Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A fluorogenic 2D glycosheet for the simultaneous identification of human- and avian-receptor specificity in influenza viruses†
Jin-Xing
Song‡
a,
Xin-Ying
Tang‡
b,
Dong-Ming
Zhou
*b,
Wenqing
Zhang
a,
Tony D.
James
 c,
Xiao-Peng
He
*a and
He
Tian
a
c,
Xiao-Peng
He
*a and
He
Tian
a
aKey Laboratory for Advanced Materials & Institute of Fine Chemicals, School of Chemistry and Molecular Engineering, East China University of Science and Technology, 130 Meilong Rd., Shanghai 200237, P. R. China. E-mail: xphe@ecust.edu.cn
bVaccine Research Center, Key Laboratory of Molecular Virology & Immunology, Institut Pasteur of Shanghai, Chinese Academy of Sciences, Shanghai 200031, China. E-mail: dmzhou@sibs.ac.cn
cDepartment of Chemistry, University of Bath, Bath, BA2 7AY, UK
First published on 7th March 2017
Abstract
The switch in glycan-receptor specificity of influenza viruses may cause their interspecies transmission. However, tools that can simultaneous unveil the different receptor specificities of a single virus strain, in a homogeneous solution, have been elusive. Here we show a simple yet effective “2D glycosheet” that is capable of simultaneously identifying the human- and avian-glycan-receptor specificity of influenza viruses. Two fluorophores with different emission colors are coupled with avian- or human-glycan receptors. Then, the glycan–fluorophore conjugates with different receptors and emission colors are co-assembled on a 2D molybdenum disulfide platform, producing a duplexed, fluorogenic 2D glycosheet. While the system shows a single emission color with viruses containing a single receptor specificity, in the presence of a virus (H7N9) containing dual (human- and avian-) receptor specificity the system produces both emission colors. Interestingly, the use of graphene oxide and carbon nanotubes as the material substrate also produces dual emission with H7N9, thus enabling a new generation of low-dimensional glycomaterials for effectively probing the switch in receptor-specificity of influenza viruses.
Conceptual insightsThe possibility of an unpredictable reoccurrence of an influenza pandemic produces a tangible social anxiety and fear worldwide. While the influenza viruses mainly circulate in birds, mutations can cause the viruses to infect, and thus circulate in, humans. Hence, an effective surveillance technique for influenza is in urgent demand. Conventional techniques for the detection of influenza are both expensive and technically demanding requiring complicated manipulations. It is clear that a simple method to simultaneously identify both the bird- and human-infection risk of a single influenza virus strain is urgently required. We have developed a simple method using a 2D glycosheet material composite that is capable of simultaneously identifying both the human- and bird-infecting ability of an influenza virus that can circulate in humans (H7N9), using two different colored emissions. We believe, this research will lead to much needed early warning systems for the surveillance and evaluation of the influenza virus and therefore prevent future global pandemics. |
Introduction
Influenza A viruses are segmented, negative-sense and single-stranded RNA viruses. They are classified into various subtypes according to the serological reactivity of their surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA).1 The HA protein is the key mediator of virus infection to the host cells with a receptor binding site (RBS) on its globular head, which determines the specificity of binding to α2,3-sialic acid (SA) or α-2,6-SA receptors on host cells.2 Generally, avian influenza viruses have a preference for α2,3-SA and attach to cells of the lower respiratory tract, type II pneumocytes and non-ciliated cells. In contrast, influenza viruses infecting mammals have a preference for α2,6-SA and attach to cells of the upper respiratory tract, primarily to ciliated cells.3,4 However, mutations and re-assortments within the RBS can switch the receptor-binding specificity of the viruses and therefore result in the interspecies transmission of influenza viruses.5–7 The traits of HA-receptor binding preference can be harnessed for rapid identification of influenza A virus.8 Although many elegant methods have been developed for the detection of influenza viruses,9–11 a tool that can concomitantly probe the avian- and human-receptor specificity of a single virus strain in a homogeneous solution has been elusive. Here we develop a 2D glycosheet by the co-assembly of two different fluorophore–glycan conjugates to a 2D material for the simultaneous identification of the dual-receptor specificity of a single influenza virus stain.The development of 2D materials has become a very topical research area in many industrial fields since these thin-layer sheets show a number of superior properties to their bulk counterparts.12–15 Of particular interest is the incorporation of 2D materials into the development of biomedical material composites.16–19 These materials have shown promise in sensing biomacromolecules, cell imaging and disease theranostics.20–29 Here we describe the use of a 2D molybdenum disulfide (2D MoS2, a graphene analogue widely used in the design of biomaterials) for the simple construction of a 2D glycosheet that can simultaneously identify the human- and avian-receptor specificity in influenza viruses.
Results and discussion
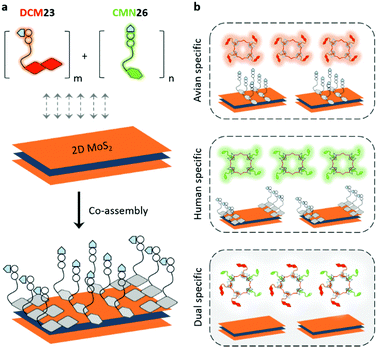
It has been reported that the human- and avian-adapted HA of influenza viruses preferentially binds to the α2,6- and α2,3-sialyl glycan receptors, respectively.2 The switch in the receptor specificity is a key factor for virus interspecies transmission.7 Therefore, we set out to conjugate two different fluorescence reporters to the regioisomeric glycans. A cyan-emitting coumarin (CMN) derivative and a red-emitting dicyanomethylene 4H-pyran (DCM) derivative were used to couple with an avian-receptor (Neu5Acα2,3Gal-β1,4Glc) and a human-receptor (Neu5Acα2,6Gal-β1,4Glc) (Fig. 1 and Scheme S1, ESI†), producing diverse glycan–fluorophore conjugates (CMN23, CMN26, DCM23 and DCM26) with different emission colors. We then took pairs of these probes (CMN23/DCM26 or CMN26/DCM23) and co-assembled them using van der Waals interactions30 with a 2D MoS2 platform to construct the fluorogenic 2D glycosheets (Fig. 2a). We predict that a single emission color would be produced for influenza viruses with a single glycan receptor specificity, whereas both emission colors should be produced in the presence of a virus strain with dual receptor specificity (Fig. 2b).The four desired glycan–fluorophore conjugates were synthesized by coupling the sialyl glycans to amino-CMN and amino-DCM intermediates (Scheme S1, ESI†). 2D MoS2 was prepared using a liquid exfoliation method, with the sheet-like morphologies confirmed by high-resolution transmission electron microscopy (Fig. S1, ESI†).31 With these materials in hand, we first tested the interaction between a single probe and 2D MoS2 using fluorescence spectroscopy (Fig. S2, ESI†). We observed concentration-dependent fluorescence quenching with all four probes containing different sialyl glycans and fluorophores with increasing 2D MoS2 (Fig. S2a, ESI†). This is in agreement with the known quenching ability of this 2D material for fluorescence species closely attached to its surface.24 Interestingly, the subsequent addition of an influenza virus strain with 2,3- (i.e. H10N8: A/Hunan/3-9/2007) or 2,6-receptor (i.e. H1N1pdm09: A/California/08/2009) specificity to the probe-material composite resulted in a concentration dependent fluorescence recovery of the probes (Fig. S2b, ESI†). This fluorescence recovery was determined to be selective in terms of receptor-specificity (i.e. H10N8 for CMN23 and DCM23 and H1N1pdm09 for CMN26 and DCM26) (Fig. S3, ESI†). These preliminary results suggest that (1) 2D MoS2 is a suitable quenching platform for assembling fluorophore-conjugated sialyl glycans and that (2) the fluorescence enhancement of the material composite formed is dependent on the receptor-specificity of the influenza viruses, irrespective of the coupled fluorophore.
Having obtained these promising results, we then set out to construct 2D glycosheets using the co-assembly of a pair of CNM/DCM probes with different glycan receptors with the 2D MoS2.20 We first determined that broad-band fluorescence was recorded for the aqueous mixture of both CMN26/DCM23 and CMN23/DCM26 at a single excitation wavelength (430 nm) (Fig. 3a). The use of a single excitation wavelength facilities the detection procedure since the majority of previous multiplexed sensing systems require the interchange of multiple excitation wavelengths to produce the different emissions.32 Then, we determined that the entire broad-band fluorescence emission could be quenched, in a concentration-dependent manner, with increasing 2D MoS2 (Fig. 3a), suggesting the powerful quenching ability of this 2D material for fluorophore mixtures.
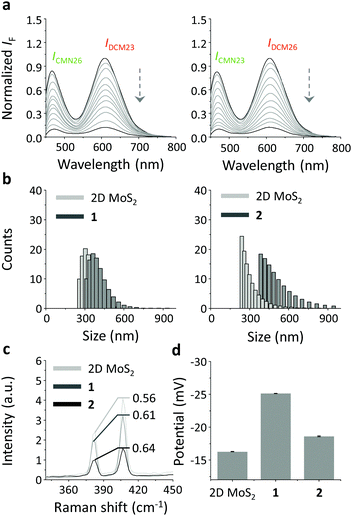
To further corroborate the glycosheet assembly, a variety of other techniques including dynamic light scattering (DLS), Raman spectroscopy and zeta potential measurements were performed. While the size distribution of the 2D MoS2 was in the low submicrometer range,31 the glycosheets (1: CMN23/DCM26 and 2: CMN26/DCM23) seemed to be larger (Fig. 3b). This suggests the attachment of the glycan–fluorophore conjugates to the 2D MoS2. We also observed typical Raman shifts for the 2D MoS2 at 404 and 378 cm−1, which could be ascribed to the A1g (out-of-plane vibration of sulfur) and E12g (in-plane relative motion between sulfur and molybdenum) modes of hexagonal MoS2 (Fig. 3c).33 However, the intensity ratio of E12g to A1g of the glycosheets (1 and 2) increased with respect to 2D MoS2 alone, suggesting that the presence of the fluorophore–glycan conjugates perturbed the in-plane relative motion between the S and Mo atoms of the material.34 In addition, the zeta potentials of the glycosheets were more negative than that of the 2D material alone (Fig. 3d). This is probably a result of the coating of the negatively charged sialyl glycans on the surface of the material.
Next, we tested the sensing performance of the duplexed 2D glycosheets for influenza viruses with different or mixed receptor specificities. The avian-specific H10N8, human specific H1N1pdm09 and an H7N9 “6+2” reassortant virus (A/Anhui/1/2013 derived HA and NA in the background of A/Puerto Rico/8/1934), with human-infecting properties (i.e. the HA can bind to 2,6-sialyl glycans in addition to the avian specific 2,3-sialyl glycans)35–37 were used. A fluorescence titration assay indicated that the presence of the viruses with single-receptor-specificity only enhanced the fluorescence of the corresponding sialyl glycan (i.e. H10N8 for CMN23 and DCM23 and H1N1pdm09 for CMN26 and DCM26) (Fig. 4 and Fig. S4, ESI†). This phenomenon was consistently observed for both CMN23/DCM26 (Fig. 4a) and CMN26/DCM23 2D glycosheets (Fig. 4b). In contrast, the presence of H7N9 caused both the 2,3- and 2,6-glycans to enhance (Fig. 4). These results are in accordance with the previously reported receptor specificity of the virus strains.36
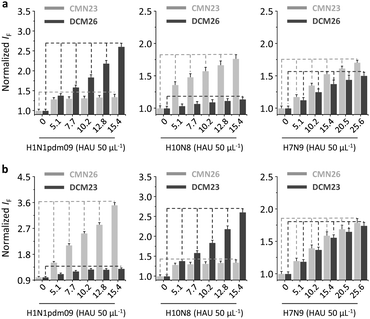 | ||
| Fig. 4 Normalized fluorescence change of (a) the CMN26/DCM23 (1/2 µM) 2D glycosheet (2D MoS2: 70 µg mL−1) and (b) the CMN23/DCM26 (1/2 µM) 2D glycosheet (2D MoS2: 70 µg mL−1) with increasing H10N8 (A/Hunan/3-9/2007), H1N1pdm09 (A/California/08/2009) and H7N9 (A/Anhui/1/2013) in phosphate buffered saline (0.01 M, pH 7.4) (excitation: 430 nm). For the original fluorescence spectra, see Fig. S4, ESI.† | ||
In addition, we observed that the quenched cyan and red fluorescence of CMN26 and DCM23 could be recovered by H1N1pdm09 and H10N8, respectively, whereas the presence of H7N9 produced a mixed color of both glycan–fluorophore conjugates (Fig. 5a). In addition, confocal laser-scanning microscopy was carried out to corroborate the selective glycan-virus binding. Incubation of the 2D glycosheet with H1N1pdm09 and H10N8 resulted in the emergence of CMN26 and DCM23 fluorescence, respectively. In contrast, both fluorescence emissions were seen upon incubation of the material with H7N9 (Fig. 5b). These results together suggest the amenability of our 2D glycosheet for the simultaneous identification of human- and avian-glycan receptors in a single influenza virus strain.
Interestingly, we also demonstrated that the use of two other widely employed low-dimensional quenching platforms,38 2D graphene oxide (GO) and 1D carbon nanotubes (CNT), instead of the 2D MoS2, as the substrate also produced a similar result (Fig. 6 and Fig. S5, ESI†). We first observed a concentration-dependent fluorescence quenching for the mixture of CMN23 and DCM26 in the presence of increasing GO (Fig. S5a) and CNT (Fig. S5e) (ESI†). Then, we observed that while a single fluorescence was selectively enhanced (Fig. S5b and c for GO and Fig. S5f and g for CNT) (ESI†) in the presence of H10N8 and H1N1pdm09, both fluorescence emissions were observed in the presence of H7N9 (Fig. 6a and Fig. S5d (ESI†) for GO and Fig. 6b and Fig. S5h (ESI†) for CNT). This suggests that the system established tolerates a wide range of low-dimensional material substrates.
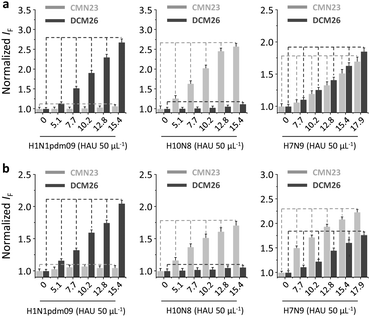 | ||
| Fig. 6 Normalized fluorescence change of (a) the CMN23/DCM26 (1/2 µM) 2D glycosheet (based on 2D graphene oxide: 40 µg mL−1) and (b) the CMN23/DCM26 (1/2 µM) 2D glycosheet (based on 1D carbon nanotube: 4.8 µg mL−1) with increasing H10N8, H1N1pdm09 and H7N9 in phosphate buffered saline (0.01 M, pH 7.4) (excitation: 430 nm). For the original fluorescence spectra, see Fig. S5, ESI.† | ||
We performed a series of experiments in order to determine the stability of the 2D glycosheet. It was found that the CMN23/DCM26 glycosheet was stable to increasing pH (Fig. S6a, ESI†) and increasing salt strength (Fig. S6b, ESI†). Also, the material did not show any fluorescence increase with a series of proteins including bovine serum albumin, IgG, lysozyme and pepsin (Fig. S6c, ESI†), suggesting that the detection system would not be adversely influenced by environmental factors. Furthermore, we used two additional viruses (the simian adenovirus AdC68 and the human adenovirus AdHu7) with the 2D glycosheet.39 Our results indicate that the material was not responsive to these two adenoviruses (Fig. S7, ESI†).
The limit of detection (LOD) for the CMN23/DCM26 glycosheet towards H1N1pdm09 and H10N8 was determined to be 0.04 and 0.02 HAU 50 µL−1, respectively, and that for H7N9 with dual receptor specificity was determined to be 0.07 (CMN23) and 0.06 (DCM26) HAU 50 µg mL−1 (Fig. S8, ESI†). We performed a fluorescence kinetic assay to determine how quickly the 2D glycosheet functions. The results indicated that a detection equilibrium was quickly established for both fluorescence emissions and within 5 min (Fig. S9, ESI†).
The most commonly used technique for influenza A virus detection is the hemagglutination assay, which is quasi-quantitative. This assay must be followed by an additional hemagglutinin inhibition assay including complicated manipulation procedures and long detection time. ELISA (enzyme-linked immunosorbent assay) is another frequently used method with good sensitivity. However, it requires the use of antibodies, which are expensive and sluggish to produce. Whereas the sensitivity of our 2D glycosheet is comparable to, or lower than, most of the competing methods developed including surface plasmon resonance,40a HPLC,40b gold nanoparticle based probes40c and quartz crystal microbalance,40d and the speed of detection appears to be the fastest (Table S1, ESI†). More importantly, our duplexed 2D glycosheet is a unique system to simultaneously determine the dual receptor specificity of a single influenza A virus in a homogeneous solution.
Conclusions
To conclude, we have developed a simple yet effective 2D glycosheet using supramolecular co-assembly of two fluorophore–glycan conjugates to a low-dimensional material platform. The fluorogenic material composite formed has proven to be effective for the simultaneous identification of the dual-receptor specificity of a single influenza virus strain. This research allows for the effective interrogation of the receptor specificity switch for seasonal influenza viruses.Acknowledgements
This research was supported by the 973 project (2013CB733700), the National Natural Science Foundation of China (21572058 and 21576088), the Science and Technology Commission of Shanghai Municipality (15540723800), the Knowledge Innovation Program (Y014P31503), the Fundamental Research Funds for the Central Universities and the 100 Talent Program (Y316P11503) from the Chinese Academy of Sciences (to D. Z.), and the Shanghai Rising-Star Program (16QA1401400) (to X.-P. H.). Miss Ya-Li Zeng is warmly thanked for her contribution to the research. The Catalysis And Sensing for our Environment (CASE) network is thanked for research exchange opportunities. T. D. J. thanks ECUST for a guest professorship.Notes and references
- N. J. Cox, G. Neumann, R. O. Donis and Y. Kawaoka, Topley & Wilson's Microbiology and Microbial Infections, John Wiley & Sons, Ltd, 2010 Search PubMed.
- Y. Suzuki, T. Ito, T. Suzuki, R. E. Holland, T. M. Chambers, M. Kiso, H. Ishida and Y. Kawaoka, J. Virol., 2000, 74, 11825–11831 CrossRef CAS PubMed.
- K. Shinya, M. Ebina, S. Yamada, M. Ono, N. Kasai and Y. Kawaoka, Nature, 2006, 440, 435–436 CrossRef CAS PubMed.
- M. N. Matrosovich, T. Y. Matrosovich, T. Gray, N. A. Roberts and H. D. Klenk, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 4620–4624 CrossRef CAS PubMed.
- C. R. Parrish and Y. Kawaoka, Annu. Rev. Microbiol., 2005, 59, 553–586 CrossRef CAS PubMed.
- J. Stevens, O. Blixt, T. M. Tumpey, J. K. Taubenberger, J. C. Paulson and I. A. Wilson, Science, 2006, 312, 404–410 CrossRef CAS PubMed.
- Y. Wang, N. Engl. J. Med., 2013, 368, 2348–2349 CrossRef CAS PubMed.
