Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A 3D microblade structure for precise and parallel droplet splitting on digital microfluidic chips†
Cheng
Dong
ab,
Yanwei
Jia
 *a,
Jie
Gao
a,
Tianlan
Chen
a,
Pui-In
Mak
ab,
Mang-I
Vai
ab and
Rui P.
Martins‡
ab
*a,
Jie
Gao
a,
Tianlan
Chen
a,
Pui-In
Mak
ab,
Mang-I
Vai
ab and
Rui P.
Martins‡
ab
aState-Key Laboratory of Analog and Mixed-Signal VLSI, University of Macau, Macao, China. E-mail: yanweijia@umac.mo
bFaculty of Science and Technology – ECE, University of Macau, Macao, China
First published on 6th February 2017
Abstract
Existing digital microfluidic (DMF) chips exploit the electrowetting on dielectric (EWOD) force to perform droplet splitting. However, the current splitting methods are not flexible and the volume of the droplets suffers from a large variation. Herein, we propose a DMF chip featuring a 3D microblade structure to enhance the droplet-splitting performance. By exploiting the EWOD force for shaping and manipulating the mother droplet, we obtain an average dividing error of <2% in the volume of the daughter droplets for a number of fluids such as deionized water, DNA solutions and DNA–protein mixtures. Customized droplet splitting ratios of up to 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80 are achieved by positioning the blade at the appropriate position. Additionally, by fabricating multiple 3D microblades on one electrode, two to five uniform daughter droplets can be generated simultaneously. Finally, by taking synthetic DNA targets and their corresponding molecular beacon probes as a model system, multiple potential pathogens that cause sepsis are detected rapidly on the 3D-blade-equipped DMF chip, rendering it as a promising tool for parallel diagnosis of diseases.
80 are achieved by positioning the blade at the appropriate position. Additionally, by fabricating multiple 3D microblades on one electrode, two to five uniform daughter droplets can be generated simultaneously. Finally, by taking synthetic DNA targets and their corresponding molecular beacon probes as a model system, multiple potential pathogens that cause sepsis are detected rapidly on the 3D-blade-equipped DMF chip, rendering it as a promising tool for parallel diagnosis of diseases.
Introduction
Parallelization and miniaturization are the clear goals of molecular diagnostics for disease detection from a minute amount of an available sample.1,2 To achieve these goals, a wide variety of materials and technologies have been investigated. To miniaturize the reaction, microfluidic systems have attracted the most attention because they only consume a few microliters of the sample and reagents, have fast reaction times, and are precise and portable.3 Most investigations have utilized channel-based flow microfluidic systems.4 However, channel-based microfluidic systems have inherent limitations that require redundant supporting equipment such as pumps and valves for the operation of the systems, which hinders their widespread application in POC diagnostics.In contrast to the channel-based microfluidic systems, electronic-based digital microfluidic (DMF) systems control microdroplets on an array of electrodes by applying actuation forces such as electrowetting on dielectric (EWOD),5–9 magnetic,10,11 or optoelectrowetting forces.12–14 The electronic driving nature of the DMF devices removes the burden of using pumps and valves. At the same time, it provides intelligent control with real-time feedback and automatic analysis. DMF systems have been successfully employed in a variety of biological and chemical assays, such as polymerase chain reaction (PCR),15–17 DNA probe hybridization,18 proteomics,19,20 immunoassays,21–23 cell-based assays,24–27 and other chemical applications.28,29 Furthermore, their small footprint is well suited for POC devices and has resulted in an increasing number of applications.
However, parallel multiple analyses in molecular diagnostics with one single droplet are still a big challenge for DMF systems. For multiple quantitative analyses, uniform on-chip droplet splitting is in high demand. The existing splitting method normally involves three consecutive electrodes. By charging the outer electrodes while keeping the middle electrode not charged, one mother droplet can be divided into two smaller daughter droplets. However, nonuniform daughter droplets have been frequently observed.30 The volume variation (the volume after splitting divided by the total volume) has been reported to be approximately 7%.30 Furthermore, the success of splitting was dependent on the size of the droplet, the size of the electrode, the actuation voltage, and the gap between the bottom plate and the top plate of the DMF device.31
Several researchers have attempted to improve the on-chip cutting process.32–35 Volume-controllable droplet splitting has been reported, but a capacitance measurement and feedback control system was required.35 Changing the arrangement of the splitting electrodes into a Y junction has shortened the splitting time from 125 to 5 ms.32 Inducing sub-electrodes with a smaller size on the DMF chip could effectively generate unequal droplets.36 However, step-by-step splitting was still needed for its applications, which required more than two reaction droplets. Dispensing droplets from a reservoir was used to generate multiple relatively uniform droplets. However, only half of the mother droplets were successfully dispensed and the remaining samples were wasted.37 A microstructure constructed from Teflon was used to generate multiple daughter droplets and make full use of the provided sample. Nevertheless, light was necessary instead of an electronic signal to actuate the droplet, which was a downside of the technique given the difficulty of patterning the light spot.33 All of these limitations of the current droplet splitting techniques have hampered the use of DMF systems for precise multiple analyses from one single drop.
