Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Biocompatible 3D printed polymers via fused deposition modelling direct C2C12 cellular phenotype in vitro†
Rowan P.
Rimington
 a,
Andrew J.
Capel
a,
Andrew J.
Capel
 ab,
Steven D. R.
Christie
ab,
Steven D. R.
Christie
 b and
Mark P.
Lewis
b and
Mark P.
Lewis
 *a
*a
aSchool of Sport, Exercise and Health Sciences, Loughborough University, Loughborough, Leicestershire LE11 3TU, UK. E-mail: M.P.Lewis@lboro.ac.uk; Tel: +44 (0)1509226430
bDepartment of Chemistry, School of Science, Loughborough University, Loughborough, Leicestershire LE11 3TU, UK
First published on 1st August 2017
Abstract
The capability to 3D print bespoke biologically receptive parts within short time periods has driven the growing prevalence of additive manufacture (AM) technology within biological settings, however limited research concerning cellular interaction with 3D printed polymers has been undertaken. In this work, we used skeletal muscle C2C12 cell line in order to ascertain critical evidence of cellular behaviour in response to multiple bio-receptive candidate polymers; polylactic acid (PLA), acrylonitrile butadiene styrene (ABS), polyethylene terephthalate (PET) and polycarbonate (PC) 3D printed via fused deposition modelling (FDM). The extrusion based nature of FDM elicited polymer specific topographies, within which C2C12 cells exhibited reduced metabolic activity when compared to optimised surfaces of tissue culture plastic, however assay viability readings remained high across polymers outlining viable phenotypes. C2C12 cells exhibited consistently high levels of morphological alignment across polymers, however differential myotube widths and levels of transcriptional myogenin expression appeared to demonstrate response specific thresholds at which varying polymer selection potentiates cellular differentiation, elicits pre-mature early myotube formation and directs subsequent morphological phenotype. Here we observed biocompatible AM polymers manufactured via FDM, which also appear to hold the potential to simultaneously manipulate the desired biological phenotype and enhance the biomimicry of skeletal muscle cells in vitro via AM polymer choice and careful selection of machine processing parameters. When considered in combination with the associated design freedom of AM, this may provide the opportunity to not only enhance the efficiency of creating biomimetic models, but also to precisely control the biological output within such scaffolds.
1. Introduction
Additive manufacture (AM), commonly known as three dimensional (3D) printing, allows the generation of physical devices within short time-periods from bespoke computer aided designs (CAD), with technologies such as laser sintering (LS), stereolithography (SL), fused deposition modelling (FDM) and polyjet printing being utilised within biochemical engineering for the fabrication of microfluidic reaction devices for flow chemistry,1 live-cell imaging2 and cellular scaffolds for tissue engineering.3 Although processes such as SL allow researchers to print complex micron-sized geometries,4,5 epoxy/acrylate based resins typically result in biologically non-compatible6 parts which require additional surface modification7 prior to cellular interaction.8 Such complex geometries can be replicated using LS, however consolidation variability of sintered particles within builds have been documented to prevent removal of un-sintered material from within micron-sized features.9 Although 3D printed cellular devices have been fabricated using polyjet printing, they have also required polymer coatings in order to improve cellular adherence.10 Typical photocurable resins, as seen in polyjet and SL, contain photoinitiators and similar acrylate/epoxy backbones that may alter intricate cellular physiological processes via the uptake and subsequent metabolism of resin derived chemical leachate within the culture medium, in addition to primary interaction with the physico-chemical parameters of the polymer. However, the exact mechanisms that drive the observed decremental cellular viability in response to photocurable resins are yet to be elucidated in mammalian cells.FDM is the most widely used, inexpensive and commercially available 3D printing process, using a temperature controlled extrusion nozzle to deposit a viscous molten thermoplastic polymer into a three-dimensional structure.11 Various commercially available FDM printable polymers have previously been utilised within cellular tissue culture: acrylonitrile butadiene styrene (ABS),12 polycarbonate (PC),13 polylactic acid (PLA)14 and polyethylene terephthalate (PET),15,16 hence are considered favorable candidate polymers for rapid prototyping of biologically receptive parts. Although FDM has previously been utilised in flow chemistry17 and the formation of acellular microfluidic channels,18 utilising FDM for microfluidic applications may result in increasing complexity when implementing this technology within biological settings due to the specificity of cell/material interactions19,20 in response to alterations in topography, surface roughness and other fundamental physical properties of the polymer.
The design freedom associated with 3D printing technology has reduced the complexity and enhanced the efficiency of the engineering methods required to create biological models,21–23 however despite the growing prevalence of 3D printing, limited research regarding cellular compatibility has been undertaken. As such, if these technologies are going to form the platforms within which complex cellular physiological processes are to be re-created, then understanding and defining the compatibility between various mammalian cell types and the printed materials is of paramount importance.24,25 Skeletal muscle is of specific interest due to its regenerative capacity via multipotent stem cells known as satellite cells,26 predicating its use within in vitro models27 as pre-clinical test beds to study the cellular and molecular mechanisms that are regulated in musculoskeletal and neuromuscular disease.28 Cell lines such as C2C12 murine skeletal myoblasts29 are frequently used within these models due to a lack of availability of primary human muscle cells, and their capacity to provide a biologically relevant, repeatable and reliable model of in vitro skeletal muscle.
Due to the potential to implement 3D printed parts to perform differing roles within varying biological systems, it is essential to determine both the in-direct and direct effects of printable polymers. Hence, this investigation sought to examine the direct (cells cultured directly on the material) and in-direct (cells cultured in chemically leached medium) effects of four commercially available and printable polymers; PLA, ABS, PET and PC on C2C12 muscle cells when 3D printed via FDM.
