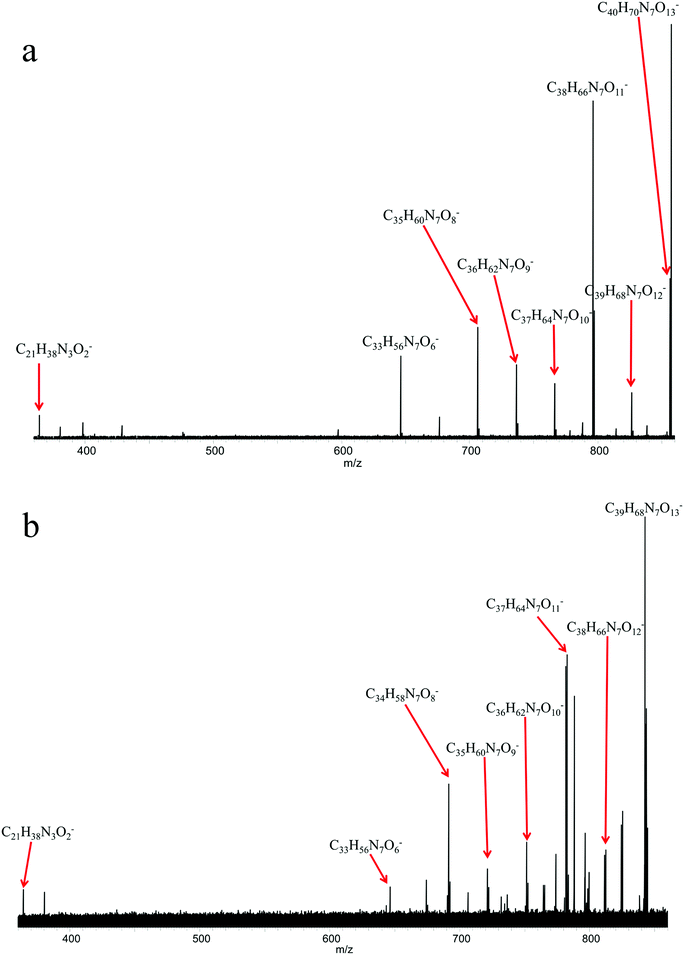
Since a number of bacterial siderophores have been detected from Vibrio cyclitrophicus 1F53 by use of database matching, molecular network analysis was performed next with a goal of identifying previously unknown siderophores. A list of m/z values was imported into Cytoscape (version 2.5.0) using the MetaNetter plug-in, and a list of 88 common chemical transformations. When network analysis was performed the first neighbors of two positively identified amphibactins D and E were selected and 11 m/z values were observed to be directly connected to these two amphibactins based on a single chemical transformation (Fig. 3). Nine of these m/z values were identified as the 56Fe peak of the known amphibactins, 54Fe peak of the positively identified amphibactins, or as the apo-amphibactin. There were two m/z values related to amphibactins D and E that were not previously known amphibactins (labeled herein as amphibactins U and V). The 54Fe peaks for these two amphibactins was not observed (due to low signal magnitude), but the 56Fe and apo-peaks of these two putative amphibactins were observable. The chemical formulas of these two new amphibactins, derived by use of accurate mass, suggest these are homologues of known amphibactins with small differences in the hydrophobic tail length and/or degree of saturation (Table 3 and Fig. 4).In order to further confirm the identity of amphibactins U and V, MS/MS was performed on the apo-species and compared to the fragmentation pattern of amphibactin E (Fig. 5). Fragmentation was performed in negative ionization mode by use of CID and a 30 V fragmentation energy. The major fragments derived from the amphibactin U corresponded to the same mass loss as for the amphibactin E (Fig. 5). The major losses were multiples of ∼30 Da. The mass difference between the corresponding fragment ions reflects the difference in hydrophobic tails of the two amphibactins (14.01565 Da, CH2). Similarly, fragment ions that do not contain the hydrophobic tails are identical for two amphibactins. These observations further confirm that these two new siderophores are structurally related to amphibactins, containing the same iron-binding head.