In this paper, we introduce a novel approach for accurate droplet splitting by constructing 3D microstructures, designated as blades, on DMF chips (Fig. 1). Compared with the currently accepted three electrodes splitting method for binary splitting, the size of the generated droplets using blades is more consistent and independent of the initial position of the mother droplet. Moreover, multiple droplets could be generated in one step by constructing multiple blades on a chip. Only four electrodes were used to simultaneously generate up to five daughter droplets, which significantly simplified the electronic control system. Using this technique, we successfully identified the DNA of a pathogen from a single droplet containing multiple potential pathogens that cause septicemia. Four kinds of pathogens were detected in one step with the same fluorescent label. The system provides a fast, precise, parallel, and point-of-care post-PCR analysis for disease diagnostics on a DMF platform.
Materials and methods
Device design and operation
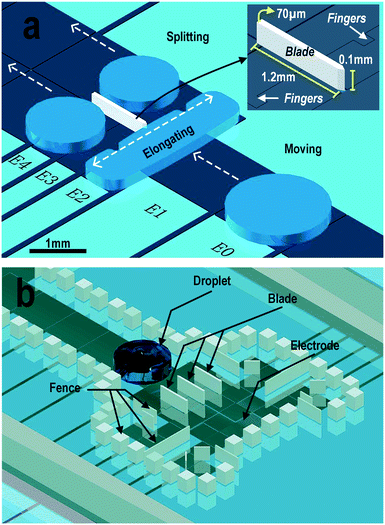
The novel design consisted of a group of electrodes cooperating with the on-chip 3D microblades to conduct multiple droplet splitting functions. Fig. 2a describes the basic design of the blade. An array of electrodes with different sizes and shapes was aligned on the droplet transportation path. A blade was located on the pathway at different locations depending on the required final splitting volume. Square-shaped 1.45 × 1.45 mm electrodes (E0 and E1) were used for droplet transportation. A narrow rectangular electrode E2 (0.725 × 2.9 mm) was placed right before the tip of the blade for droplet elongation. Rectangular electrodes E3 and E4 (0.54 mm × 2.9 mm) with fingers (inset of Fig. 2a) pointing to E2 were beneath the blade to achieve splitting of the droplets.During the splitting process, electrodes were charged in sequence from E1 to E4 using a technology called cooperative electrodes (CE).38 Each splitting operation began with droplet actuation on E1. After that, electrode E2 was charged to elongate the droplet to a slender rectangular shape by the EWOD force. CE was applied at this moment by overlapping the actuation duration of E2 and E3. With the little “fingers” inserted into E2, the elongated droplet was grasped by E3 synchronously at each section to prevent an unbalanced driving force, which is a common reason for nonuniform dissection. Accompanied by droplet movement towards E3, cut-off points appeared at the tip of each on-chip blade. The splitting operation was accomplished by releasing E2 and actuating E3 and E4.
During the splitting process, the mother droplet was cut by the tip of the blades and the daughter droplets were separated by the body of the blades. The blades were positioned depending on the required application. The tips of the blades were equally spaced over electrode E2 to achieve an equal volume division. Unequal volume droplet splitting was performed by positioning the blades at the appropriate locations. The lengths of the blades were designed to be long enough to prevent the daughter droplets from re-merging. In our design, the body of the blades extended from E2 to E4. Therefore, the daughter droplets had an elliptical shape and were restricted between the blades. As the blades were made of SU8 photoresist, their width was set to be tens of micrometers to guarantee a robust adhesion of the blade to the substrate. The effect of the blade height on the splitting performance is discussed in detail in the following sections. In our experiments, for the generation of multiple droplets, fences with the same height as the blades were fabricated around the electrodes to prevent droplet drifting on uneven coated surfaces, as shown in Fig. 2b. When integrating with other upstream on-chip functional design, the fences could be redesigned to prevent interference in the sample loading and droplet transportation. Detection reagents could be preloaded on the electrodes and secured with fences to remain untouched before the experiments.