2. Materials and methods
2.1. 3D printing and sample design
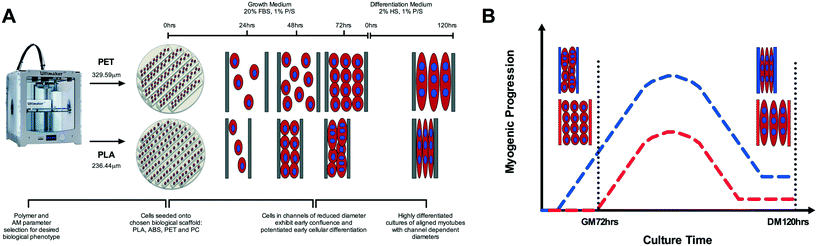
All 3D printing was performed via fused deposition modelling (Fig. 1) utilising a commercially available Ultimaker 2 system. 3D CAD modelling was performed using Siemens NX 8.5 software; with completed .stl files verified using Materialise MiniMagics. Completed .stl files were processed using the in-house Cura Software for Ultimaker 2. | ||
| Fig. 1 (A) CAD model of sample prior to printing. (B & C) Ultimaker 2 during the printing of sample discs. | ||
Prior to printing of ABS and PC parts, the build platform was coated in an ABS/acetone solution (0.1 g ml−1) in order to facilitate material adhesion. All other polymers were extruded onto the standard glass build plate. Printing parameters were fixed at 0.4 mm nozzle size, 50 mm s−1 print speed, 0.1 mm layer height and 100% fill density, with polymer dependent extrusion settings (Table 1). Each sample scaffold had a diameter of 30 mm and thickness of 1 mm, designed to cover the majority of the culture area of a six-well plate. All 3D printed scaffolds were sterilised under UV light for ≥30 minutes prior to each experiment.
| Polymer | Nozzle temperature (°C) | Build plate temperature (°C) | Fan (%) | Flow (%) |
|---|---|---|---|---|
| Polylactic acid | 210 | 60 | 100 | 100 |
| Acrylonitrile butadiene styrene | 260 | 90 | 100 | 107 |
| Polyethylene terephthalate | 210 | 75 | 100 | 100 |
| Polycarbonate | 260 | 90 | 100 | 100 |
2.2. Cell culture
C2C12 murine skeletal myoblasts, (ECACC, Sigma, UK) all below passage 10, were cultured in T80 flasks (Nunc™, Fisher Scientific, UK) and incubated in a 5% CO2 humidified atmosphere at 37 °C (HERA cell 240i, Thermo Fisher, UK) in growth medium (GM); composed of 79% DMEM (Sigma), 20% FBS (Pan Biotech) and 1% penicillin–streptomycin (Fisher) until 80% confluence was obtained. Incubated cells GM was changed every 24 h until confluence was attained, prior to transfer into differentiation medium (DM); 97% DMEM, 2% horse serum (Sigma) and 1% penicillin–streptomycin for 120 h. C2C12 cell DM was changed once after 72 h, prior to remaining in culture for a further 48 h.2.3. Experimental treatments
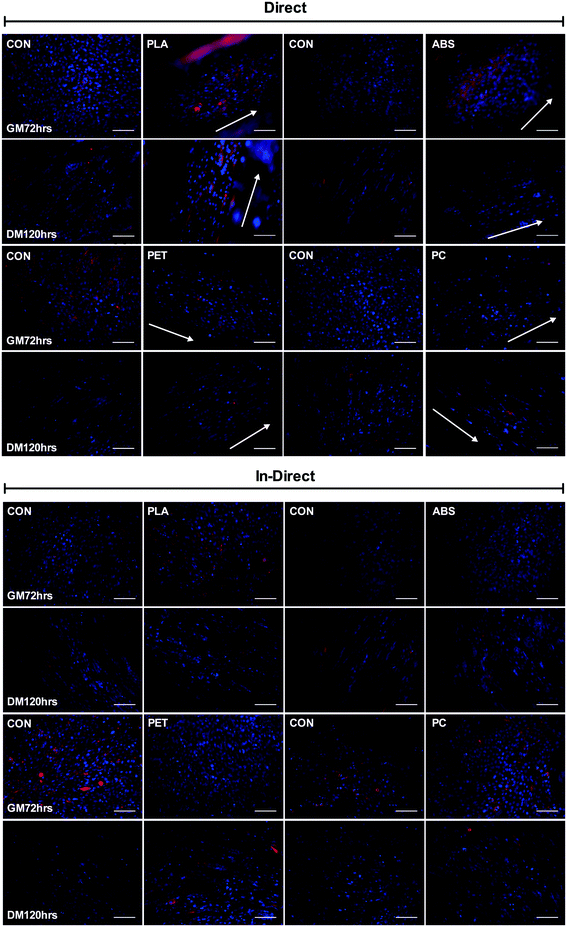
Experiments were designed to simultaneously assess both the direct and in-direct effects of each polymer on cellular viability, morphology and myogenin expression as an indication of skeletal muscle differentiation, after 3 days in GM (GM72 h) and 5 days in DM (DM120 h). Experiments yielded n = 9 per condition (control/polymer), totalling n = 18 per analyses at each time-point (GM72 h and DM120 h), derived from n = 3 experimental repeats. This protocol was completed separately for each polymer (PLA, ABS, PET and PC). One experimental repeat was conducted to assess the biocompatibility of the adhesive (aquarium glue) used to facilitate the attachment of 3D printed discs to the surface of each well. Immunofluorescence staining indicated comparable morphology in C2C12 cells cultured in both direct and in-direct conditions after DM120 h, outlining the biocompatibility of this adhesive (Fig. S1 and S2†).Three 13 mm cover slips (VWR International), added per well in control conditions (direct/in-direct at GM72 h and DM120 h time-points) for fluorescence analysis, were pre-coated with 1 ml of 0.2% gelatin solution (Sigma) and incubated for 15 min at room temperature, prior to seeding cells at 1 × 105 in 2 ml GM per well for all analyses across each experimental repeat.
To assess the direct effects on cellular behaviour, 1 × 105 cells were seeded directly onto the 3D printed scaffolds that had been glued to three wells per six-well plate and left for ≥1 hour to allow for initial cellular adhesion. Once adhered, 2 ml GM was added to each well. Control (CON) wells were also seeded at 1 × 105 cells per well in 2 ml GM. To assess the in-direct cellular response to the chemical leachate from each polymer, 3D printed scaffolds were attached to the culture area of three wells per six-well plate that contained only media (MEDIA ONLY). Each six-well plate was used for the transfer of chemical leachate medium to its corresponding experimental well during each repeat, ensuring that transferred medium was directly representative of either cumulative polymer degradation or CON at each specific time-point. Each MEDIA ONLY well was pre-incubated with 2 ml GM for 24 h prior to the start of each experiment, to ensure cells were seeded within media that had been exposed to the chemical leachate of each polymer. Once medium had been transferred to experimental wells, this was replenished with 2 ml GM for 24 h incubation prior to each subsequent media change. During the last 24 h in which the cells were cultured in GM, DM was added to the MEDIA ONLY wells in preparation to induce differentiation.
Cells were fixed for fluorescence imaging and analysed for cell viability at GM72 h and DM120 h time-points. After cellular viability analysis was conducted, cells were then washed twice with 2 ml PBS, prior to RNA isolation with cells being immersed and homogenised in 0.5 ml TRIzol per well.
2.4. Fluorescence staining and microscopy
Myotubes were washed twice in 2 ml PBS per well, fixed (3.7% paraformaldehyde), permeabilised (Triton X-100, Fisher, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) and stained using f-actin molecular probe rhodamine phalloidin (1
500) and stained using f-actin molecular probe rhodamine phalloidin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200; Life Technologies, Molecular Probes). Dapi nuclear stain (1
200; Life Technologies, Molecular Probes). Dapi nuclear stain (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000; Life Technologies, Molecular Probes) was used to stain nuclei. Rhodamine phalloidin was excited at 540 nm and emitted at 565 nm and appears red in fluorescence images. Dapi was excited at 358 nm and emitted at 461 nm and appears blue.
2000; Life Technologies, Molecular Probes) was used to stain nuclei. Rhodamine phalloidin was excited at 540 nm and emitted at 565 nm and appears red in fluorescence images. Dapi was excited at 358 nm and emitted at 461 nm and appears blue.
Microscopy images were captured on a Leica DMIL LED light microscope. Fluorescence images were visualised using a Leica DM2500 fluorescence microscope (20×) with manufacturer's software (Leica Application Suite X). Images were analysed using IMAGE J 1.50a/Fiji (Java 1.6.0_24) software (National Institute of Health, Bethesda, MD, USA). Image inclusion criteria (n = 1) were set at ≥5 images taken at random locations per well. Myotube inclusion criteria were defined as containing ≥3 nuclei per myotube. Cumulative frequencies were calculated to determine the number of myotubes necessitated per image for each analysis. Myotube alignment was measured by drawing a line through the centre, spanning the entire length of each myotube in Image-J. Topographical channel angle was determined by drawing a line through the centre of each channel in Image-J. Myotube alignment data was then normalised to the angle of each material channel or 90° (CON), sorted by numerical value and analysed using an interquartile range. Orientation values expressed are the difference in alignment between polymer channel and myotube orientation, with lower values indicative of greater alignment. Analysis of nuclei alignment was performed using an in-house macroinstruction designed for Fiji (Java 1.6.0_24) image analysis software (Image J 1.50a).
2.5. Cell viability alamarBlue® assay
Cell viability reagent; alamarBlue® diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 DMEM, was used to assess cell viability in C2C12 at 72 and 120 h experimental time-points. alamarBlue® is non-toxic, stable in culture and permeable through cell membranes allowing continuous monitoring of live cells in culture. Cells were washed twice with 2 ml PBS prior to being treated with 2 ml per well alamarBlue® stock solution and humidified 5% C02 at 37 °C for 4 h. 100 μl per well of solution was then added to a black well 96-well plate and analysed for fluorescence intensity. alamarBlue® fluorescence signal was excited at 540–570 nm (peak excitation: 570 nm) and emitted at 580–610 nm (peak emission: 585 nm).
10 DMEM, was used to assess cell viability in C2C12 at 72 and 120 h experimental time-points. alamarBlue® is non-toxic, stable in culture and permeable through cell membranes allowing continuous monitoring of live cells in culture. Cells were washed twice with 2 ml PBS prior to being treated with 2 ml per well alamarBlue® stock solution and humidified 5% C02 at 37 °C for 4 h. 100 μl per well of solution was then added to a black well 96-well plate and analysed for fluorescence intensity. alamarBlue® fluorescence signal was excited at 540–570 nm (peak excitation: 570 nm) and emitted at 580–610 nm (peak emission: 585 nm).
2.6. RNA extraction and quantitative RT-PCR
RNA was extracted using the TRIzol method, according to manufacturer's instructions (Sigma). RNA concentration and purity were obtained by UV spectroscopy at ODs of 260 and 280 nm using a Nanodrop 2000 (Fisher, Rosklide, Denmark). All RNA samples were analysed in duplicate. Twenty nanograms of RNA were used per RT-PCR reaction for RPII-β & myogenin (Table 2).| Target mRNA | Primer sequence (5′–3′) | Product length | NCBI reference sequence |
|---|---|---|---|
| Myogenin | F: CCAACTGAGATTGTCTGTC | 173 | NM_153789.2 |
| R: GGTGTTAGCCTTATGTGAAT | |||
| RPII-β | F: GGTCAGAAGGGAACTTGTGGTAT | 197 | NM_031189.2 |
| R: GCATCATTAAATGGAGTAGCGTC |
RT-PCR amplifications were carried out using Power SYBR Green RNA-to-CT 1 step kit (Qiagen) on a ViiA™ Real-Time PCR System (Applied Biosystems, Life Technologies), analysed using ViiA™ 7RUO software. RT-PCR procedure was as follows: 50 °C, 10 min (for cDNA synthesis), 95 °C, 5 min (transcriptase inactivation), followed by 95 °C, 10 s (denaturation), 60 °C, 30 s (annealing/extension) for 40 cycles. Relative gene expressions were calculated using the comparative CT (ΔΔCT) equation for normalised expression ratios; relative expression calculated as 2−ΔΔCT, where CT is representative of the threshold cycle. RPII-β was used as the housekeeping gene in all RT-PCR assays. In order to compare conditions, one control sample from each experimental repeat (n = 3) was used as the calibrator condition in the CT (ΔΔCT) equation. RT-PCR data is presented as relative gene expression level, determined by the ΔΔCT equation.
2.7. Scanning electron microscopy (SEM)
All scanning electron micrographs were obtained using a LEO 1530 VP FE-SEM at 5 kV. Before loading into the SEM samples were coated with 20% Au (gold) and 80% Pd (palladium) in an argon atmosphere, using a Q 150R S/E/ES sputter coater for 30 seconds at a current of 20 mA. Images were obtained across 3 printed scaffolds for each polymer (n = 3) with inclusion criteria set at ≥3 images/scaffold. Once acquired, images were analysed using IMAGE J 1.50a/Fiji (Java 1.6.0_24) software for mean channel width across ≥3 channels/image totalling ≥27 channels per polymer, with ≥3 measures obtained per channel.2.8. Statistical analysis
Statistical analyses and significance of data were determined using IBM© SPSS© Statistics version 22. Mauchly's test of sphericity and Shapiro–Wilk tests were used to confirm homogeneity of variance and normal distribution of data respectively. Where parametric assumptions were met, a 2 × 2 ANOVA was used for alamarBlue®, gene expression and nuclei alignment analyses. One-way ANOVA was used to analyse morphological data only concerned with DM120 h time-point; myotube number, myotube width, myotube alignment, in addition to channel width and all inter-polymer differences. Where significant interactions were observed, independent t-tests (t) were used to analyse differences between conditions at specific time-points. Non-parametric Kruskal–Wallis (H) analysis was undertaken where data violated parametric assumptions. All data is reported as mean ± standard deviation (SD). Significance was assumed at P ≤ 0.05.3. Results
3.1. SEM topographical characterisation of 3D printed polymers
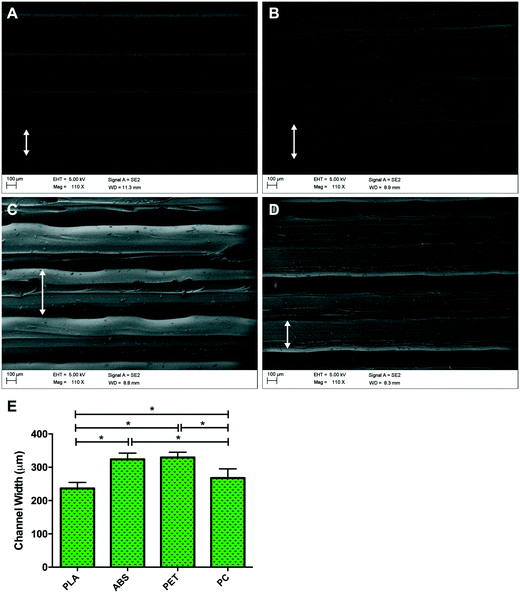
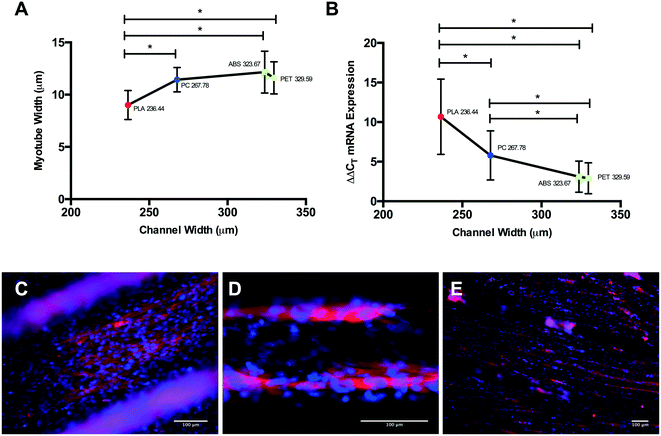
Topographical observations of each polymer (Fig. 2) outlined different channel widths when polymers were 3D printed using FDM, despite standardised nozzle diameter and print mechanics. Channel width was defined as the central raised ‘plateau’ of polymer on each channel, that allowed improved fluorescent micrographic focussing and image analysis compared with the polymer groove on either side of it (Fig. 2). Channel widths (Fig. 2E) observed in PLA had significantly reduced diameter than ABS (P ≤ 0.0005), PET (P ≤ 0.0005) and PC (P = 0.005), however no variation was observed between ABS and PET (P = 0.270). Despite comprising larger channels than PLA, channels with significantly reduced width were also observed in 3D printed PC samples when compared to PET (P = 0.001) and ABS (P = 0.001), outlining potential polymer dependent properties during post extrusion layer cooling. This variability in surface topography between material types is most likely to be a consequence of polymer dependent expansion upon leaving the print head nozzle. This phenomenon, known as the Barus effect or “die swell”, is typically associated with polymer extrusion processes.30 As the molten bead of polymer passes through the nozzle it is constrained with this energy being stored elastically. However, when the polymer melt leaves the print head and is no longer constrained this energy is released, leading to a radial expansion of the melt referred to as die swelling. The phenomenon plays a significant role in determining the resolution achieved in FDM processes. In addition each polymer bead experiences conductive cooling upon hitting the FDM build plate. This cooling process will lead to a reduction in bead width as it solidifies into the final build part. Both of these heating and cooling phases are highly polymer dependent. FDM filaments often contain a variable mixture of raw material and processing aids, leading to some variability between FDM reels of the same polymer. Typically however these reels contain ≥98% of the defined native polymer, making trends in material topography regular and predictable between suppliers and machine manufacturers.313.2. Cell viability
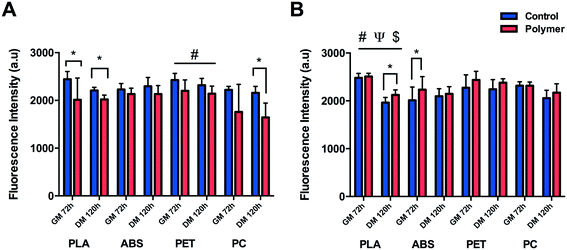