Application to other metal-containing siderophores
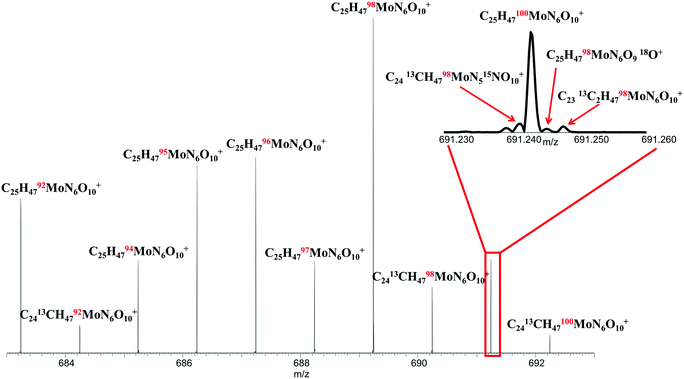
Siderophores are also known to complex metals other than Fe, including Zn, Mn, Mo, and Cu.37 To investigate the ability of the FTICR MS approach to characterize siderophores containing other metals, we substituted Mo for Fe in the ferrioxamine B sample. Molybdenum, with 7 stable isotopes, provides a multiplicity of metal-molecular peaks on which to establish supremely confident identification and analysis and to further validate the FTICR MS approach. The spectrum acquired for the Mo-saturated siderophore is shown in Fig. 6. All 7 Mo isotopes are clearly observed in the spectrum showing the C25H47MoN6O10 molecular ions, in the correct signal magnitude ratios relative to isotopic natural abundance as shown in Table 4. The level of detail obtainable with this approach is also illustrated in the main spectrum where single 13C substitutions can been observed, as well as in the inset spectrum, where 13C, 15N, 18O, and 98,100Mo isotopologues can all be distinguished by use of the ultrahigh resolution approach. The ability to discern such isotopic detail enables not only near-unequivocal identification of molecular species but also provides the opportunity for standardless, absolute quantitation of such complex metal–organic species by use of isotope dilution MS techniques.38| Formula | Measured m/z | Calculated m/z | Error (ppm) | Natural Mo isotope ratio | Measured Mo isotope ratio | Error (%) |
|---|---|---|---|---|---|---|
| [C25H4792MoN6O10]+ | 683.24218 | 683.24218 | 0.003 | 0.148 | 0.135 | −9.08 |
| [C25H4794MoN6O10]+ | 685.24050 | 685.24046 | 0.066 | 0.093 | 0.081 | −12.4 |
| [C25H4795MoN6O10]+ | 686.24119 | 686.24121 | −0.027 | 0.159 | 0.162 | 2.05 |
| [C25H4796MoN6O10]+ | 687.24007 | 687.24007 | 0.035 | 0.167 | 0.169 | 1.27 |
| [C25H4797MoN6O10]+ | 688.24149 | 688.24149 | 0.148 | 0.096 | 0.079 | −17.2 |
| [C25H4798MoN6O10]+ | 689.24070 | 689.24077 | −0.109 | 0.241 | 0.292 | 21.0 |
| [C25H47100MoN6O10]+ | 691.24295 | 691.24295 | 0.153 | 0.096 | 0.082 | −15.2 |
Conclusions
The majority of known siderophores in model systems were identified by use of a simple direct injection ESI 21 T FTICR MS approach that affords ultra-high resolution needed to confidently detect iron isotopic signatures and high mass accuracy needed for confident matching to our custom siderophore database. This approach offers simple sample preparation, rapid data acquisition and analysis. The compiled database contains all currently known siderophores and was verified for the correct protonation state of the siderophores by use of five different acquired siderophores and an exo-metabolome sample derived from a known siderophore producing organism (Synechococcus). This then allowed for the novel identification of the siderophores present in the exo-metabolome of a Vibrio species previously shown to contain amphibactins.25 These results indicated that a single Vibrio species can produce different, closely related siderophores and, in conjunction with network analysis, enabled previously unknown siderophores to be identified.Knowledge of siderophore structure is largely limited by our ability to isolate and culture various bacterial strains. While earlier studies under laboratory conditions have shown that some bacterial and fungal species can produce multiple types of siderophores, more research is needed in particular regarding siderophore production and function in natural environments since a majority (>99%) of trillions of microbial species have never been isolated.12,21,22,39 This study suggests that a more complete suite of siderophores produced by microbes in the marine and terrestrial systems can be revealed by use of ultra-high resolution FTICR MS.
