vhfRNAi: a web-platform for analysis of host genes involved in viral infections discovered by genome wide RNAi screens†
Abstract
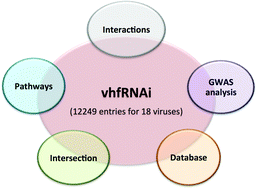
Knockdown of host genes using high-throughput genome-wide RNA interference screens has identified numerous host factors that affect viral infections, which would be helpful in understanding host–virus interactions. We have developed a vhfRNAi web resource based on genome-wide RNAi experiments for viruses. It contains experimental details of 12 249 entries (host factors + restriction factors) for 18 viruses. Simultaneously, this resource encompasses analysis of overlapping genes, genome wide association studies, gene ontology (GO), pathogen interacting proteins, interaction networks and pathway enrichment. Using overlap analysis, it was found that Influenza A virus shared overlapping host genes with the majority of viruses including Hepatitis C virus and Dengue virus 2. In the genome wide association studies analysis, 429 diseases/traits were mapped, of which obesity-related traits were the most common. GO analysis revealed that the major categories belonged to metabolic processes, molecule transport, signal transduction, proteolysis, etc. In the pathogen interacting protein analysis, protein interaction data from different resources can be explored for further understanding of host–virus biology. By pathway enrichment analysis, a total of 8955 genes were mapped on 303 pathways with most of the hits coming from metabolic pathways. We have found 491 genes that are not essential for the host but essential for the virus and can be targeted to inhibit the virus. These may be explored as potential candidates for drug targets. The resource is freely accessible at http://bioinfo.imtech.res.in/manojk/vhfrnai and will be useful in understanding host–virus biology as well as identification of targets for the development of antiviral therapeutics.
- This article is part of the themed collection: Coronavirus articles - free to access collection


 Please wait while we load your content...
Please wait while we load your content...