Device fabrication
The fabrication of the DMF device used in this work was similar to that described previously.39 Briefly, the device had a sandwiched structure in which an aqueous droplet of the sample was immersed in the medium oil (silicone oil, 1 Cst, Clearco, USA) between a bottom glass plate patterned with chromium (Cr) electrodes and a top plate made of indium tin oxide (ITO) glass. A 10 μm layer of SU8 photoresist (Microchem, Newton, Massachusetts, USA) was coated on the electrode array as the dielectric layer. Microblades to perform droplet splitting, fences to prevent the droplets from drifting and spacers to form the working chamber of the DMF device were patterned on top of the dielectric layer with a 100 μm layer of SU8. The height of the blades was the same as the spacers, unless specified otherwise. Soft lithography of a double layer SU8 photoresist coating was employed for precise patterning of the blades and fences at the right positions using a mask aligner (ABM, California, USA). After the first layer of SU8 photoresist was coated and exposed, the second layer of 100 μm SU8 was coated on top. A mask with patterns of the blades, fences, spacers and alignment markers was used for the patterning. The high resolution optical system of the aligner ensured the alignment markers on the Cr layer of the chip and the markers on the mask to match each other at the appropriate position. The resolution of the pattern alignment was less than 5 μm. The bottom plate with microstructures matching its Cr pattern was obtained after a sequence of routine processes of exposure, baking and development in soft lithography. Teflon AF (Dupont, USA) with a thickness of 100 nm was coated on both plates to prevent sample adhesion on the surfaces. The detailed fabrication process and chip assembly can be found in Fig. S2 (ESI†).System setup
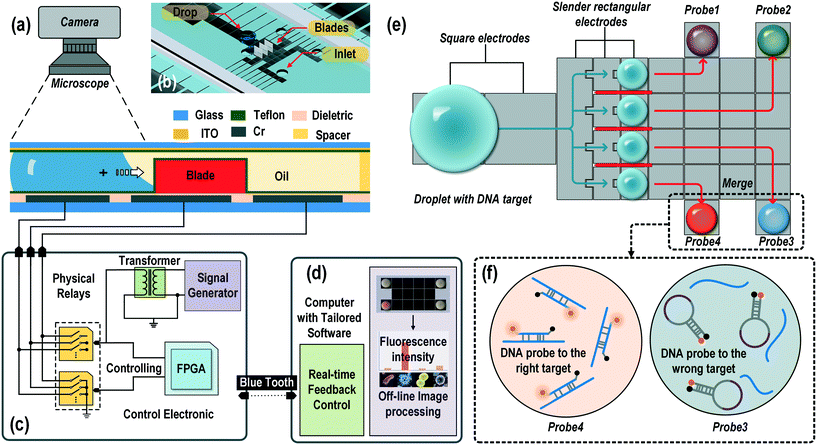
The experimental setup for the on-chip droplet splitting involved four parts: a DMF device fixed on a 3D-printed device holder, the control electronics on a printed circuit broad (PCB), custom software, and a fluorescence microscope mounted with a high-speed camera. Briefly, the DMF chip was held in position by the device holder and test clips. The holder and clips connected the electrodes on the DMF chip to the switches on the PCB for droplet actuation. A computer program, written in C# and compiled in Microsoft Visual Studio©, was developed to automate the acquisition of the droplet position and the execution of the droplet manipulation by controlling the power switches, which were physical relays (HRS2H-S-DC5V) driven by bipolar transistors (2N3904), on the PCB via a pre-configured field-programmable gate array (FPGA). An AC actuation signal (0.5–2 Vrms, 2.4 kHz, sinusoidal wave) was generated by a high-resolution signal generator and amplified to 80–300 Vrms by a transformer for charging the electrodes. The DMF chip and the chip holder were placed on the stage of a fluorescence microscope (Olympus BX63, Japan) and monitored by a high-speed camera. The videos and images were obtained by the software cellSens© accompanied by an Olympus microscope. Details of the system setup can be found in Fig. S1 (ESI†).Data analysis
Image processing software, Image J, was used to analyze pictures to calculate the droplet volume after splitting of the droplet and the fluorescence intensity in DNA probe–target analysis. For the former, as the height of the droplet was uniform on one chip, the volume of the droplet was proportional to the area of the droplet. We determine the size of the droplets by counting the number of pixels after automatically recognizing their profiles. The ratio between the sizes of the mother droplet and the daughter droplets was calculated through the ratio of the total number of pixels. The mean error was defined by the difference between the expected volume and the mean volume in a series of experiments. The standard deviation was the variation of the observed volume from the mean volume. In the DNA probe–target analysis, the fluorescence intensity of each droplet was obtained by calculating the mean value of the grayscale intensity across the droplet. Detailed image processing can be found in Fig. S3 (ESI†).DNA probe–target testing
The molecular beacon probes and target DNA oligonucleotides were designed in house and ordered from Genewiz (Suzhou, China) with the following sequences:S. aureus probe:
5′-Cy3-AATGGGTCATCTTTTAAGCTTTGGTT-BHQ2-3′;
S. aureus target:
5′-CCAAAGCTTAAAAGATGACCCA-3′;
K. pneumoniae probe:
5′-Cy3-TAAAGAACGCGAACAAGCTGGTA-BHQ2-3′;
K. pneumoniae target:
5′-CCAGCTTGTTCGCGTTCTT-3′;
Coag. negative probe:
5′-Cy3-AACTGTTACTGGTGTAGAATT-BHQ2-3′;
Coag. negative target:
5′-TTCTACACCAGTAACAG-3′;
L. lactis probe:
5′-Cy3-ATAAACCTTTCTTAAAAT-BHQ2-3′;
L. lactis target:
5′-TTTAAGAAAGGTTT-3′.