The 21 T FTICR MS approach offers the resolving power, mass accuracy, and sensitivity that are needed to study these metabolites that are present in small quantities. The ability to discern the different and unique isotopes of Fe (54Fe, 56Fe, 57Fe) in the spectra facilitates confident identification of the different siderophores. This approach can also be applied to other multi-isotopic metal-containing siderophores. Similar samples were run on a lower field 12 T FTICR MS and the detection limit, resolving power and mass accuracy made it more difficult to positively identify siderophores based on the iron isotopic ratio.
The methodology described herein provides a means to confidently detect and measure masses of even minor siderophores in a biological extract. By use of this approach, a wide range of known siderophores were characterized, and the presence of two additional homologues, present at low concentrations, were confirmed. As the libraries of known siderophores grow, our ability to characterize the role and function of new siderophores will improve. The new 21 T FTICR MS approach will also make studying other complex organic samples easier and faster with greater measurement accuracy. This will sharpen our ability to pose and answer new questions about metallo-organic matter associations in environmental and biological systems.
Acknowledgements
We thank Drs David Wunschel of PNNL and Craig McLean and Rene Boiteau of Woods Hole Oceanographic Institute for providing us with siderophore samples. The vibrio strain sample originates form the collection of Martin Polz at the Massachusetts Institute of Technology. This research was performed at EMSL, a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research (BER) and located at Pacific Northwest National Laboratory, using EMSL intramural capability development funds.References
- E. Ahmed and S. J. Holmstrom, Siderophores in environmental research: roles and applications, Microb. Biotechnol., 2014, 7, 196–208 CrossRef CAS PubMed.
- J. W. Kloepper, J. Leong, M. Teintze and M. N. Schroth, ENhanced Plant Growth by Siderophores Produced by Plant Growth-Promoting Rhizobacteria, Nature, 1980, 286, 885–886 CrossRef CAS.
- A. A. Gust, F. Brunner and T. Nurnberger, Biotechnological concepts for improving plant innate immunity, Curr. Opin. Biotechnol., 2010, 21, 204–210 CrossRef CAS PubMed.
- E. Gamalero and B. R. Glick, in Bacteria in Agrobiology, ed. D. K. Maheshwari, Springer Verlag, Berlin, Heidelberg, Germany, 2011, pp. 17–46 Search PubMed.
- V. C. Verma, S. K. Singh and S. Prakash, Bio-control and plant growth promotion potential of siderophore producing endophytic Streptomyces from Azadirachta indica A. Juss, J. Basic Microbiol., 2011, 51, 550–556 CrossRef CAS PubMed.
- P. M. Schenk, L. C. Carvalhais and K. Kazan, Unraveling plant-microbe interactions: can multi-species transcriptomics help?, Trends Biotechnol., 2012, 30, 177–184 CrossRef CAS PubMed.
- I. J. Schalk, M. Hannauer and A. Braud, New roles for bacterial siderophores in metal transport and tolerance, Environ. Microbiol., 2011, 13, 2844–2854 CrossRef CAS PubMed.
- Q. Wang, D. Xiong, P. Zhao, X. Yu, B. Tu and G. Wang, Effect of applying an arsenic-resistant and plant growth-promoting rhizobacterium to enhance soil arsenic phytoremediation by Populus deltoides LH05-17, J. Appl. Microbiol., 2011, 111, 1065–1074 CrossRef CAS PubMed.
- C. Xu, S. Zhang, D. I. Kaplan, Y.-F. Ho, K. A. Schwehr, K. A. Roberts, H. Chen, N. DiDonato, M. Athon, P. G. Hatcher and P. H. Santschi, Evidence for Hydroxamate Siderophores and Other N-Containing Organic Compounds Controlling 239,240Pu Immobilization and Remobilization in a Wetland Sediment, Environ. Sci. Technol., 2015, 49, 11458–11467 CrossRef CAS PubMed.