C2C12 cells were shown to display reduced metabolic activity (Fig. 3A) after 72 h in GM (P = 0.047) and 120 h in DM (P = 0.001) when cultured directly onto 3D printed transparent PLA in comparison to control. Reduced cellular activity was also documented in cells cultured on PET with a main effect for condition (P = 0.004), however no main effect for time (P = 0.129) or interaction (P = 0.673) were documented outlining that although metabolic activity appears reduced in the PET condition, this remained constant over eight days in culture. C2C12 cells cultured on PC appeared to display diminished viability after 72 h in GM, however this effect was not significant (P = 0.145) due to the high variability across repeats. This variability was however reduced after 120 h in DM resulting in significantly reduced metabolic activity (P = 0.001) at this time-point. Reduced metabolic activity was also not observed in cells cultured on ABS with no significant difference between condition (P = 0.089), time (P = 0.327) or interaction (P = 0.324).Significant main effects for time (P ≤ 0.0005), condition (P = 0.024) and interaction (P = 0.036) were documented in cells cultured to analyse the potential effect of leached chemical constituents (Fig. 3B) of PLA. No significant differences in metabolic activity were observed between conditions after 72 h in GM, however cells cultured in PLA leachate appeared to be more metabolically active than CON after 120 h in DM (P = 0.008). Significant increases in cellular activity were also documented in ABS after 72 h in GM (P = 0.036) compared to CON, however this was not evident after 120 h in DM (P = 0.462). No significant difference was documented in metabolic activity after 72 h in GM (P = 0.208) or 120 h in DM (P = 0.141) in cells cultured in chemical constituents of PET. No difference in cellular viability was observed in C2C12 cells cultured in chemical leachate of PC after either 72 h in GM (P = 0.848) or 120 h in DM (P = 0.225).
3.3. Morphological analyses
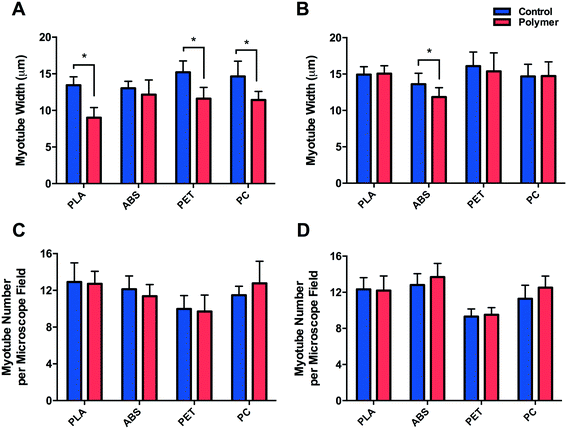
Morphological observations (Fig. 4) outlined significantly reduced myotube width (Fig. 5A) documented in cells cultured on PLA (P ≤ 0.0005), PET (P ≤ 0.0005) and PC (P = 0.008) when compared to their respective controls, however no difference (P = 0.262) was observed between myotubes cultured on ABS and CON. Myotubes did not differ in number between PLA (P = 0.812), ABS (P = 0.091), PET (P = 0.741) or PC (P = 0.246) and their respective controls (Fig. 5C) after 120 h in DM.C2C12 cells cultured in chemically leached medium showed no significant difference in myotube width between CON and PLA (P = 0.784), PET (P = 0.512) or PC (P = 0.957), however thinner myotubes were documented (Fig. 5B) when cultured in ABS chemical leachate (P = 0.016). Myotube number again remained consistent between polymer conditions (Fig. 5D) with no difference documented between CON and PLA (P = 0.848), ABS (P = 0.195), PET (P = 0.609) or PC (P = 0.081).
Despite the reduced myotube width observed in cells cultured in ABS chemical constituents, it is likely that these results may primarily be indicative of biological variability considering no significant difference was observed when cells were cultured directly onto this material; simultaneously interacting with both the physical and chemical characteristics of the polymer. However, due to the homogeneity of myotube widths of cells cultured in PLA, PET and PC chemical leachate compared to their respective controls, it may be plausible that the reduction in myotube width documented in cells cultured directly onto the aforementioned polymers provides an indication of cellular interaction with the physical/physico-chemical characteristics of each polymer.
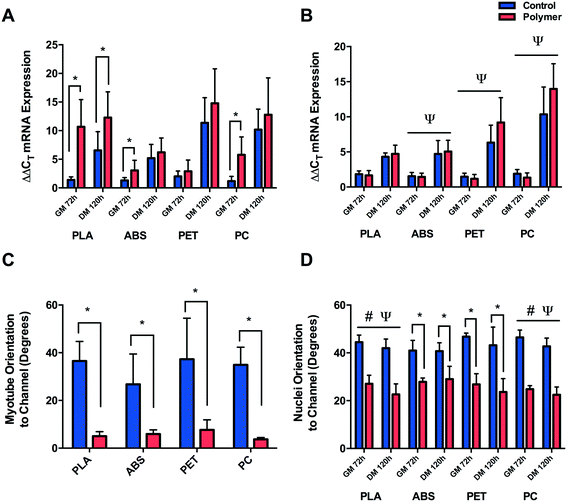
3.4. Myogenin mRNA expression
Myogenin ΔΔCT expression (Fig. 6A) was significantly up-regulated after 72 h in GM (P ≤ 0.0005) and 120 h in DM (P = 0.015) when cultured on PLA compared to CON, suggesting early and potentiated activation of this gene. This response was partially replicated when cells were cultured on ABS and PC, with significant increases in myogenin documented after 72 h in GM (P = 0.046, P ≤ 0.0005), however the early activation observed did not remain up-regulated after 120 h in DM (P = 0.674, P = 0.354) compared to CON. Cells cultured on PET displayed no difference in early myogenin expression after 72 h in GM (P = 0.233) or after 120 h in DM (P = 0.133).No significant difference was observed in cells cultured in PLA chemical constituents at either GM72 (P = 0.294) or DM120 h (P = 0.563) compared to CON. Main effects for time were documented in cells cultured in ABS (P ≤ 0.0005), PET (P ≤ 0.0005) and PC (P ≤ 0.0005) chemical leachate, however no main effects for condition (P = 0.899, P = 0.186, P = 0.354) or interaction were observed (P = 0.743, P = 0.076, P = 0.147) outlining consistent increases in myogenin ΔΔCT expression in both conditions between GM72 h and DM120 h time-points (Fig. 6B).
3.5. Nuclear and myofibrillar alignment
Significant myofibrillar alignment was documented in cells cultured on PLA (P ≤ 0.0005), ABS (P = 0.006), PET (P = 0.001) and PC (P = 0.004) compared to CON after 120 h in DM (Fig. 6C). Nuclear alignment was also documented in cells cultured on PLA and PC with significant main effects for time (P = 0.009, P = 0.008) and condition (P ≤ 0.0005, P ≤ 0.0005), however no significant interaction effects were observed (P = 0.425, P = 0.469). C2C12 myoblast nuclei appeared to exhibit enhanced alignment when cultured on both materials after 72 h in GM and 120 h in DM with the nuclei displaying increasing alignment over time when cultured on both PLA and PC, however this effect of time was not significantly different between conditions on either polymer (Fig. 6D). Enhanced alignment of nuclei cultured on ABS and PET was also evident after both 72 h in GM (P = 0.004, P = 0.001) and 120 h in DM (P = 0.006, P = 0.002) when compared to CON. As significant nucleic and myofibrillar alignment appears to be consistent across polymers, and is not observed within in-direct morphological alignment analyses (data not shown), it may be postulated that C2C12 cells are responding to the topographical stimuli elicited as a consequence of the extrusion based process of FDM.4. Discussion
The capability to 3D print bespoke biologically receptive parts within short time periods has driven the growing prevalence of AM technology within biological settings.21–23 This research demonstrates the capability to specifically tailor desired biological phenotype within the printed microenvironment (Fig. 8A) via polymer selection, utilising time and cost effective commercially available AM instrumentation. In addition, there remains little compatibility data with regard to cellular interaction with 3D printed polymers;25 a potentially decisive factor of critical importance when creating platforms for the examination of physiologically relevant cellular processes. In this work, we used skeletal muscle cells in order to ascertain critical evidence of cellular behaviour in response to 3D printed polymers.The biological interaction with the physical properties of 3D printed polymers is an important aspect when considering the architecture of scaffolds for implantation3 or the creation of physiologically representative in vitro pharmaceutical screening models;27 processes which are currently being revolutionised by the design freedom of AM. As such, it is imperative that the cells utilised within these models display viable cellular physiological phenotypes. Here we observed polymer dependent viability of C2C12 cells when compared to respective controls, with decreases documented in PLA, PET and PC polymer conditions; however it is important to outline that surfaces associated with 3D printing, will often yield lower viability when compared to the industrially optimised equivalent of tissue culture plastic. Hence, despite the statistical significant difference observed in PLA and PET, it would be plausible to suggest cells cultured on PLA, PET and ABS all maintained a viable metabolic phenotype within culture (alamarBlue assay readings ≥2000 a.u).
We observed no difference in cell viability in PET and PC leachate conditions, in addition to increases in viability compared to control in PLA and ABS, further outlining polymer chemical bio-inertness. It is likely that increases in PLA and ABS were as a consequence of assay variability opposed to each polymer chemically stimulating metabolic activity, as this would have also been evident when cells were cultured simultaneously with both the chemical constituents and physical parameters of each polymer in the direct condition. Such chemical bio-inertness would also suggest that the significant decrements observed in viability of cells cultured directly on 3D printed polymers, are indicative primarily of cellular interaction with the physical/physico-chemical characteristics of the polymer and not that of any potential chemical leachate within the culture medium. This may hold specific relevance within flow reactor applications of 3D printing, where it is of paramount importance to understand the effects of polymer chemical leachate on cellular behaviour, with many parts designed where polymer interaction is often confined to perfused medium, such as live-cell imaging systems.2
Differential myotube phenotypes were observed in cells cultured on 3D printed polymers, with reductions in width documented in PLA, PET and PC conditions (Fig. 5A). Homogeneous myotube numbers across conditions suggests that differences in width observed were consequential of reduced total protein accretion, as opposed to hyperplasic phenotype. Such reductions demonstrate correlation with polymer channel width (Fig. 7A); elicited due to the extrusion process of FDM, with tighter channels appearing to stimulate thinner myotube formation. Myotubes cultured on PLA exhibited the significantly thinnest phenotype within the tightest channel, when compared to myotubes within the incrementally wider channels of PC, ABS and PET (Fig. 7A). The aforementioned, in addition to the homogeneous myotube widths observed in chemical polymer leachate conditions, suggests a potential polymer dependent threshold at which 3D printed polymer scaffolds may cease to modulate myotube morphology.
Myogenin expression at GM72 h appeared more sensitive to varying polymer channel widths, with cells cultured within channels of PLA displaying significantly up-regulated myogenin expression when compared to PC, ABS and PET conditions at this time-point (Fig. 7B). In addition, cells within the second thinnest channel of PC also displayed significantly up-regulated expression of this gene compared to ABS and PET. Despite documented increases in myogenin expression in ABS compared to CON, no difference was observed between this polymer and the comparable channel width of PET at this time-point, outlining potential differential thresholds of where 3D printed polymer scaffolds cease to modulate morphology (≥236.44 ± 18.04 μm) and early potentiated myogenin expression (≥267.78 ± 27.39 μm), when 3D printing with PLA, PC, ABS and PET polymers.
In addition polymer dependent myotube widths and gene expression, we observed significantly enhanced nuclear alignment after 72 h in GM, outlining consistent cellular alignment across polymers. Cells cultured on all 3D printed polymers also displayed significantly enhanced nuclear and myofibrillar alignment after 120 h in DM, however only cells cultured within the tightest channels of PLA exhibited statistically significant potentiated myogenin expression at this time-point. The aforementioned consistency in nuclear and myofibrillar alignment would appear to indicate that C2C12 cellular alignment is being stimulated by a consistent feature across scaffolds, suggesting that C2C12 cells display no preferential channel diameter (Fig. 2), or physico-chemical polymer features when morphological alignment is desired.
Main effects of time observed in ABS, PET and PC chemical leachate conditions demonstrated that although myogenin expression increased between 72 h in GM and 120 h in DM, this was homogenous across conditions outlining negligible polymer chemical leachate stimulatory effects. The aforementioned response of myogenin expression to reducing channel width, in combination with the consistent level of nuclear alignment documented across polymers at GM72 h, would suggest that early cellular confluence within channels is driving augmented myogenin transcription as opposed to enhanced cellular alignment, with thinner channels achieving confluence quicker, hence resulting in increased time for the transcriptional up-regulation of this gene (Fig. 8A). This would also suggest that reductions in myotube diameter documented in correlation with decreasing channel width, are primarily due to reduced cell number within each channels specific local environment. The aforementioned significant up-regulation of myogenin in PLA at DM120 h also indicates that such enhanced early transcriptional activity may remain potentiated throughout myogenesis, eliciting greater peak expression of this gene (Fig. 8B), simultaneously resulting in highly differentiated cultures of reducing myotube width within decreasing channel diameters (Fig. 7E).
Although C2C12 cells have previously been shown to exhibit enhanced morphological alignment in response to topographical cues, often coinciding with the enhanced differentiation of skeletal myoblasts,34 within this research it is important to caveat the potential for polymer dependent physico-chemical signals to have also influenced the various aspects of observed cellular phenotypes. As such, future investigations should be undertaken to ascertain whether a within polymer linear relationship between a range of 3D printed topographies and C2C12 cellular phenotype is evident, and whether this response is replicated across polymers. A variety of polymer manipulatory techniques32,33 have been utilised to fabricate topographies to enhance the biomimicry of skeletal muscle cell cultures, however such procedures are designed with the primary intention of manipulating morphological alignment, in addition to often being the product of complex time-consuming soft/photolithographic processes. Here we observed the aforementioned desired cellular physiological benefits being elicited through simple time efficient 3D printing methods, outlining the potential not only to achieve cellular alignment but also to modulate desired biological phenotype via polymer selection at the press of a button (Fig. 8A). When considered in combination with the associated design freedom and ever advancing technology of 3D printing, this may provide the opportunity to not only enhance the efficiency of creating biomimetic models, but also to control the biological output within such scaffolds. This approach would be of significant interest across a diverse range of tissue engineering applications, pertinently in models whereby controlled and reproducible cellular alignment is necessary to produce a more biomimetic output.
5. Conclusions
This investigation observed biocompatible polymers 3D printed via FDM which exhibit morphological alignment and early differentiation of C2C12 cells when cultured directly on the 3D printed scaffold, providing rationale for use when re-creating platforms for cellular physiological processes. The consequential benefits of 3D printing biologically receptive parts observed in this work, in addition to the design freedom to rapidly produce and reproduce design iterations, also appears to hold the potential to modulate biological phenotype and enhance the biomimicry of skeletal muscle cells in vitro, factors of essential importance when creating biological scaffolds. Future research should focus on the capabilities of simple extrusion based processes such as FDM; which now encompasses the commercial availability to manipulate nozzle diameter (250–800 μm), in addition to more complex AM techniques in the context of controlling biological responses. This should be explored while simultaneously generating a comprehensive knowledge base of compatibility data with regard to candidate materials from varying 3D printing processes across a diverse range of mammalian cell type populations.Acknowledgements
This research was undertaken within a mini-centre for doctoral training (CDT) funded by Loughborough University. This work was supported in part by EPSRC Grant REF: EP/L02067X/2.References
- K. B. Anderson, S. Y. Lockwood, R. S. Martin and D. M. Spence, A 3D printed fluidic device that enables integrated features, Anal. Chem., 2013, 85, 5622–5626 CrossRef CAS PubMed
.
- S. Balakrishnan,
et. al., A Scalable Perfusion Culture System with Miniature Peristaltic Pumps for Live-Cell Imaging Assays with Provision for Microfabricated Scaffolds, BioRes. Open Access, 2015, 4, 343–357 CrossRef CAS PubMed
.
- S. Lohfeld,
et. al., A method to fabricate small features on scaffolds for tissue engineering via selective laser sintering, J. Biomed. Sci. Eng., 2010, 3, 138–147 CrossRef
.
- F. P. W. Melchels, J. Feijen and D. W. Grijpma, A review on stereolithography and its applications in biomedical engineering, Biomaterials, 2010, 31, 6121–6130 CrossRef CAS PubMed
.
- A. J. Capel,
et. al., 3D printed fluidics with embedded analytic functionality for automated reaction optimisation, Beilstein J. Org. Chem., 2017, 13, 111–119 CrossRef CAS PubMed
.
- N. P. Macdonald,
et. al., Assessment of biocompatibility of 3D printed photopolymers using zebrafish embryo toxicity assays, Lab Chip, 2016, 16, 291–297 RSC
.
- A. Díaz-Lantada, A. Mosquera, J. L. Endrino and P. Lafont, Design and rapid prototyping of DLC coated fractal surfaces for tissue engineering applications, J. Phys.: Conf. Ser., 2010, 252, 12003 CrossRef
.
- A. D. Lantada,
et. al., Tissue engineering using novel rapid prototyped diamond-like carbon coated scaffolds, Plasma Processes Polym., 2012, 9, 98–107 CrossRef CAS
.
- A. J. Capel,
et. al., Design and additive manufacture for flow chemistry, Lab Chip, 2013, 13, 4583–4590 RSC
.
- B. C. Gross, K. B. Anderson, J. E. Meisel, M. I. McNitt and D. M. Spence, Polymer Coatings in 3D-Printed Fluidic Device Channels for Improved Cellular Adherence Prior to Electrical Lysis, Anal. Chem., 2015, 87, 6335–6341 CrossRef CAS PubMed
.
- S. H. Masood, Intelligent rapid prototyping with fused deposition modelling, Rapid Prototyp. J., 1996, 2, 24–33 CrossRef
.
- D. H. Rosenzweig,
et. al., 3D-Printed ABS and PLA Scaffolds for Cartilage and Nucleus Pulposus Tissue Regeneration, Int. J. Mol. Sci., 2015, 16, 15118–15135 CrossRef CAS PubMed
.
- P. M. Van Midwoud, A. Janse, M. T. Merema, G. M. M. Groothuis and E. Verpoorte, Comparison of biocompatibility and adsorption properties of different plastics for advanced microfluidic cell and tissue culture models, Anal. Chem., 2012, 84, 3938–3944 CrossRef CAS PubMed
.
- L. Ricotti,
et. al., Adhesion and proliferation of skeletal muscle cells on single layer poly(lactic acid) ultra-thin films, Biomed. Microdevices, 2010, 12, 809–819 CrossRef CAS PubMed
.
- P. Zhang,
et. al., Local delivery of controlled-release simvastatin to improve the biocompatibility of polyethylene terephthalate artificial ligaments for reconstruction of the anterior cruciate ligament, Int. J. Nanomed., 2016, 11, 465–478 CrossRef CAS PubMed
.
- Y. Jiang, Y. Liang, H. Zhang, W. Zhang and S. Tu, Preparation and biocompatibility of grafted functional β-cyclodextrin copolymers from the surface of PET films, Mater. Sci. Eng., C, 2014, 41, 1–7 CrossRef CAS PubMed
.
- P. J. Kitson, M. H. Rosnes, V. Sans, V. Dragone and L. Cronin, Configurable 3D-Printed millifluidic and microfluidic ‘lab on a chip’ reactionware devices, Lab Chip, 2012, 12, 3267 RSC
.
- V. Saggiomo and A. H. Velders, Simple 3D Printed Scaffold-Removal Method for the Fabrication of Intricate Microfluidic Devices, Adv. Sci., 2015, 2, 1500125 CrossRef PubMed
.
- X. Yao, R. Peng and J. Ding, Cell-material interactions revealed via material techniques of surface patterning, Adv. Mater., 2013, 25, 5257–5286 CrossRef CAS PubMed
.
- N. M. Alves, I. Pashkuleva, R. L. Reis and J. F. Mano, Controlling cell behavior through the design of polymer surfaces, Small, 2010, 6, 2208–2220 CrossRef CAS PubMed
.
- H. Y. He, J. Y. Zhang, X. Mi, Y. Hu and X. Y. Gu, Rapid prototyping for tissue-engineered bone scaffold by 3D printing and biocompatibility study, Int. J. Clin. Exp. Med., 2015, 8, 11777–11785 Search PubMed
.
- J. A. Inzana,
et. al., 3D printing of composite calcium phosphate and collagen
scaffolds for bone regeneration, Biomaterials, 2014, 35, 4026–4034 CrossRef CAS PubMed
.
- J. Korpela,
et. al., Biodegradable and bioactive porous scaffold structures prepared using fused deposition modeling, J. Biomed. Mater. Res., Part B, 2013, 101, 610–619 CrossRef PubMed
.
- S. M. Oskui,
et. al., Assessing and Reducing the Toxicity of 3D-Printed Parts, Environ. Sci. Technol. Lett., 2016, 3, 1–6 CrossRef CAS
.
- C. M. B. Ho, S. H. Ng, K. H. H. Li and Y.-J. Yoon, 3D printed microfluidics for biological applications, Lab Chip, 2015, 15, 3627–3637 RSC
.
- A. S. Brack and T. A. Rando, Tissue-specific stem cells: lessons from the skeletal muscle satellite cell, Cell Stem Cell, 2012, 10, 504–514 CrossRef CAS PubMed
.
- A. S. T. Smith, S. Passey, L. Greensmith, V. Mudera and M. P. Lewis, Characterization and optimization of a simple, repeatable system for the long term in vitro culture of aligned myotubes in 3D, J. Cell. Biochem., 2012, 113, 1044–1053 CrossRef CAS PubMed
.
- A. P. Sharples,
et. al., Modelling in vivo skeletal muscle ageing in vitro using three-dimensional bioengineered constructs, Aging Cell, 2012, 11, 986–995 CrossRef CAS PubMed
.
- A. P. Sharples, N. Al-Shanti and C. E. Stewart, C2 and C2C12 murine skeletal myoblast models of atrophic and hypertrophic potential: Relevance to disease and ageing?, J. Cell. Physiol., 2010, 225, 240–250 CrossRef CAS PubMed
.
- B. N. Turner, R. Strong and S. A. Gold, A review of melt extrusion additive manufacturing processes: I. Process design and modeling, Rapid Prototyp. J., 2014, 20, 192–204 CrossRef
.
-
A. M. Innofil for, http://www.desktop3dprinter.com/user/pdf/Material%20Data%20Sheets/Innofil3D%20Material%20Safety%20Data%20Sheet%20ABS%20150313.pdf, 2008, pp. 1–8
.
- C. Monge,
et. al., Engineering Muscle Tissues on Microstructured Polyelectrolyte Multilayer Films, Tissue Eng., Part A, 2012, 18, 1664–1676 CrossRef CAS PubMed
.
- L. Altomare, N. Gadegaard, L. Visai, M. C. Tanzi and S. Farè, Biodegradable microgrooved polymeric surfaces obtained by photolithography for skeletal muscle cell orientation and myotube development, Acta Biomater., 2010, 6, 1948–1957 CrossRef CAS PubMed
.
- P.-Y. Wang, H.-T. Yu and W.-B. Tsai, Modulation of alignment and differentiation of skeletal myoblasts by submicron ridges/grooves surface structure, Biotechnol. Bioeng., 2010, 106, 285–294 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7lc00577f |
| This journal is © The Royal Society of Chemistry 2017 |