In the on-chip probe–target binding experiments, the dried DNA oligonucleotides were dissolved in 10 mM Tris-HCl buffer (Sigma, USA; with a conductivity of 1.52 mS cm−1, a viscosity of 0.9307 mPa s and a surface tension of 73 mN m−1). The mother droplet contained 1 μM of one type of target or a mixture of certain types of DNA targets. After splitting, a daughter droplet containing 1 μM of a certain type of probe specified to only one target was mixed with a droplet of the same size to obtain a final concentration of approximately 500 nM of each DNA oligonucleotide and probe.
Results and discussion
Comparison with the three consecutive electrodes method
For multiplex biological/chemical analysis on a chip from one single droplet, uniform droplet splitting would provide more reliable and reproducible experimental results. To test the performance of our microblade structure for droplet splitting, we compared the uniformity of the daughter droplets with those split by the widely accepted three consecutive electrodes splitting method (Fig. 3a) under the same coating and actuation conditions using DI water with a conductivity of 5.5 μS m−1, a viscosity of 1.0016 mPa s and a surface tension of 72.75 mN m−1. Fig. 3b shows the results of droplet splitting of one mother droplet to two equally sized daughter droplets. The volume of the droplets split by the three consecutive electrodes method was quite scattered with an overall variation of 6% (taking into account all the data obtained under all tested splitting voltages). On the other hand, the on-chip blade method gave a narrow range of droplet sizes with a variation of less than 2%. This could be attributed to the arrangement of the electrodes and the path of the droplet splitting in each method. With the three consecutive electrodes splitting method, the mother droplet was dragged at the two ends by the two outer electrodes depicted in Fig. 3a. The off-center placement of the mother droplet would cause unequal initial coverage on the two sides, giving a bias during the splitting. Actuating the middle electrode could not centralize the mother droplet due to its big size. Conversely, using our on-chip blade splitting method, the droplet was elongated using a rectangular electrode just before cutting. This would set the cutting point to a more reproducible position compared with the three electrodes method.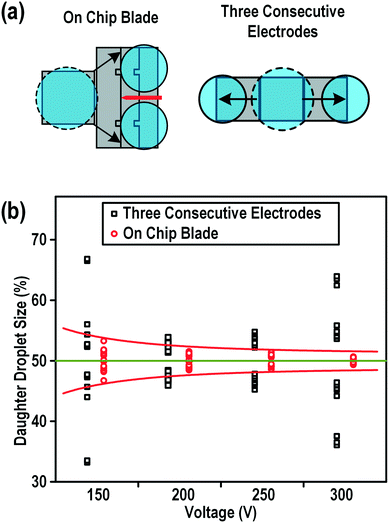 | ||
| Fig. 3 Comparison of the splitting performance between the three consecutive electrodes splitting method and the on-chip blade method. (a) Schematics of the splitting principles of the on-chip blade method and the three consecutive electrodes method. (b) The splitting performance of these two methods in terms of the daughter droplet size as a percentage of the mother droplet size. Ten trails had been performed at each of the four voltages for both methods. The mother droplet was DI water with a diameter of approximately 1.7 mm and a height of 100 μm. Videos of the typical splitting can be found in the ESI.† | ||
Moreover, as shown in Fig. 3b, the size variation of the blade splitting method could be further narrowed by increasing the actuation voltage. The volume variance decreased four times from 1.7% to 0.4% when the actuation voltage doubled from 150 Vrms to 300 Vrms. The performance of the three consecutive electrodes splitting method was unpredictable and totally independent of the splitting voltage. Therefore, although the chip fabrication became more complicated due to the construction of the on-chip blades, the on-chip blade structure using a higher splitting voltage would still be a better option for a more uniform droplet splitting.
In order to find out the effect of the switching rate of the actuation signal to the success rate of the splitting, the electrodes from E1 to E4 were actuated in sequence with a charging time for each electrode from 12 ms to 500 ms with a fixed actuation voltage of 250 Vrms. It was observed that the droplet was not able to split with a charging period of 12 ms (83.3 Hz switching rate). For the charging period of 25 ms (40 Hz switching rate), the success rate of splitting was around 30%. With the charging period equal to or longer than 50 ms (20 Hz switching rate), the droplet splitting always succeeded. The main reason for the success or failure was because the droplet needed a certain time to entirely move onto the actuated electrode and touch the next electrode for splitting.