- O. X. Cordero, L.-A. Ventouras, E. F. DeLong and M. F. Polz, Public good dynamics drive evolution of iron acquisition strategies in natural bacterioplankton populations, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 20059–20064 CrossRef CAS PubMed.
- J. M. Vraspir and A. Butler, Chemistry of Marine Ligands and Siderophores, Ann. Rev. Mar. Sci., 2009, 1, 43–63 CrossRef PubMed.
- S. W. Wilhelm and C. G. Trick, Iron-limited growth of cyanobacteria: Multiple siderophore production is a common response, Limnol. Oceanogr., 1994, 39, 1979–1984 CrossRef CAS.
- Y. Ito and A. Butler, Structure of synechobactins, new siderophores of the marine cyanobacterium Synechococcus sp. PCC 7002, Limnol. Oceanogr., 2005, 50, 1918–1923 CrossRef CAS.
- E. Mawji, M. Gledhill, J. A. Milton, G. A. Tarran, S. Ussher, A. Thompson, G. A. Wolff, P. J. Worsfold and E. P. Achterberg, Hydroxamate Siderophores: Occurrence and Importance in the Atlantic Ocean, Environ. Sci. Technol., 2008, 42, 8675–8680 CrossRef CAS PubMed.
- M. P. Kem, H. K. Zane, S. D. Springer, J. M. Gauglitz and A. Butler, Amphiphilic siderophore production by oil-associating microbes, Metallomics, 2014, 6, 1150–1155 RSC.
- K. Barbeau, G. Zhang, D. H. Live and A. Butler, Petrobactin, a Photoreactive Siderophore Produced by the Oil-Degrading Marine Bacterium Marinobacter hydrocarbonoclasticus, J. Am. Chem. Soc., 2002, 124, 378–379 CrossRef CAS PubMed.
- M. P. Schmitt and R. K. Holmes, Iron-dependent regulation of diphtheria toxin and siderophore expression by the cloned Corynebacterium diphtheriae repressor gene dtxR in C. diphtheriae C7 strains, Infect. Immun., 1991, 59, 1899–1904 CAS.
- G. M. Rodriguez, M. I. Voskuil, B. Gold, G. K. Schoolnik and I. Smith, ideR, An essential gene in mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response, Infect. Immun., 2002, 70, 3371–3381 CrossRef CAS PubMed.
- V. Braun, A. Pramanik, T. Gwinner, M. Koberle and E. Bohn, Sideromycins: tools and antibiotics, BioMetals, 2009, 22, 3–13 CrossRef CAS PubMed.
- U. Mollmann, L. Heinisch, A. Bauernfeind, T. Kohler and D. Ankel-Fuchs, Siderophores as drug delivery agents: application of the “Trojan Horse” strategy, BioMetals, 2009, 22, 615–624 CrossRef PubMed.
- J. Granger and N. M. Price, The importance of siderophores in iron nutrition of heterotrophic marine bacteria, Limnol. Oceanogr., 1999, 44, 541–555 CrossRef CAS.
- A. Das, R. Prasad, A. Srivastava, P. H. Giang, K. Bhatnagar and A. Varma, in Microbial Siderophores, ed. A. Varma and S. B. Chincholkar, Springer Berlin Heidelberg, Berlin, Heidelberg, 2007 DOI:10.1007/978-3-540-71160-5_1, pp. 1–42.
- J. B. Shaw, T.-Y. Lin, F. E. Leach III, A. V. Tolmachev, N. Tolić, E. W. Robinson, D. W. Koppenaal and L. Paša-Tolić, 21 Tesla Fourier Transform Ion Cyclotron Resonance Mass Spectrometer Greatly Expands Mass Spectrometry Toolbox, J. Am. Soc. Mass Spectrom., 2016, 27, 1929–1936 CrossRef CAS PubMed.
- R. M. Boiteau and D. J. Repeta, An extended siderophore suite from Synechococcus sp. PCC 7002 revealed by LC-ICPMS-ESIMS, Metallomics, 2015, 7, 877–884 RSC.