- A. García-Sastre and M. Schmolke, Lancet, 2014, 383, 676–677 CrossRef.
- S. Fukui, T. Feizi, C. Galustian, A. M. Lawson and W. Chai, Nat. Biotechnol., 2002, 20, 1011–1017 CrossRef CAS PubMed.
- T. Feizi and W. Chai, Nat. Rev. Mol. Cell Biol., 2004, 5, 582–588 CrossRef CAS PubMed.
- C. Lee, M. A. Gaston, A. A. Weiss and P. Zhang, Biosens. Bioelectron., 2013, 42, 236–241 CrossRef CAS PubMed.
- Q. H. Wang, K. Kalantar-Zadeh, A. Kis, J. N. Coleman and M. S. Strano, Nat. Nanotechnol., 2012, 7, 699–712 CrossRef CAS PubMed.
- M.-R. Gao, Y.-F. Xu, J. Jiang and S.-H. Yu, Chem. Soc. Rev., 2013, 42, 2986–3017 RSC.
- H. Wang, H. Feng and J. Li, Small, 2014, 10, 2165–2181 CrossRef CAS PubMed.
- Y. Sun, S. Gao, F. Lei, C. Xiao and Y. Xie, Acc. Chem. Res., 2015, 48, 3–12 CrossRef CAS PubMed.
- M. Pumera and A. H. Loo, Trends Anal. Chem., 2014, 61, 49–53 CrossRef CAS.
- Y. Chen, C. Tan, H. Zhang and L. Wang, Chem. Soc. Rev., 2015, 44, 2681–2701 RSC.
- D. Chimene, D. L. Alge and A. K. Gaharwar, Adv. Mater., 2015, 27, 7261–7284 CrossRef CAS PubMed.
- (a) X.-P. He and H. Tian, Small, 2016, 12, 144–160 CrossRef CAS PubMed; (b) H.-L. Zhang, X.-L. Wei, Y. Zang, J.-Y. Cao, S. Liu, X.-P. He, Q. Chen, Y.-T. Long, J. Li, G.-R. Chen and K. Chen, Adv. Mater., 2013, 25, 4097–4101 CrossRef CAS PubMed; (c) X.-P. He, Q. Deng, L. Cai, C.-Z. Wang, Y. Zang, J. Li, G.-R. Chen and H. Tian, ACS Appl. Mater. Interfaces, 2014, 6, 5379–5382 CrossRef CAS PubMed; (d) X. Sun, B. Zhu, D.-K. Ji, Q. Chen, X.-P. He, G.-R. Chen and T. D. James, ACS Appl. Mater. Interfaces, 2014, 6, 10078–10082 CrossRef CAS PubMed; (e) X.-P. He, B.-W. Zhu, Y. Zang, J. Li, G.-R. Chen, H. Tian and Y.-T. Long, Chem. Sci., 2015, 6, 1996–2001 RSC; (f) D.-K. Ji, Y. Zhang, X.-P. He and G.-R. Chen, J. Mater. Chem. B, 2015, 3, 6656–6661 RSC; (g) D.-K. Ji, Y. Zhang, Y. Zang, W. Liu, X. Zhang, J. Li, G.-R. Chen, T. D. James and X.-P. He, J. Mater. Chem. B, 2015, 3, 9182–9185 RSC; (h) D.-K. Ji, Y. Zhang, Y. Zang, J. Li, G.-R. Chen, X.-P. He and H. Tian, Adv. Mater., 2016, 28, 9356–9363 CrossRef CAS PubMed; (i) D. Xie, D.-K. Ji, Y. Zhang, J. Cao, H. Zheng, L. Liu, Y. Zang, J. Li, G.-R. Chen, T. D. James and X.-P. He, Chem. Commun., 2016, 52, 9418–9421 RSC; (j) M. Wahiba, X.-Q. Feng, Y. Zang, T. D. James, J. Li, G.-R. Chen and X.-P. He, Chem. Commun., 2016, 52, 11689–11692 RSC; (k) S. Guo, J. Chen, B.-Y. Cai, W.-W. Chen, Y.-F. Li, X. Sun, G.-R. Chen, X.-P. He and T. D. James, Mater. Chem. Front., 2017, 1, 61–64 RSC; (l) Y.-H. Ma, W.-T. Dou, Y.-F. Pan, L.-W. Dong, Y.-X. Tan, X.-P. He, H. Tian and H.-Y. Wang, Adv. Mater., 2017, 29, 1604253 CrossRef PubMed.