Factors affecting the splitting performance
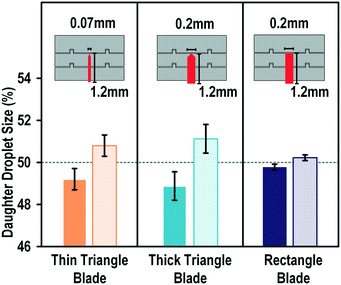
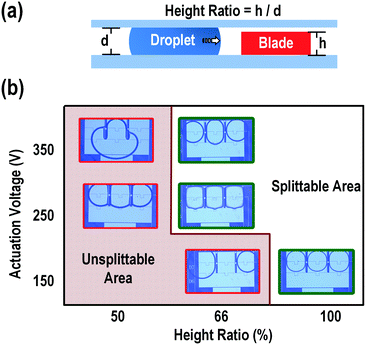
Besides the splitting voltage, we suspected that the blade shape and height and the size of the mother droplet may also affect the splitting performance. Therefore, we fabricated blades with different cutting tip shapes, namely, thin triangle, thick triangle, and rectangle, and investigated their performance in terms of splitting uniformity. The dimensions of the shapes are shown in Fig. 4. The variation of the droplet size for all these shapes was within 1%, in the range of the experimental error. These results indicated that the shapes of the blade did not affect the splitting uniformity.The dependence of the splitting success on the height of the blade was also studied. We defined the “height ratio” as the ratio of the height of the blade to the height of the spacer to indicate how much the droplet encountered the blade (Fig. 5a). The blades fabricated on the chip were 100 μm high and the spacers used in our experiments were 100, 150, 200, 250, and 300 μm, giving height ratios of 100, 66, 50, 40, and 33%, respectively. For each height ratio, the applied actuation signal was scanned from 150 to 300 Vrms. Fig. 5b shows the dependence of the splitting capability on the height ratio. At a height ratio of 100%, the droplet splitting was always successful, from the lowest tested voltage (150 Vrms) to the highest voltage (300 Vrms). When the height ratio was reduced to 66%, the success of the splitting was dependent on the splitting voltage. Droplet splitting failed at voltages lower than 250 Vrms. When the height ratio was 50% or less, the droplet could not be cut regardless of the applied splitting voltage. This might be because the height of the blade was less than half of that of the droplet, which would allow the droplet to climb up the blade and cover it. The surface energy of the big droplet would be lower than if it was split into multiple droplets.
The size of the mother droplet was also found to have an impact on the droplet splitting performance. The droplet splitting only succeeded in a certain range with a specific geometric design. The minimum size required to generate uniform daughter droplets was determined by the size of E2 (Fig. 2) because smaller droplets would not be able cover the electrode during elongation. That would lead to a non-uniform droplet generation or a failure to reliably transport the mother droplet to the next electrode. The maximum size was the sum of the areas of E3 and E4 as a larger droplet would envelop all the blades as a big drop instead of splitting into several daughter droplets. Detailed calculation and discussion can be found in the ESI.†
In summary, the splitting capability was influenced by the size of the mother droplet and the blade structure, in particular its height rather than its shape.
Unequal droplet splitting
In quantitative analysis, a specific percentage of a sample instead of exactly half of its initial volume may be required to be mixed with another sample, for example, in multiplex biological analysis and chemical titrations. We investigated the capability and accuracy of the on-chip blade method for unequal droplet splitting. As shown in Fig. 6, the blades were intentionally placed at different locations on the splitting electrode to generate daughter droplets with 50/50, 30/70, and 20/80 ratios. As shown, the blade demonstrated a high splitting accuracy with a less than 1% division error and standard deviation. We also tried but failed to split the droplet at a ratio of 10/90. This may be because of the overwhelming EWOD force on the side with a larger electrode, which was so dominant that it dragged the whole droplet away before the splitting neck emerged.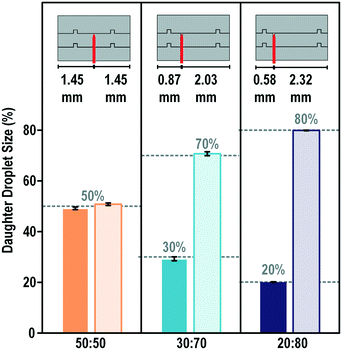 | ||
| Fig. 6 On-chip blade for unequal droplet splitting. Ten trails were performed for each design with the same actuation voltage. The mother droplet was DI water with a diameter of approximately 1.7 mm and a height equal to 100 μm. The splitting images are provided in the ESI.† | ||
One-step multiple-droplet generation
With precious biopsy samples, multiple analyses under the same conditions with the same droplet are in high demand. However, only two droplets can be generated in a single splitting process with the current widely used three electrodes splitting method. Although a series of splitting procedures could be performed to generate more droplets, the final volumes and number of daughter droplets are limited by the number and size of the electrodes. Furthermore, multiple steps slow down the splitting process. Using the proposed blade-splitting system, we could generate multiple droplets with various droplet volumes and number of droplets in one-step splitting.As shown in Fig. 7, multiple blades were fabricated on the cutting electrodes. The tips of the blades were equally separated from each other. Blades with thin triangular tips were used in this investigation. We first tested the splitting of deionized water. One mother droplet could be split into three to five identical daughter droplets depending on the number of blades used. The mean error was less than 2% with a standard deviation of less than 1%. Compared to the droplet dispensers commonly used on DMF devices, the downside of the on-chip blade for the multiple droplet generation method was that it would be difficult to be scaled up to a very large number of droplets. However, it showed big advantages that the entire mother droplet could be used and the daughter droplets were generated simultaneously which would be very welcome in biological applications.
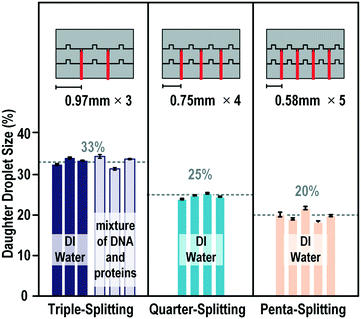 | ||
| Fig. 7 Generation of multiple equal volume daughter droplets in one splitting step with on-chip blades. The mother droplets used in these experiments were DI water or a mixture of DNA and proteins with a diameter of approximately 1.7 mm and a height of 100 μm. The videos are provided in the ESI.† | ||
In real biochemical applications, most of the samples contain DNA, proteins, buffers, or other biological materials, which may increase the viscosity of the samples. Fragmentation has frequently been observed during transportation, which requires a lower moving and splitting voltage. A low voltage may provide a challenge for effective droplet splitting.
To investigate the capability of our system for splitting biochemical samples, we took the master mixture of reagents used for Loop-mediated isothermal amplification (LAMP) assays as a model system for one-step multiple droplet generation from biochemical samples. As shown in Fig. 7, the performance observed for splitting of the viscous sample was consistent with that observed for deionized water. The overall variance of the volume was approximately 1.4% with a maximum volume error of approximately 2.4%. Such a performance makes it possible for the application of this system for on-chip fast biological sample splitting for multiple analyses in parallel.
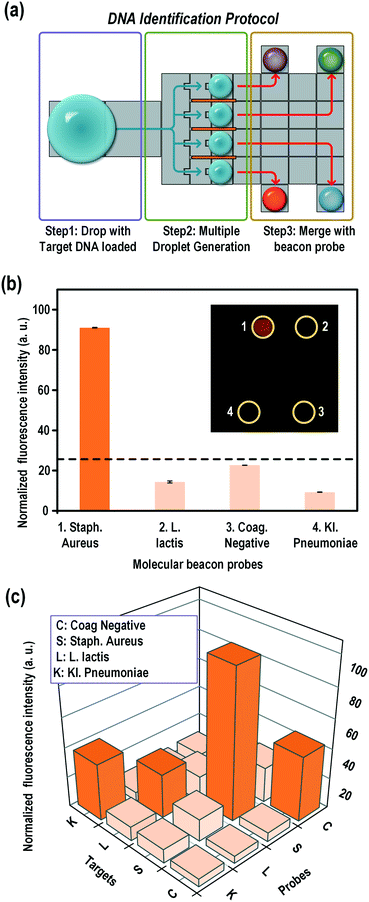
DNA identification on a DMF chip
Sepsis is a life-threatening condition that can be triggered by an infection of a variety of bacteria, fungi, mycobacteria, rickettsiae, viruses, or protozoans.40 Because of the high mortality rate of patients becoming septic, fast and precise determination of the pathogen is required to save a patient's life. Nucleic-acid-based DNA identification provides most of the genetic information on infectious species and mutations for drug resistance. The current DNA identification methods for septicemia are either performed in a large array of tubes containing different sets of probes and primers or in a single tube containing complicated fluorescent probes labelled with different fluorophores. Some groups innovatively used single fluorescence to simplify the detection system by designing probes with different probe–target melting temperatures.41 In that setup, a melting curve analysis needs to be performed after PCR amplification to identify the pathogen target. The requirement for multichannel fluorescence reading or precise temperature control has prevented the application of these methods for precise and convenient tests in a POC device.We tested our novel one-step multiple-droplet generation with the 3D microblade structure for fast portable DNA identification. Common pathogens causing sepsis (S. aureus, Coag. negative, L. lactis, and K. pneumoniae) were chosen as the detection DNA targets. Each target contained the specific sequence of a particular organism. Specific probes were designed for each of the targets and labelled with the same fluorophore (Cy3). All the probe–target melting temperatures, Tm, were approximately 60 °C. These Tm values ensured an instant fluorescence light-up under the most simplified detection settings, i.e. at room temperature in one fluorescence channel.
In this fast pathogen DNA identification system, one single droplet containing a large amount of different pathogen DNAs was loaded on the DMF chip, as shown in Fig. 8a. The droplet was split into four identical daughter droplets by the on-chip blades and mixed with individual DNA molecular beacon probes preloaded on the chip. The droplet splitting, moving, and mixing processes were automatically accomplished in an appropriate sequence using self-developed intelligent electronic control software. Fig. 8b shows the fluorescence detection after the probes for Coag. negative, S. aureus, L. lactis, and K. pneumoniae were mixed with the incoming sample droplet containing only the S. aureus target. As shown, only the probe for S. aureus lit up, whereas all the other mixtures were dark. We also tested samples containing different targets. As shown in Fig. 8c, the corresponding probes lit up solely for the specific pathogen.
In summary, this system provided a simple platform for the detection of different septicemia pathogens. The system is fast and portable and can detect pathogens in a single fluorescence channel with excellent specificity and precision. These desirable features would make this system appropriate for PCR based point-of-care disease diagnostics in the future.
Conclusions
We developed a novel droplet splitting method based on a 3D microblade structure. This simple, robust, and accurate splitting method allowed us to perform multiple analysis from a minute amount of sample. The performance of different on-chip blade configurations was investigated in terms of actuation voltage, splitting accuracy, and reproducibility. This technology was used to prepare DNA molecular beacon probes labelled with the same fluorophore for the detection of multiple targets. The significantly simplified detection system is very difficult to achieve using conventional off-chip assays. This system has high potential for practical applications in a range of multistep biochemical applications including post-PCR molecular detection for point-of-care disease diagnostics.Acknowledgements
We appreciate the collaboration of Ho Cheung Shum and Matthew Tang from the University of Hong Kong for technical support for the fabrication and characterization of SU8. This work was supported through the Macao Science and Technology Development Fund (FDCT) – 047/2014/A1, University of Macau SRG2016-00072-AMSV and the State Key Laboratory on Analog and Mixed-Signal VLSI Funds.References
- M. Baker, Nat. Methods, 2010, 7, 351–356 CrossRef CAS.
- P. K. Drain, E. P. Hyle, F. Noubary, K. A. Freedberg, D. Wilson, W. R. Bishai, W. Rodriguez and I. V. Bassett, Lancet Infect. Dis., 2014, 14, 239–249 CrossRef PubMed.
- S. Park, Y. Zhang, S. Lin, T.-H. Wang and S. Yang, Biotechnol. Adv., 2011, 29, 830–839 CrossRef CAS PubMed.
- W. G. Lee, Y.-G. Kim, B. G. Chung, U. Demirci and A. Khademhosseini, Adv. Drug Delivery Rev., 2010, 62, 449–457 CrossRef CAS PubMed.
- W. C. Nelson and C. J. Kim, J. Adhes. Sci. Technol., 2012, 26, 1747–1771 CAS.
- M. J. Schertzer, R. Ben Mrad and P. E. Sullivan, Sens. Actuators, B, 2012, 164, 1–6 CrossRef CAS.
- C. Peng, Z. Zhang, C. J. Kim and Y. S. Ju, Lab Chip, 2014, 14, 1117–1122 RSC.
- S. Park, P. A. L. Wijethunga, H. Moon and B. Han, Lab Chip, 2011, 11, 2212–2221 RSC.
- M. J. Schertzer, R. Ben-Mrad and P. E. Sullivan, Sens. Actuators, B, 2010, 145, 340–347 CrossRef CAS.
- Z. G. Guo, F. Zhou, J. C. Hao, Y. M. Liang, W. M. Liu and W. T. S. Huck, Appl. Phys. Lett., 2006, 89, 081911 CrossRef.
- A. A. García, A. Egatz-Gómez, S. A. Lindsay, P. Domínguez-García, S. Melle, M. Marquez, M. A. Rubio, S. T. Picraux, D. Yang, P. Aella, M. A. Hayes, D. Gust, S. Loyprasert, T. Vazquez-Alvarez and J. Wang, J. Magn. Magn. Mater., 2007, 311, 238–243 CrossRef.
- P. Y. Chiou, A. T. Ohta and M. C. Wu, Nature, 2005, 436, 370–372 CrossRef CAS PubMed.
- P. Y. Chiou, S. Y. Park and M. C. Wu, Appl. Phys. Lett., 2008, 93, 22–24 CrossRef.
- P. Y. Chiou, H. Moon, H. Toshiyoshi, C. J. Kim and M. C. Wu, Sens. Actuators, A, 2003, 104, 222–228 CrossRef CAS.
- H. Norian, R. M. Field, I. Kymissis and K. L. Shepard, Lab Chip, 2014, 14, 4076–4084 RSC.
- P. Neuzil, C. Zhang, J. Pipper, S. Oh and L. Zhuo, Nucleic Acids Res., 2006, 34, e77 CrossRef PubMed.
- W. A. Schell, J. L. Benton, P. B. Smith, M. Poore, J. L. Rouse, D. J. Boles, M. D. Johnson, B. D. Alexander, V. K. Pamula, A. E. Eckhardt, M. G. Pollack, D. K. Benjamin, J. R. Perfect and T. G. Mitchell, Eur. J. Clin. Microbiol. Infect. Dis., 2012, 31, 2237–2245 CrossRef CAS PubMed.
- L. Malic, T. Veres and M. Tabrizian, Biosens. Bioelectron., 2009, 24, 2218–2224 CrossRef CAS PubMed.
- M. J. Jebrail and A. R. Wheeler, Anal. Chem., 2009, 81, 330–335 CrossRef CAS PubMed.
- H. Moon, A. R. Wheeler, R. L. Garrell, J. A. Loo and C. J. Kim, Lab Chip, 2006, 6, 1213–1219 RSC.
- M. H. Shamsi, K. Choi, A. H. C. Ng and A. R. Wheeler, Lab Chip, 2014, 14, 547–554 RSC.
- A. H. C. Ng, K. Choi, R. P. Luoma, J. M. Robinson, A. R. Wheeler, A. Diagnostics, A. P. Road, A. Park and U. States, Anal. Chem., 2012, 84, 8805–8812 CrossRef CAS PubMed.
- K. Choi, A. H. C. Ng, R. Fobel, D. A. Chang-Yen, L. E. Yarnell, E. L. Pearson, C. M. Oleksak, A. T. Fischer, R. P. Luoma, J. M. Robinson, J. Audet and A. R. Wheeler, Anal. Chem., 2013, 85, 9638–9646 CrossRef CAS PubMed.
- S. M. Imaad, N. Lord, G. Kulsharova and G. L. Liu, Lab Chip, 2011, 11, 1448–1456 RSC.
- V. Lecault, A. K. White, A. Singhal and C. L. Hansen, Curr. Opin. Chem. Biol., 2012, 16, 381–390 CrossRef CAS PubMed.
- D. Bogojevic, M. D. Chamberlain, I. Barbulovic-Nad and A. R. Wheeler, Lab Chip, 2012, 12, 627–634 RSC.
- D. Witters, N. Vergauwe, S. Vermeir, F. Ceyssens, S. Liekens, R. Puers and J. Lammertyn, Lab Chip, 2011, 11, 2790–2794 RSC.
- G. J. Shah, H. Ding, S. Sadeghi, S. Chen, C. J. Kim and R. M. van Dam, Lab Chip, 2013, 13, 2785–2795 RSC.
- Y. Wang, W.-Y. Lin, K. Liu, R. J. Lin, M. Selke, H. C. Kolb, N. Zhang, X.-Z. Zhao, M. E. Phelps, C. K. F. Shen, K. F. Faull and H.-R. Tseng, Lab Chip, 2009, 9, 2281–2285 RSC.
- H. Ren, Ph.D. Thesis, Duke University, 2004 Search PubMed.
- S. K. Cho, H. Moon and C. J. Kim, J. Microelectromech. Syst., 2003, 12, 70–80 CrossRef.
- N. Y. J. B. Nikapitiya, S. M. You and H. Moon, in 2014 IEEE 27th International Conference on Micro Electro Mechanical Systems (MEMS), IEEE, 2014, pp. 955–958 Search PubMed.
- S. N. Pei and M. C. Wu, in 16th International Conference on Miniaturized Systems for Chemistry and Life Sciences, Okinawa, Japan, 2012, pp. 341–343 Search PubMed.
- J. H. Song, R. Evans, Y. Y. Lin, B. N. Hsu and R. B. Fair, Microfluid. Nanofluid., 2009, 7, 75–89 CrossRef CAS.
- J. Gong and C. J. Kim, Lab Chip, 2008, 8, 898–906 RSC.
- E. Samiei and M. Hoorfar, J. Micromech. Microeng., 2015, 25, 55008 CrossRef.
- S. Kalsi, M. Valiadi, M.-N. Tsaloglou, L. Parry-Jones, A. Jacobs, R. Watson, C. Turner, R. Amos, B. Hadwen, J. Buse, C. Brown, M. Sutton and H. Morgan, Lab Chip, 2015, 15, 3065–3075 RSC.
- T. Chen, C. Dong, J. Gao, Y. Jia, P.-I. Mak, M.-I. Vai and R. P. Martins, AIP Adv., 2014, 4, 47129 Search PubMed.
- T. Chen, Y. Jia, C. Dong, J. Gao, P.-I. Mak and R. P. Martins, Lab Chip, 2016, 16, 743–752 RSC.
- J. E. Tintinalli, J. S. Stapczynski, O. J. Ma, D. M. Cline, R. K. Cydulka and G. D. Meckler, Tintinalli's Emergency Medicine: A Comprehensive Study Guide, 7th edn, 2011 Search PubMed.
- L. M. Rice, A. H. Reis, B. Ronish, R. K. Carver-Brown, J. W. Czajka, N. Gentile, G. Kost and L. J. Wangh, J. Appl. Microbiol., 2013, 114, 457–469 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Supplementary figures and videos. See DOI: 10.1039/c6lc01539e |
| ‡ On leave from Instituto Superior Técnico, Universidade de Lisboa, Portugal. |
| This journal is © The Royal Society of Chemistry 2017 |