- R. M. Boiteau, N. J. Hawco, D. R. Mende, M. R. McIlvin, J. N. Fitzsimmons and P. N. Sedwick, et al., Siderophore-based microbial adaptations to iron scarcity across the eastern Pacific Ocean, Proc. Natl. Acad. Sci. U. S. A. DOI:10.1073/pnas.1608594113; R. M. Boiteau, Molecular determination of marine ligands by mass spectrometry, PhD thesis, 2016, DOI:10.1575/1912/7837, http://hdl.handle.net/1912/7837.
- R. C. Hider and X. Kong, Chemistry and biology of siderophores, Nat. Prod. Rep., 2010, 27, 637–657 RSC.
- M. E. Smoot, K. Ono, J. Ruscheinski, P.-L. Wang and T. Ideker, Cytoscape 2.8: new features for data integration and network visualization, Bioinformatics, 2011, 27, 431–432 CrossRef CAS PubMed.
- R. Breitling, S. Ritchie, D. Goodenowe, L. M. Stewart and P. M. Barrett, Ab initio prediction of metabolic networks using Fourier transform mass spectrometry data, Metabolomics, 2006, 2, 155–164 CrossRef CAS PubMed.
- O. Baars, F. M. M. Morel and D. H. Perlman, ChelomEx: Isotope-Assisted Discovery of Metal Chelates in Complex Media Using High-Resolution LC-MS, Anal. Chem., 2014, 86, 11298–11305, DOI:10.1021/ac503000e.
- H. Waska, A. Koschinsky, M. J. Ruiz Chancho and T. Dittmar, Investigating the potential of solid-phase extraction and Fourier-transform ion cyclotron resonance mass spectrometry (FT-ICR-MS) for the isolation and identification of dissolved metal–organic complexes from natural waters, Mar. Chem., 2015, 173, 78–92 CrossRef CAS.
- S. Kodani, J. Bicz, L. Song, R. J. Deeth, M. Ohnishi-Kameyama, M. Yoshida, K. Ochi and G. L. Challis, Structure and biosynthesis of scabichelin, a novel tris-hydroxamate siderophore produced by the plant pathogen Streptomyces scabies 87.22, Org. Biomol. Chem., 2013, 11, 4686–4694 CAS.
- J. M. Gauglitz and A. Butler, Amino acid variability in the peptide composition of a suite of amphiphilic peptide siderophores from an open ocean Vibrio species, JBIC, J. Biol. Inorg. Chem., 2013, 18, 489–497 CrossRef CAS PubMed.
- D. J. Grimes, C. N. Johnson, K. S. Dillon, A. R. Flowers, N. F. Noriea and T. Berutti, What Genomic Sequence Information Has Revealed About Vibrio Ecology in the Ocean—A Review, Microb. Ecol., 2009, 58, 447–460 CrossRef CAS PubMed.
- J. S. Martinez, J. N. Carter-Franklin, E. L. Mann, J. D. Martin, M. G. Haygood and A. Butler, Structure and membrane affinity of a suite of amphiphilic siderophores produced by a marine bacterium, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 3754–3759 CrossRef CAS PubMed.
- M. E. Hibbing, C. Fuqua, M. R. Parsek and S. B. Peterson, Bacterial competition: surviving and thriving in the microbial jungle, Nat. Rev. Microbiol., 2010, 8, 15–25 CrossRef CAS PubMed.
- J. Gobin and M. A. Horwitz, Exochelins of Mycobacterium tuberculosis remove iron from human iron-binding proteins and donate iron to mycobactins in the M. tuberculosis cell wall, J. Exp. Med., 1996, 183, 1527–1532 CrossRef CAS PubMed.
- T. C. Johnstone and E. M. Nolan, Beyond iron: non-classical biological functions of bacterial siderophores, Dalton Trans., 2015, 44, 6320–6339 RSC.
- K. Heumann, Isotope dilution mass spectrometry, in Inorganic Mass Spectrometry, ed. F. Adams, R. Gijbels and R. Van Grieken, Chemical Analysis Series, Wiley-Interscience, New York, 1989, vol. 95, ch. 7, pp. 301–376 Search PubMed.
- K. J. Locey and J. T. Lennon, Scaling laws predict global microbial diversity, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 5970–5975 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |