Open Access Article
Open Access ArticleNonomuraea sp. ATCC 55076 harbours the largest actinomycete chromosome to date and the kistamicin biosynthetic gene cluster†‡
Behnam
Nazari
 a,
Clarissa C.
Forneris
a,
Clarissa C.
Forneris
 a,
Marcus I.
Gibson
a,
Kyuho
Moon
a,
Kelsey R.
Schramma
a and
Mohammad R.
Seyedsayamdost
a,
Marcus I.
Gibson
a,
Kyuho
Moon
a,
Kelsey R.
Schramma
a and
Mohammad R.
Seyedsayamdost
 *ab
*ab
aDepartment of Chemistry, Princeton University, Princeton, NJ 08544, USA. E-mail: mrseyed@princeton.edu
bDepartment of Molecular Biology, Princeton University, Princeton, NJ 08544, USA
First published on 30th January 2017
Abstract
Glycopeptide antibiotics (GPAs) have served as potent clinical drugs and as an inspiration to chemists in various disciplines. Among known GPAs, complestatin, chloropeptin, and kistamicin are unique in that they contain an unusual indole–phenol crosslink. The mechanism of formation of this linkage is unknown, and to date, the biosynthetic gene cluster of only one GPA with an indole–phenol crosslink, that of complestatin, has been identified. Here, we report the genome sequence of the kistamicin producer Nonomuraea sp. ATCC 55076. We find that this strain harbours the largest actinobacterial chromosome to date, consisting of a single linear chromosome of ∼13.1 Mbp. AntiSMASH analysis shows that ∼32 biosynthetic gene clusters and ∼10% of the genome are devoted to production of secondary metabolites, which include 1,6-dihydroxyphenazine and nomuricin, a new anthraquinone-type pentacyclic compound that we report herein. The kistamicin gene cluster (kis) was identified bioinformatically. A unique feature of kis is that it contains two cytochrome P450 enzymes, which likely catalyze three crosslinking reactions. These findings set the stage for examining the biosynthesis of kistamicin and its unusual indole–phenol crosslink in the future.
1. Introduction
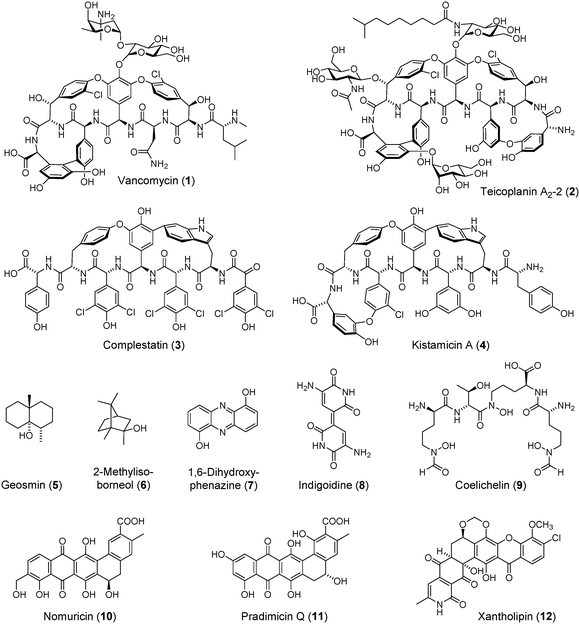
Glycopeptides antibiotics (GPAs) represent one of our most medicinally important and structurally beautiful cures.1–4 Since the discovery of vancomycin (1, Fig. 1),5,6 the founding member of this class, GPAs have served as indispensible weapons against Gram positive bacterial infections and recently as drugs-of-last resort against methicillin-resistant Staphylococcus aureus (MRSA) and Clostridium difficile. The World Health Organization lists vancomycin as one of 200 essential medicines. The importance of GPAs in a clinical setting cannot be overstated.The remarkable bioactivity of GPAs, which include teicoplanin (2),7 another clinically deployed member of this class, is based on inhibition of cell wall biosynthesis by tight binding and sequestration of the D-Ala-D-Ala terminus of the growing muropeptide portion of the peptidoglycan.4,8,9 This mode of action is made possible by the architecturally complex structure of GPAs, which contain a rigid, cup-shaped topology that is locked in its structure via several biaryl and bisaryl ether crosslinks. Installation of these unusual modifications is of great biochemical interest and their mechanism of formation has yet to be fully elucidated.10,11 Pioneering studies by Wohlleben, Süssmuth, and Robinson have shown that cytochrome P450 enzymes catalyze these transformations.12–17 Specifically, the reaction of OxyB, which installs the first bisaryl ether crosslink in vancomycin, has been reconstituted in vitro.16,17 However, only a few examples of these reactions have been demonstrated, and the enzymatic semi-synthesis of GPAs remains a long-standing and unaccomplished goal. The recent discovery of a so-called X-domain that recruits P450 enzymes during bisaryl ether bond formation in teicoplanin shows that GPAs still bear many surprises and lessons in biosynthesis.18–20
GPAs are categorized into five classes based on structural considerations.2 Classes I–IV contain either biaryl or bisaryl ether crosslinks involving tyrosine, 4-hydroxphenylglycine (Hpg), 3,5-dihydroxyphenylglycine (Dpg), or chlorinated derivates of Tyr and Hpg. Class V GPAs are rare in that they incorporate tryptophan into the peptidic scaffold and contain characteristic indole–phenol crosslinks (Fig. 1). The only available biosynthetic gene cluster for a class V GPA is that of complestatin (3).21,22 The P450 enzymes involved in side-chain crosslink formation in complestatin biosynthesis have yet to be characterized. While numerous class V GPA gene clusters can be seen in the available bacterial genome data base, they have not yet been linked to a cognate GPA. Conversely, kistamicin (4),23 another prominent member of the class V GPAs, has not yet been linked to its biosynthetic genes.
In contrast to complestatin, kistamicin contains two bisaryl ether bonds and an indole–phenol crosslink (Fig. 1). To begin to understand the production of the class V GPAs as well as expand the genomic basis of these fascinating molecules, we herein report the complete genome sequence of Nonomuraea sp. ATCC 55076 (hereafter ATCC 55076), a kistamicin producer previously isolated from soil near the Kistna River in India. Aside from identifying and characterizing the kistamicin gene cluster (kis) in silico, we find that ATCC 55076 harbours the largest actinobacterial genome to date, with a single linear chromosome consisting of ∼13.1 Mbp. In addition to kis, ATCC 55076 contains more than 30 secondary metabolite biosynthetic gene clusters that bear little homology to known ones, suggesting that this strain will serve as an attractive target for future genome mining strategies in the search for new and useful small molecules.
2. Results and discussion
2.1 Kistamicin production by Nonomuraea sp. ATCC 55076
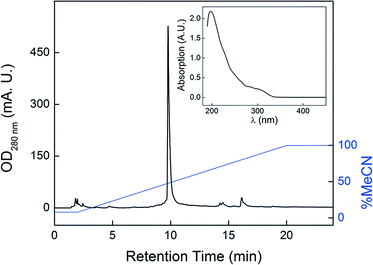
Kistamicin was originally isolated from Microtetraspora pervosata in the 1980s.23 Like its relative complestatin, it exhibited potent antiviral activity as well as mild antibiosis against Gram positive bacteria. The producing strain was re-classified as a member of the Actinomadura genus, then a member of the rare actinomycete, Nonomuraea. We obtained Nonomuraea sp. ATCC 55076 and examined its ability to produce kistamicin using HPLC-MS. Production of kistamicin was not observed in the typical ISP2 actinomycete growth medium. We thus conducted an OSMAC-type approach,24 in which we monitored kistamicin production in culture supernatants and mycelial extracts in six distinct media. Kistamicin was observed under only one condition, a fish meal-based medium, which we designed according to the original reports.23 Acetone extraction of the mycelial pellet yielded nearly pure kistamicin (Fig. 2), thus obviating the extensive purification protocol originally employed by Naruse et al.23 Consistent with previous reports, ATCC 55076 proved to be a prolific producer when cultured in the appropriate medium delivering kistamicin yields of >3 mg L−1 culture. The identity of kistamicin was verified by high-resolution MS (HR-MS, [M + H]+calc 1171.3241, [M + H]+obs 1171.3256) as well as by comparison with reported 1D/2D NMR data (Fig. S1, S2 and Table S1‡). That kistamicin is biosynthesized only in certain culture media suggests that an as-of-yet unknown trace element, component, or culture condition is required for its production.2.2 Whole genome sequencing of Nonomuraea sp. ATCC 55076
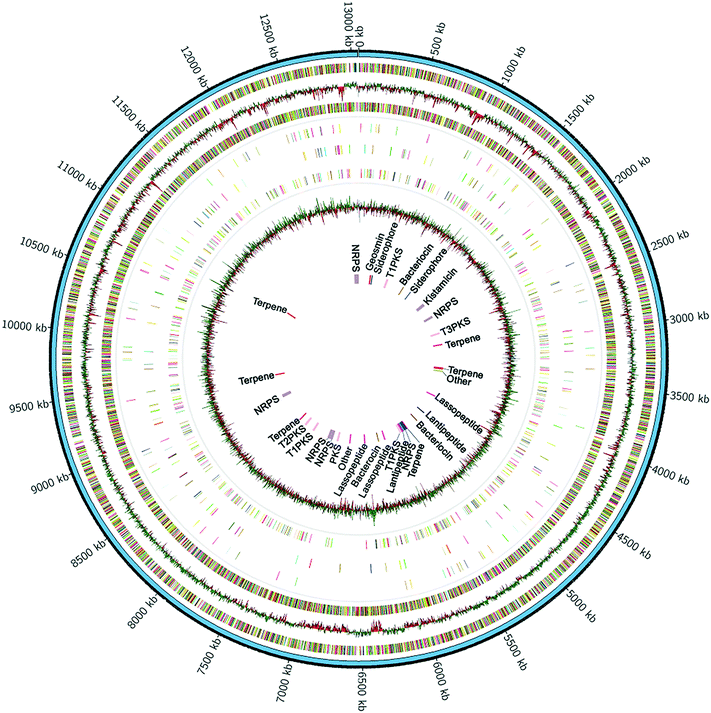
To find the biosynthetic gene cluster of kistamicin, we used next-generation genome sequencing and bioinformatic methods. Genomic DNA was isolated and its sequence determined using the PacBio single molecule real-time (SMRT) sequencing technology.25 A long insert library of ∼8 kb was prepared and sequences generated from 3 SMRT cells. A total of ∼227![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 reads from an average fragment size of 7.8 kb were used to construct the genome sequence. The hierarchical genome assembly process (HGAP)26 allowed de novo assembly of the entire chromosome resulting in a single contig of ∼13.1 Mbp with 130-fold coverage and an average GC content of 72.1%, consistent with the reported GC content for this genus (Fig. 3).27 This marks the first complete genome sequence for a member of the genus Nonomuraea. The largest actinobacterial genome to date is that of Streptomyces bingchinggensis at 11.9 Mbp and 10
000 reads from an average fragment size of 7.8 kb were used to construct the genome sequence. The hierarchical genome assembly process (HGAP)26 allowed de novo assembly of the entire chromosome resulting in a single contig of ∼13.1 Mbp with 130-fold coverage and an average GC content of 72.1%, consistent with the reported GC content for this genus (Fig. 3).27 This marks the first complete genome sequence for a member of the genus Nonomuraea. The largest actinobacterial genome to date is that of Streptomyces bingchinggensis at 11.9 Mbp and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 023 open reading frames (ORFs).28 In a phylum known for its large genomes, the single, 13.1 Mbp linear chromosome of Nonomuraea sp. ATCC 55076 is the largest of any sequenced actinobacterium to date. The genome sequence has been deposited at GenBank (BioProject ID PRJNA348567, accession CP017717) and IMG (ID 98346).
023 open reading frames (ORFs).28 In a phylum known for its large genomes, the single, 13.1 Mbp linear chromosome of Nonomuraea sp. ATCC 55076 is the largest of any sequenced actinobacterium to date. The genome sequence has been deposited at GenBank (BioProject ID PRJNA348567, accession CP017717) and IMG (ID 98346).
Genome annotation using RAST29 predicted a record >11600 coding sequences, about 50% each on the leading and lagging strands. Among these, we found a staggering 96 predicted sigma factors, 187 two-component signalling systems and 536 transcriptional regulators (Fig. 3). Consistent with the saprophytic nature of ATCC 55076, we also detected over ∼400 secreted proteins and enzymes, as well as ∼1100 proteins with a predicted transport function. The large numbers of predicted regulatory proteins, transporters, sigma factors, and secondary metabolite biosynthetic genes (see below), in part, explain the large genome size of ATCC 55076.
2.3 Secondary metabolome analysis
To assess the potential of ATCC 55076 for secondary metabolite synthesis, antiSMASH30 analysis was carried out, revealing ∼32 biosynthetic gene clusters, with an average length of 38 kb (Table 1). Thus, this strain devotes a remarkable ∼9.6% of its genome to secondary metabolite production compared to 4.5% and 6% for the representative actinomycetes Streptomyces coelicolor or Streptomyces avermitilis, respectively, both of which have significantly smaller chromosomes (approx. 8.5 Mbp).31 The genome of ATCC 55076 is especially rich in biosynthetic gene clusters for terpenes (6 total) and bacteriocins (8 total), including lantipeptides and lasso peptides. Only three of the 32 clusters can be confidently assigned to a known secondary metabolite, that of geosmin (5) and 2-methylisoborneol (6) – both of which show complete similarity to previously characterized gene clusters30,32 – as well as kistamicin, which bears high sequence similarity to the cluster of A40926, a teicoplanin-type GPA (Table 1). Biosynthetic gene clusters for indigoidine (8) and coelichelin (9) also share high similarities (80% and 63%, respectively) with previously characterized relatives.33,34 However, these molecules were not detected in cultures of ATCC 55076.| Cluster | Location | Structural type | Predicted product | Similarity to characterized biosynthetic gene clusters |
|---|---|---|---|---|
| 1 | BKM31_280108-302390 | Terpene | Geosmin | Geosmin gene cluster (100%) |
| 2 | BKM31_329031-342275 | Siderophore | Unknown | |
| 3 | BKM31_692756-738554 | Type I PKS | Thiotetroamide gene cluster (11%) | |
| 4 | BKM31_1149280-1160095 | Bacteriocin | ||
| 5 | BKM31_1361626-1374873 | Siderophore | ||
| 6 | BKM31_1780967-1852209 | NRPS-type III PKS | Kistamicin | A40926 gene cluster (43%) |
| 7 | BKM31_2151947-2205643 | NRPS | Spinosad gene cluster (8%) | |
| 8 | BKM31_2596957-2638933 | Type III PKS | Alkyl-resorcinol | Alkylresorcinol gene cluster (66%) |
| 9 | BKM31_2891146-2912186 | Terpene | ||
| 10 | BKM31_3470395-3514790 | Terpene-lantipeptide | Catenulipeptin gene cluster (60%) | |
| 11 | BKM31_3542307-3586182 | Other | ||
| 12 | BKM31_4190659-4213196 | Lasso peptide | ||
| 13 | BKM31_4679144-4706144 | Lantipeptide | ||
| 14 | BKM31_4941385-4961402 | Bacteriocin-proteusin | Daptomycin gene cluster (3%) | |
| 15 | BKM31_5249766-5270908 | Terpene | 2-Methyl-isoborneol | 2-Methylisoborneol gene cluster (100%) |
| 16 | BKM31_5273165-5342461 | NRPS | Coelichelin | Coelichelin gene cluster (63%) |
| 17 | BKM31_5358340-5379605 | Lantipeptide | Rishirilide B gene cluster (7%) | |
| 18 | BKM31_5380350-5422832 | Lasso peptide | Indigoidine | Indigoidine gene cluster (80%) |
| 19 | BKM31_5421197-5467325 | Type I PKS | Tiacumicin B gene cluster (9%) | |
| 20 | BKM31_5861156-5883651 | Lasso peptide | ||
| 21 | BKM31_6026507-6038816 | Bacteriocin | BE-14106 gene cluster (7%) | |
| 22 | BKM31_6364194-6386746 | Lasso peptide | ||
| 23 | BKM31_7031092-7072453 | Other | ||
| 24 | BKM31_7183678-7226695 | NRPS | ||
| 25 | BKM31_7211748-7316683 | Phenazine | 1,6-Dihydroxy-phenazine | Marinophenazines gene cluster (34%) |
| 26 | BKM31_7681311-7727480 | Type I PKS | Maklamicin gene cluster (6%) | |
| 27 | BKM31_7918324-7969091 | Type II PKS | Xantholipin | Xantholipin gene cluster (42%) |
| 28 | BKM31_8119872-8140816 | Terpene | ||
| 29 | BKM31_8820493-8889698 | NRPS | Pyridomycin gene cluster (7%) | |
| 30 | BKM31_9392432-9413598 | Terpene | Chlortetracycline gene cluster (5%) | |
| 31 | BKM31_10970078-10991184 | Terpene | ||
| 32 | BKM31_12946245-13047416 | NRPS-aryl-polyene | WS9326 gene cluster (25%) |
In an effort to assign additional molecules, we carried out an MS/MS network analysis of the secreted metabolome of ATCC 55076. This led to identification of 1,6-dihydroxy-phenazine, a bioactive molecule that has previously been isolated from bacteria (7, Fig. 1, S3 and Table S2‡).35 A gene cluster coding for a phenazine-like molecule can also be spotted in the genome of ATCC 55076 and is likely responsible for production of 7 (see Table 1, cluster 25). An aromatic secondary metabolite, identified by its UV-vis spectrum, was also produced at appreciable levels in ATCC 55076 cultures. Comparison of its high-resolution mass ([M + H]+obs = 463.1020) to a library of known natural products showed that it was a new compound; we have given it the trivial name nomuricin. It was purified to homogeneity and analyzed by UV-vis spectroscopy and 1D/2D NMR. Nomuricin's key UV-vis feature, a broad absorption peak at ∼460 nm, is consistent with a hydroxylated anthraquinone skeleton, as revealed by comparisons with di-hydroxy-anthraquinone and other hydroxy-anthraquinone-containing natural products (Fig. S4 and Table S3‡). Further analysis by 1D/2D NMR revealed that nomuricin contains a benz[a]anthracycline scaffold with a benzoic acid moiety and an unusual hydroxymethyl substituent (10, Fig. 1, Fig. S4–S6 and Table S3 and S4‡). This scaffold is shared with the pradimicin aglycone (11), which has been isolated from Actinomadura hibiscus (Fig. S6‡),36,37 a relative of ATCC 55076. Mosher ester analysis revealed the R stereoconfiguration for the chiral carbon bearing the hydroxyl group (Fig. 1 and S7‡), and thus completed the structural assignment of nomuricin.
Anthracycline scaffolds are typically biosynthesized by type II polyketide synthases (PKS). The genome of ATCC 55076 contains only one such gene cluster, which shares significant homology with the xantholipin biosynthetic genes (12, Fig. 1, Table 1).38 We suspect that this type II PKS cluster is responsible for nomuricin biosynthesis, though additional studies are necessary to test this hypothesis. Nomuricin did not exhibit growth-inhibitory activity when tested against S. aureus, Bacillus subtilis, Escherichia coli, Pseudomonas aeruginosa or Saccharomyces cerevisiae at concentrations up to 50 μg mL−1. Broader screens are necessary to characterize the biological activity of nomuricin.
2.4 The kistamicin biosynthetic gene cluster
The kistamicin gene cluster was among those that could be assigned bioinformatically because of its homology to known GPA clusters. A comparison of the complestatin and kistamicin gene clusters is shown (Fig. 4A and Table S5‡). Like complestatin, the kis cluster contains four large non-ribosomal peptide synthetases (NRPSs, KisI–KisL). The predicted A-domain specificities are consistent with the amino acid sequence of kistamicin. An X-domain can also be found at the C-terminal end of the ultimate NRPS (KisL), which is likely important for orchestrating the aryl coupling reactions.18 Interestingly, the kis cluster only encodes two P450 enzymes (KisN and KisO), suggesting that one of these catalyzes formation of two crosslinks, possibly the two biaryl ether linkages, leaving the second P450 enzyme for the phenol–indole modification. Alternatively, another P450 enzyme could be encoded elsewhere in the genome; however, a search of the remainder of the ATCC 55076 genome did not reveal a P450 enzyme with sufficiently homology. Accordingly, kis is the first GPA cluster in which a bifunctional P450 enzyme is implicated. In all other GPA clusters, a distinct P450 enzyme has been proposed for each aromatic crosslinking reaction.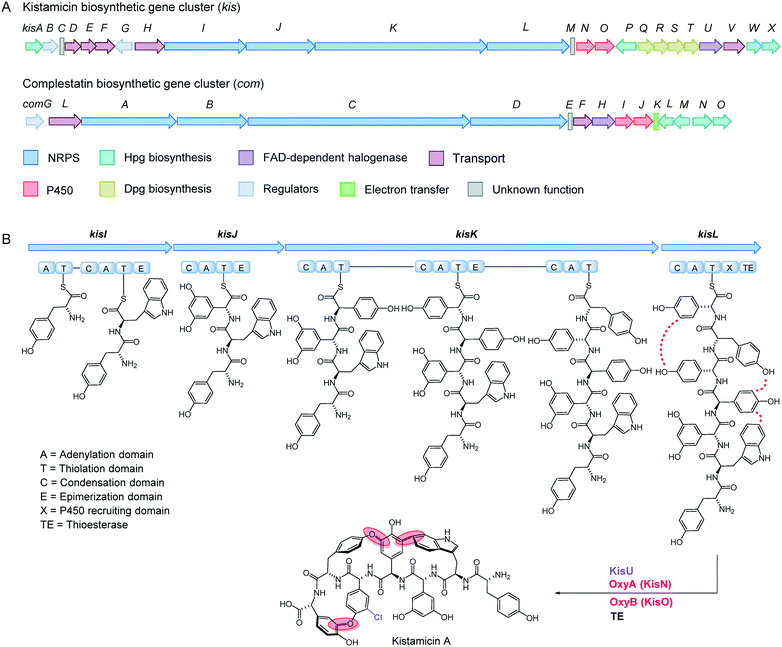 | ||
| Fig. 4 Gene cluster and biosynthetic model for kistamicin. (A) Comparison of the complestatin and kistamicin biosynthetic gene clusters. Genes are colour-coded as indicated (Table S5‡). (B) Biosynthetic model for kistamicin. Four NRPS enzymes, KisI–KisL, give rise to a peptidyl-carrier protein (PCP)-bound 7mer. Chlorination of the fifth residue, followed by three aromatic cross-coupling reactions, catalyzed by KisN and KisO, and TE-catalyzed cleavage from the terminal PCP yields kistamicin A. | ||
In addition to the NRPS and P450 enzymes, the genes required for synthesis of Hpg (kisA, kisP, kisW, kisX) and Dpg (kisQ–kisT) are also contained in the kis cluster (Fig. 4A).10 An FAD-dependent chlorinase (KisU), probably responsible for generating the 3-chloro-4-hydroxy-phenylglycine at the 5th residue of the peptide chain, can also be identified. The sequence of kis allows us to propose a biosynthetic pathway for kistamicin, which will guide future studies (Fig. 4B). As with vancomycin and complestatin, we propose production of the 7mer peptide as a thioester-conjugate in a typical NRPS assembly line fashion. The 7mer contains six D-amino acids and one L-amino acid (6th L-Tyr residue). Five epimerization domains can be seen in the four NRPS enzymes suggesting that one perhaps catalyzes two epimerization reactions. In our proposed pathway, the timing of chlorination is based on the recent results by Park et al.,39 in which the complestatin gene cluster was heterologously expressed in S. lividans. In this case, chlorination was observed to precede the cyclization reactions, though the timing of chlorination with respect to synthesis of the 7mer peptide remains to be determined. Upon formation of the chlorinated 7mer peptide, we propose that one P450 enzyme catalyzes both aryl ether cyclization reactions, while the second completes kistamicin synthesis by installing the phenol–indole crosslink, again in accordance with recent results with complestatin. The lack of a resistance operon in the kis cluster is consistent with a natural function for kistamicin that excludes antibacterial activity or with immunity imparted by other aspects of this strain's physiology.
2.5 Expression and purification of OxyAkis and OxyBkis
As mentioned above, aromatic cross-coupling P450 enzymes are one of the hallmarks of GPA biosynthetic gene clusters. To further characterize this aspect of the kis cluster, we cloned and successfully expressed both P450 enzymes (KisN and KisO) with an N-terminal His6-tag in E. coli (Table S6‡). Subsequent isolation via immobilized metal affinity chromatography followed by thrombin-mediated cleavage of the His6-tag yielded purified enzyme (Fig. 5A). UV-vis analysis of both as-purified enzymes showed that they contain the expected heme Soret bands with λmax of 360 and 415 nm, as well as the so-called Q-bands at 535 and 565 nm, indicative of a typical low-spin FeIII-heme (Fig. 5B and S8‡). The UV-vis spectrum revealed substoichiometric levels of heme, with approx. 40–60% of the protein purified in the apo-form. In vitro reconstitution with the cofactor led to stoichiometric heme content within both proteins. The P450 enzymes in the kistamicin biosynthetic pathway bind heme and can be purified in high yields setting the stage for future studies with the synthetic substrate. Enzymatic activity of only a small number of GPA P450 enzymes has been reconstituted in vitro.16–20 These assays are complicated by the laborious procedures needed to prepare the substrate, a complex 7mer peptide that is PCP-tethered via a pendant phosphopantetheine. Nonetheless, the identification of the kis gene cluster and its P450 enzymes provides additional opportunities to examine the fascinating aromatic cross-coupling enzymes.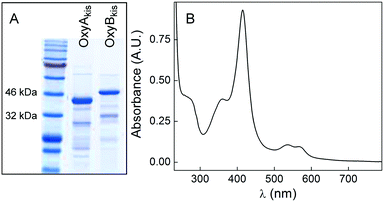 | ||
| Fig. 5 Purification of KisN (OxyAkis) and KisO (OxyBkis). (A) SDS-PAGE gel of Ni-NTA-purified OxyA (exp. 43 kDa) and OxyB (exp. 47 kDa). (B) UV-vis spectrum of purified OxyAkis exhibiting spectral properties typical for low-spin FeIII-heme. A similar spectrum was obtained for OxyBkis (see Fig. S8‡). | ||
3. Conclusions
We have determined the first complete genome sequence of a Nonomuraea species and the sequence of the kistamicin biosynthetic gene cluster. In addition to their remarkable bioactivities, GPAs also boast complex structures that have inspired synthetic and biosynthetic chemists alike. Heroic efforts have led to the total synthesis of a number of GPAs but their biosyntheses, especially installation of the unusual bis-phenol and phenol–indole crosslinks, remain puzzling.40–42 Our findings will allow future examination of these facets of kistamicin biosynthesis, especially the putative bifunctional P450 enzyme, which may catalyze two crosslinking reactions. Purification of both P450 enzymes reported herein is a first step toward that goal.We also report that Nonomuraea sp. ATCC 55076 contains the largest actinobacterial genome sequenced to date. This strain has a great potential for secondary metabolite biosynthesis with a large amount of genome real estate dedicated to this process. The products of five gene clusters can be putatively assigned in this report. Aside from the kistamicin, geosmin, and 2-methylisoborneol clusters, we also identified 1,6-dihydroxyphenazine as well as nomuricin, a new anthracycline-type secondary metabolite. Collectively, our results suggest that ATCC 55076 will serve as a fruitful target for future genome mining efforts in the hopes of discovering new, bioactive natural products.
4. Experimental
4.1 Strain, media, and general procedures
Nonomuraea sp. ATCC 55076 was obtained from the American Type Culture Collection (ATCC). Glucose-fish meal medium (GFM) was used for production of kistamicin and other secondary metabolites as previously reported.23 Fish meal was obtained from Petco. Oatmeal agar (ISP Medium 3) was used for sporulation and preparation of mycelial stocks. A modified R4 liquid medium was used as pre-cultures, which were used to inoculate GFM medium (see below). The modified R4 medium contains (per litter) 5 g glucose, 0.5 g of yeast extract, 50 mg casamino acids, 1.5 g L-proline, 1.18 g L-valine, 5 g MgCl2 hexahydrate, 2 g CaCl2 dihydrate, 0.1 g K2SO4, 2.8 g TES and 1 mL of trace elements. It was adjusted to pH 7.2 using NaOH.43 HPLC separations were carried out on an Agilent 1260 Infinity Series analytical or preparative HPLC system equipped with a photodiode array detector and an automated fraction collector. High-resolution HPLC-MS was performed on an Agilent UHD Accurate Mass Q-tof LC-MS system, equipped with a 1260 Infiinty series HPLC, a JetStream ESI source, and the 6540 Q-tof. Samples were typically resolved on an Agilent Poroshell 120 EC-C18 column (3 × 50 mm, 2.7 μm) at 0.4 mL min−1 using water and MeCN (both containing 0.1% formic acid). NMR spectra were acquired at the Princeton University Department of Chemistry NMR Facilities. Spectra were collected in MeOD-d4 in the triple resonance cryoprobe of a Bruker A8 Avance III HD 800 MHz NMR spectrometer.4.2 Whole genome sequencing
Genomic DNA from Nonomuraea sp. ATCC 55076 was extracted from mycelia grown in modified R4 medium using standard protocols previously described.44 Briefly, the strain was grown in modified R4 medium at 30 °C and 250 rpm for 48 h. Then genomic DNA was isolated using lysozyme and sodium dodecyl sulphate for cell disruption followed by a phenol-chloroform extraction. The genomic DNA was submitted to the UCI Genomics High-Throughput Facility (GHTF) for library preparation and sequencing. A total of 227![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 454 reads, each consisting of 7850 bp, were derived from 3 SMRT cell. These were de novo assembled using HGAP 3.0.26 Genome annotation was carried out using the RAST29 server and graphical mapping was performed with the CIRCOS visualization engine.
454 reads, each consisting of 7850 bp, were derived from 3 SMRT cell. These were de novo assembled using HGAP 3.0.26 Genome annotation was carried out using the RAST29 server and graphical mapping was performed with the CIRCOS visualization engine.
4.3 Production and purification of kistamicin
Kistamicin production was not observed in ISP2, ISP3, ISP4, and R4 media. To obtain kistamicin, ATCC 55076 was pre-cultured on oatmeal agar for 3 days. The collected mycelium and spores were used to inoculate 0.2 L of modified R4 liquid medium. The pre-cultures were incubated at 250 rpm and 30 °C. After 3 days, the mycelium was collected by centrifugation and used to inoculate 6 × 1 L flasks (each containing 200 mL GFM) at a concentration of 1% (w/v). The GFM cultures were grown at 250 rpm and 30 °C for 9 days. Mycelia were isolated by centrifugation, combined, mixed with 30 mL acetone and vortexed for one minute. Subsequently, the acetone extract was filtered off and the process was repeated. The acetone extracts were combined, dried in vacuo, and the residue dissolved in MeOH for analysis by NMR (Fig. S1 and S2‡) and HPLC-Qtof-MS ([M + H]+calc 1171.3241, [M + H]+obs 1171.3256).4.4 Production and purification of 1,6-dihydroxyphenazine (7)
A total of 4 L of Nonomuraea sp. ATCC 55076 were grown in GFM medium at 250 rpm and 30 °C for 9 days, as described above. Cells were removed by centrifugation and the supernatant extracted twice with acidified ethyl acetate (supplemented with 0.1% formic acid). The extract was dried over Na2SO4, then dried completely in vacuo. The residue was resuspended in MeOH and resolved on a preparative Agilent HPLC on a Phenomenex Luna C18 column (5 μm, 21.2 × 250 mm) at a flow rate of 12 mL min−1. The solvent system consisted of water and MeCN, each containing 0.1% formic acid. A gradient from 20% to 100% MeCN over 40 min was used to purify compound 7, which eluted at 65% MeCN. Compound 7 was purified further on an Agilent Eclipse C8 column (5 μm, 4.6 × 150 mm), a Supelco C18-PEG column (4 μm, 4.6 × 250 mm), and finally a Phenomenex Kinetex Phenyl-hexyl column (5 μm, 4.6 × 100 mm). Elution on each of these columns was performed isocratically at 40% MeCN at 1 mL min−1 by monitoring the signal at 280 nm. Pure 7 was characterized by NMR (Fig. S3 and Table S2‡) and HR-MS, which gave [M + H]+obs 213.0654 ([M + H]+calc 213.0664).4.5 Production and purification of nomuricin (10)
A total of 4 L of Nonomuraea sp. ATCC 55076 were grown in GFM medium at 250 rpm and 30 °C for 9 days, as described above. Cells were removed by centrifugation and the supernatant extracted twice with acidified ethyl acetate (0.1% formic acid). After drying over Na2SO4, the extract was evaporated in vacuo, and the resulting residue resuspended in acetone. Purification of 10 was performed on an Agilent preparative HPLC using a Phenomenex Luna C18 column (5 μm, 21.2 × 250 mm) operating at 12 mL min−1. A gradient from 20% to 100% MeCN over 40 min was used to purify compound 10, which eluted at 75% MeCN. Compound 10 was purified further on a Phenomenex Phenyl-hexyl column (5 μm, 4.6 × 100 mm), a Supelco C18-PEG column (4 μm, 4.6 × 250 mm), and finally an Agilent Eclipse C8 column (5 μm, 4.6 × 150 mm). Elution on each of these columns was performed isocratically at 45% MeCN at 1 mL min−1. Pure 10 was characterized by UV-vis spectroscopy, NMR (Fig. S4–S6 and Table S3 and S4‡) and HR-MS, which gave [M + H]+obs 463.1035 ([M + H]+calc 463.1029).4.6 Cloning, expression, and purification of OxyAkis and OxyBkis
The oxyA and oxyB genes from the kistamicin gene cluster in Nonomuraea sp. ATCC 55076 were amplified by PCR using kisN–F/kisN–R and kisO–F/kis-O–R primer sets (see Table S6‡) and cloned separately into the NdeI and HindIII restriction sites of a pET-28b expression vector, which contains an N-terminal His-tag. The resulting plasmids were transformed into E. coli KRX cells for expression of OxyA and OxyB.
E. coli KRX cells containing pET-28b_oxyA or pET-28b_oxyB were cultured in TB media at 37 °C, shaking at 200 rpm until the cultures reached an OD600nm between 0.4 and 0.6. Cultures were then induced for 24 hours at 18 °C in the presence of 0.2 mM IPTG and 0.5 mM δ-aminolevulinic acid. Following induction, cells were harvested by centrifugation at 8000 g for 20 minutes and then resuspended in 2 mL lysis buffer (50 mM Tris, pH 7.8, 50 mM NaCl, 10 mM imidazole, 5% glycerol, 1 mM β-mercaptoethanol, 1 mg mL−1 lysozyme, 1 μL mL−1 protease cocktail) per gram of cell pellet. Following a 1 hour incubation time, the cells were lysed by sonication, and the lysate cleared of cell debris by centrifugation at 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 1 hour.
000g for 1 hour.
OxyA and OxyB were purified from the soluble fractions of respective lysates by Ni-NTA chromatography. Upon loading, elution was carried out with lysis buffer + 100 mM imidzole. The proteins were then desalted on a Sephadex G-25 column. Both OxyA and OxyB exhibited incomplete heme loading following purification as inferred from the UV-visible spectrum (A408![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) A280 ratios of 1
A280 ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for OxyA and 1
1 for OxyA and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 for OxyB). Both enzymes were incubated with 10-fold excess hemin on ice for 1 hour. Following reconstitution, excess hemin was removed and the buffer exchanged by Sephadex G-25 chromatography (for 50 mM HEPES, pH 6.8, 50 mM KCl). UV-visible spectra of the reconstituted OxyA and OxyB showed A408
2 for OxyB). Both enzymes were incubated with 10-fold excess hemin on ice for 1 hour. Following reconstitution, excess hemin was removed and the buffer exchanged by Sephadex G-25 chromatography (for 50 mM HEPES, pH 6.8, 50 mM KCl). UV-visible spectra of the reconstituted OxyA and OxyB showed A408![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) A280 ratios of approximately 2
A280 ratios of approximately 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for both enzymes, indicative of complete heme loading.
1 for both enzymes, indicative of complete heme loading.
Acknowledgements
We thank Dr. Fei Xu for assistance with nomuricin bioactivity assays, as well as the Pew Biomedical Scholars Program and the Princeton Environmental Institute Innovative Research Award for generous support of this work. C. F. and K. R. S. were supported by a Edward C. Taylor 3rd year Fellowship and an Eli Lily-Edward C. Taylor Fellowship in Chemistry, respectively.References
- D. P. Levine, Clin. Infect. Dis., 2006, 42, 55 CrossRef PubMed.
- K. C. Nicolaou, C. N. Boddy, S. Bräse and N. Winssinger, Angew. Chem., Int. Ed., 1999, 38, 2096 CrossRef PubMed.
- D. Kahne, C. Leimkuhler, W. Lu and C. Walsh, Chem. Rev., 2000, 105, 425 CrossRef PubMed.
- J. Pootoolal, J. Neu and G. D. Wright, Annu. Rev. Pharmacol. Toxicol., 2002, 42, 381 CrossRef CAS PubMed.
- M. H. McCormick, J. M. McGuire, G. E. Pittenger, R. C. Pittenger and W. M. Stark, Antibiot. Annu., 1955, 3, 606 Search PubMed.
- J. M. McGuire, R. N. Wolfe and D. W. Ziegler, Antibiot. Annu., 1955, 3, 612 Search PubMed.
- M. R. Bardone, M. Paternoster and C. Coronelli, J. Antibiot., 1978, 31, 170 CrossRef CAS PubMed.
- T. D. H. Bugg, G. D. Wright, S. Dutka-Malen, M. Arthur, P. Courvalin and C. T. Walsh, Biochemistry, 1991, 30, 10408 CrossRef CAS PubMed.
- T. D. H. Bugg, S. Dutka-Malen, P. Arthur, P. Courvalin and C. T. Walsh, Biochemistry, 1991, 30, 2017 CrossRef CAS PubMed.
- B. K. Hubbard and C. T. Walsh, Angew. Chem., Int. Ed., 2003, 42, 730 CrossRef CAS PubMed.
- G. Yim, M. N. Thaker, K. Koteva and G. Wright, J. Antibiot., 2014, 67, 31 CrossRef CAS PubMed.
- D. Bischoff, B. Bister, M. Bertazzo, V. Pfeifer, E. Stegmann, G. J. Nicholson, S. Keller, S. Pelzer, W. Wohlleben and R. D. Süssmuth, ChemBioChem, 2005, 6, 267 CrossRef CAS PubMed.
- D. Bischoff, S. Pelzer, B. Bister, G. J. Nicholson, S. Stockert, M. Schirle, W. Wohlleben, G. Jung and R. D. Süssmuth, Angew. Chem., Int. Ed., 2001, 40, 4688 CrossRef CAS PubMed.
- D. Bischoff, S. Pelzer, A. Höltzel, G. J. Nicholson, S. Stockert, W. Wohlleben, G. Jung and R. D. Süssmuth, Angew. Chem., Int. Ed., 2001, 40, 1693 CrossRef CAS PubMed.
- R. D. Süssmuth, S. Pelzer, G. Nicholson, T. Walk, W. Wohlleben and G. Jung, Angew. Chem., Int. Ed., 1999, 38, 1976 CrossRef.
- K. Zerbe, K. Woithe, D. B. Li, F. Vitali, L. Bigler and J. A. Robinson, Angew. Chem., Int. Ed., 2004, 43, 6709 CrossRef CAS PubMed.
- K. Woithe, N. Geib, K. Zerbe, D. B. Li, M. Heck, S. Fournier-Rousset, O. Meyer, F. Vitali, N. Matoba, K. Abou-Hadeed and J. A. Robinson, J. Am. Chem. Soc., 2007, 129, 6887 CrossRef CAS PubMed.
- K. Haslinger, M. Peschke, C. Brieke, E. Maximowitsch and M. J. Cryle, Nature, 2015, 521, 105 CrossRef CAS PubMed.
- K. Haslinger, E. Maximowitsch, C. Brieke, A. Koch and M. J. Cryle, ChemBioChem, 2014, 15, 2719 CrossRef CAS PubMed.
- M. Peschke, C. Brieke and M. J. Cryle, Sci. Rep., 2016, 6, 35584 CrossRef CAS PubMed.
- H. T. Chiu, B. K. Hubbard, A. N. Shah, J. Eide, R. A. Fredenburg, C. T. Walsh and C. Khosla, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 8548 CrossRef CAS PubMed.
- T. Fujioka, K. Furihata, I. Kaneko, S. Takahashi and H. Seto, J. Antibiot., 1988, 41, 606 CAS.
- N. Naruse, M. Oka, M. Konishi and T. Oki, J. Antibiot., 1993, 46, 1812 CrossRef CAS PubMed.
- H. B. Bode, B. Bethe, R. Höfs and A. Zeeck, ChemBioChem, 2002, 3, 619 CrossRef CAS PubMed.
- J. Eid, et al. , Science, 2009, 323, 133 CrossRef CAS PubMed.
- C. S. Chin, D. H. Alexander, P. Marks, A. A. Klammer, J. Drake, C. Heiner, A. Clum, A. Copeland, J. Huddleston, E. E. Eichler, S. W. Turner and J. Korlach, Nat. Methods, 2013, 10, 563 CrossRef CAS PubMed.
- R. Sungthong and N. Nakaew, J. Basic Microbiol., 2015, 55, 554 CrossRef PubMed.
- X. J. Wang, et al. , J. Bacteriol., 2010, 192, 4526 CrossRef CAS PubMed.
- R. K. Aziz, et al. , BMC Genomics, 2008, 9, 75 CrossRef PubMed.
- T. Weber, et al. , Nucleic Acids Res., 2015, 43, W237 CrossRef PubMed.
- G. L. Challis and D. A. Hopwood, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 14555 CrossRef CAS PubMed.
- M. H. Medema, et al. , Nat. Chem. Biol., 2015, 11, 625 CrossRef CAS PubMed.
- D. Yu, F. Xu, J. Valiente, S. Wang and J. Zhan, J. Ind. Microbiol. Biotechnol., 2013, 40, 159 CrossRef CAS PubMed.
- S. Lautru, R. J. Deeth, L. M. Bailey and G. L. Challis, Nat. Chem. Biol., 2005, 1, 265 CrossRef CAS PubMed.
- H. Akabori and M. Nakamura, J. Antibiot., 1959, 12, 17 CAS.
- T. Oki, M. Konishi, K. Tomatsu, K. Tomita, K. Saitoh, M. Tsunakawa, M. Nishio, T. Miyaki and H. Kawaguchi, J. Antibiot., 1988, 41, 1701 CrossRef CAS PubMed.
- Y. Sawada, T. Tsuno, T. Ueki, H. Yamamoto, Y. Fukagawa and T. Oki, J. Antibiot., 1993, 46, 507 CrossRef CAS PubMed.
- W. Zhang, L. Wang, L. Kong, T. Wang, Y. Chu, Z. Deng and D. You, Chem. Biol., 2012, 19, 422 CrossRef CAS PubMed.
- O. K. Park, H. Y. Choi, G. W. Kim and W. G. Kim, ChemBioChem, 2016, 17, 1725 CrossRef CAS PubMed.
- D. A. Evans, M. R. Wood, B. W. Trotter, T. I. Richardson, J. C. Barrow and J. L. Katz, Angew. Chem., Int. Ed., 1998, 37, 2700 CrossRef CAS.
- K. C. Nicolaou, H. J. Mitchell, N. F. Jain, N. Winssinger, R. Hughes and T. Bando, Angew. Chem., Int. Ed., 1999, 38, 240 CrossRef CAS.
- D. L. Boger, S. Miyazaki, S. H. Kim, J. H. Wu, S. L. Castle, O. Loiseleur and Q. Jin, J. Am. Chem. Soc., 1999, 121, 10004 CrossRef CAS.
- J. Shima, A. Hesketh, S. Okamoto, S. Kawamoto and K. Ochi, J. Bacteriol., 1996, 178, 7276 CrossRef CAS PubMed.
- T. Kieser, M. J. Bibb, M. J. Buttner, K. F. Chater and D. A. Hopwood, Practical Streptomyces Genetics, John Innes Foundation, Norwich (UK), 2000, pp. 4161–4171 Search PubMed.
Footnotes |
| † The authors declare no competing interests. |
| ‡ Electronic supplementary information (ESI) available. See DOI: 10.1039/c6md00637j |
| This journal is © The Royal Society of Chemistry 2017 |