- Y. Zhang, B. Zheng, C. Zhu, X. Zhang, C. Tan, H. Li, B. Chen, J. Yang, J. Chen, Y. Huang, L. Wang and H. Zhang, Adv. Mater., 2015, 27, 935–939 CrossRef CAS PubMed.
- L. Cheng, J. Liu, X. Gu, H. Gong, X. Shi, T. Liu, C. Wang, X. Wang, G. Liu, H. Xing, W. Bu, B. Sun and Z. Liu, Adv. Mater., 2014, 26, 1886–1893 CrossRef CAS PubMed.
- T. Liu, C. Wang, X. Gu, H. Gong, L. Cheng, X. Shi, L. Feng, B. Sun and Z. Liu, Adv. Mater., 2014, 26, 3433–3440 CrossRef CAS PubMed.
- W. Yin, L. Yan, J. Yu, G. Tian, L. Zhou, X. Zheng, X. Zhang, Y. Yong, J. Li, Z. Gu and Y. Zhao, ACS Nano, 2014, 8, 6922–6933 CrossRef CAS PubMed.
- C. Zhu, Z. Zeng, H. Li, F. Li, C. Fan and H. Zhang, J. Am. Chem. Soc., 2013, 135, 5998–6001 CrossRef CAS PubMed.
- B. L. Li, H. L. Zou, L. Lu, Y. Yang, J. L. Lei, H. Q. Luo and N. B. Li, Adv. Funct. Mater., 2015, 25, 3541–3550 CrossRef CAS.
- H. Fan, Z. Zhao, G. Yan, X. Zhang, C. Yang, H. Meng, Z. Chen, H. Liu and W. Tan, Angew. Chem., Int. Ed., 2015, 54, 4801–4805 CrossRef CAS PubMed.
- S. S. Chou, B. Kaehr, J. Kim, B. M. Foley, M. De, P. E. Hopkins, J. Huang, C. J. Brinker and V. P. Dravid, Angew. Chem., Int. Ed., 2013, 52, 4160–4164 CrossRef CAS PubMed.
- L. Cheng, C. Yuan, S. Shen, X. Yi, H. Gong, K. Yang and Z. Liu, ACS Nano, 2015, 9, 11090–11101 CrossRef CAS PubMed.
- J. Nam, W.-G. La, S. Hwang, Y. S. Ha, N. Park, N. Won, S. Jung, S. H. Bhang, Y.-J. Ma, Y.-M. Cho, M. Jin, J. Han, J.-Y. Shin, E. K. Wang, S. G. Kim, S.-H. Cho, J. Yoo, B.-S. Kim and S. Kim, ACS Nano, 2013, 7, 3388–3402 CrossRef CAS PubMed.
- Y. Ling, Z. Gu, S.-G. Kang, J. Luo and R. Zhou, J. Phys. Chem. C, 2016, 120, 6796–6803 CAS.
- J. N. Coleman, M. Lotya, A. O'Neill, S. D. Bergin, P. J. King, K. Young, A. Gaucher, S. De, R. J. Smith, I. V. Shvets, S. K. Arora, G. Stanton, H.-Y. Kim, K. Lee, G. T. Kim, G. S. Duesberg, T. Hallam, J. J. Boland, J. J. Wang, J. F. Donegan, J. C. Grunlan, G. Moriarty, A. Shmeliov, R. J. Nicholls, J. M. Perkins, E. M. Grieveson, K. Theuwissen, D. W. McComb, P. D. Nellist and V. Nicolosi, Science, 2011, 311, 568–571 CrossRef PubMed.
- D.-K. Ji, G.-R. Chen, X.-P. He and H. Tian, Adv. Funct. Mater., 2015, 25, 3483–3487 CrossRef CAS.
- C. Lee, H. Yan, L. E. Brus, T. F. Heinz, J. Hone and S. Ryu, ACS Nano, 2010, 4, 2695–2700 CrossRef CAS PubMed.
- P. T. K. Loan, W. Zhang, C.-T. Lin, K.-H. Wei, L.-J. Li and C.-H. Chen, Adv. Mater., 2014, 26, 4838–4844 CrossRef CAS PubMed.
- Y. Shi, W. Zhang, F. Wang, J. Qi, Y. Wu, H. Song, F. Gao, Y. Bi, Y. Zhang, Z. Fan, C. Qin, H. Sun, J. Liu, J. Haywood, W. Liu, W. Gong, D. Wang, Y. Shu, Y. Wang, J. Yan and G. F. Gao, Science, 2013, 342, 243–247 CrossRef CAS PubMed.
- (a) X. Xiong, S. R. Martin, L. F. Haire, S. A. Wharton, R. S. Daniels, M. S. Bennett, J. W. McCauley, P. J. Collins, P. A. Walker, J. J. Skehel and S. J. Gamblin, Nature, 2013, 499, 496–499 CrossRef CAS PubMed; (b) M. Wang, W. Zhang, J. Qi, F. Wang, J. Zhou, Y. Bi, Y. Wu, H. Sun, J. Liu, C. Huang, X. Li, J. Yan, Y. Shu, Y. Shi and G. F. Gao, Nat. Commun., 2015, 6, 5600 CrossRef CAS PubMed; (c) T. R. Maines, A. Jayaraman, J. A. Belser, D. A. Wadford, C. Pappas, H. Zeng, K. M. Gustin, M. B. Pearce, K. Viswanathan, Z. H. Shriver, R. Raman, N. J. Cox, R. Sasisekharan, J. M. Katz and T. M. Tumpey, Science, 2009, 325, 484–487 CAS.
- S. G. Vachieri, X. Xiong, P. J. Collins, P. A. Walker, S. R. Martin, L. F. Haire, Y. Zhang, J. W. McCauley, S. J. Gamblin and J. J. Skehel, Nature, 2014, 511, 475–477 CrossRef CAS PubMed.
- (a) X.-P. He, Y. Zang, T. D. James, J. Li and G.-R. Chen, Chem. Soc. Rev., 2015, 44, 4239–4248 RSC; (b) X.-P. He, Y. Zang, T. D. James, J. Li, G.-R. Chen and J. Xie, Chem. Commun., 2017, 53, 82–90 RSC.
- D. Zhou, X. Zhou, A. Bian, H. Li, H. Chen, J. C. Small, Y. Li, W. Giles-Davis, Z. Xiang and H. C. J. Ertl, Nat. Protoc., 2010, 5, 1775–1785 CrossRef CAS PubMed.
- (a) H. Bai, R. Wang, B. Hargis, H. Lu and Y. Li, Sensors, 2012, 12, 12506–12518 CrossRef CAS PubMed; (b) L. Urbas, B. Košir, M. Peterka, B. Pihlar, A. Štrancar and M. Barut, J. Chromatogr. A, 2011, 1218, 2432–2437 CrossRef CAS PubMed; (c) Y. Liu, L. Zhang, W. Wei, H. Zhao, Z. Zhou, Y. Zhang and S. Liu, Analyst, 2015, 140, 3989–3995 RSC; (d) R. Wang and Y. Li, Biosens. Bioelectron., 2013, 42, 148–155 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Additional figures, Table S1 and experimental section. See DOI: 10.1039/c6mh00571c |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2017 |