Open Access Article
Open Access ArticleA simple and low-cost chip bonding solution for high pressure, high temperature and biological applications†
M.
Serra‡
abc,
I.
Pereiro‡
 *abc,
A.
Yamada
abc,
J.-L.
Viovy
abc,
S.
Descroix
abc and
D.
Ferraro
abc
*abc,
A.
Yamada
abc,
J.-L.
Viovy
abc,
S.
Descroix
abc and
D.
Ferraro
abc
aLaboratoire Physico Chimie Curie, Institut Curie, PSL Research University, CNRS UMR168, 75005, Paris, France. E-mail: iago.pereiro@curie.fr
bSorbonne Universités, UMPC Univ. Paris 06, 75005 Paris, France
cInstitut Pierre-Gilles de Gennes, 75005, Paris, France
First published on 16th January 2017
Abstract
The sealing of microfluidic devices remains a complex and time-consuming process requiring specific equipment and protocols: a universal method is thus highly desirable. We propose here the use of a commercially available sealing tape as a robust, versatile, reversible solution, compatible with cell and molecular biology protocols, and requiring only the application of manually achievable pressures. The performance of the seal was tested with regards to the most commonly used chip materials. For most materials, the bonding resisted 5 bars at room temperature and 1 bar at 95 °C. This method should find numerous uses, ranging from fast prototyping in the laboratory to implementation in low technology environments or industrial production.
Introduction
Most microfluidic applications rely on the efficient bonding of two different parts to create enclosed channels. However, this fabrication step is still often complex, time-consuming and represents a factor that limits the spread of lab-on-a-chip systems in non-technical environments. A variety of techniques have been proposed over the years for the bonding of microfluidic chips fabricated with different materials and, in general, all these methods require either specific surface chemistries, high temperature, extremely clean surfaces or specialized tools.1 For example, polydimethylsiloxane (PDMS), the most widely used material by the microfluidic community, is most often bonded to a glass slide or to another PDMS layer by previous oxygen plasma2 or corona discharge3 activation. Alternatively, techniques based on chemical modifications have been proposed.4–6 Thermoplastics, on the other hand, are often sealed by applying pressure and temperature either above their glass transition temperature (Tg) by thermal bonding or below Tg by a solvent assisted approach,7,8 although recently the possibility of bonding cyclic olefin copolymer (COC) by applying pressure at room temperature has been reported with the right mixture of solvents.9 These approaches require a delicate tuning to avoid the deformation of the initial structures, especially if they are small or present a high aspect ratio. Then, glass to glass bonding requires chemical procedures at high temperature10 or highly corrosive acids.11 Furthermore, in principle, 3D printing allows the production of directly closed devices, but open microfluidic channels are often preferred to easily remove the support material or the uncrosslinked resin, especially when high printing resolutions are needed.12 Therefore, the bonding strategy for 3D printed devices must be customized depending on the microfabrication strategy and on the resin material.Alternative approaches have been reported, notably with the use of pre-polymerized adhesive layers to obtain a more versatile bonding. For example, UV curing adhesives (e.g. NOA 81)13,14 have been reported as a possible solution to allow patterning directly in the adhesive followed by its bonding by sticking at room temperature; however, homogeneous thickness, patterning and possible channel clogging remain a challenge. Thin layers of SU-8 have also been proposed as a bonding material, allowing a more precise patterning, but needing high temperature or UV activation to obtain good bond strengths.1,15 Furthermore, the use of a PDMS prepolymer film as an adhesive layer has been shown for PDMS/PDMS, glass/glass and PDMS/glass bonding.16 This method has also been extended for the bonding of PDMS devices to photoresist, metallic17 and plastic substrates.18
Microfabrication based on pressure sensitive adhesive (PSA) has also been reported for the fast and easy prototyping of entire microfluidic devices manufactured by desktop craft cutters.19,20 Some reported works combine this type of adhesive with more commonly employed materials. In particular, double-coated adhesive tape has been proposed for the fabrication of PDMS-tape composites that can be bound to other materials,21 but this still implies elaborate fabrication protocols. More recently, it was suggested that another type of adhesive tape could be directly used to seal PDMS devices,22 however it required long baking time (around 2 h) to obtain a resistant bonding, and it is still limited to working pressures below 0.5–1 bars. Beyond this pressure limit, Saffman–Taylor fingers appear around the inlet region. Furthermore, the resistance to high or cycling temperature was not investigated, and bio-compatibility tests were limited to confined droplets, limiting the possible applications of the resulting devices.
In this work, we propose the use of a commercially available PCR adhesive tape, based on the PSA concept, as a multi-purpose bonding solution for microfluidic channels patterned in a wide variety of materials commonly used by the microfluidic community, i.e. PDMS, COC, poly(methyl-methacrylate) (PMMA), polycarbonate (PC), glass and stereolithographic 3D printer resin. We demonstrate that this PCR tape sealing is able to sustain high pressures (up to 5 bar) directly after bonding without any further strengthening steps, even working at relatively high temperatures (ca. 100 °C). The deformation of the tape under high pressure was characterized, and its impact on high-frequency droplet generation was investigated, proving that the bonding covers the required pressure range without any instability. Furthermore, we explored the biocompatibility of the tape for cell culture and molecular biology. In particular, its relatively inert surface allowed successful cell culture and on-chip PCR, whereas its transparency enabled high-resolution optical microscopy observations. Finally, this tape bonding strategy also presents an easy reversible bonding, making expensive to fabricate or difficult to clean microfluidic devices readily available for reuse.
Results
A straight channel (200 μm in width and depth, 15 mm in length) was patterned in PDMS (Dow Corning, Sylgard 184 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
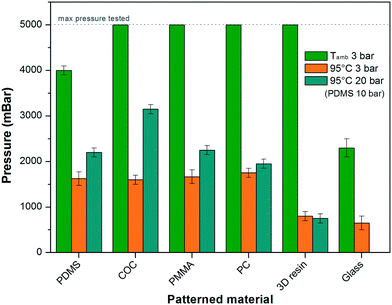
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, by replica molding of a brass micromilled mold), COC (Topas 6013, by hot embossing of the same mold), PMMA (OTMT), PC (Radiospares) (by direct material micromilling), glass slide (ROTH, with laser engraver C180II from GCC LaserPro) and stereolithographic 3D printer resin (DS2000, by DWS). The PCR sealing tape (ThermalSeal RTS™, Excel Scientific) having a thickness of 100 μm, a contact angle of 101.9 ± 1.1° and a roughness of Ra = 131.9 ± 3.4 nm (see ESI† Fig. S1a), was cut with a size of 20 × 35 mm and manually placed over the pattern, pressing directly with fingers or by simple tweezers. We estimated the local force applied manually to be between 1 and 5 kg, corresponding to 1–5 bars by direct finger pressing (surface of ∼1 cm2) or 10–50 bars with a tweezer tip (surface of ∼1 mm2). However, for a user-independent characterization of the bond strength, the protocol was performed by applying pressure with a hydraulic press at 3 bar, corresponding to the pressure range applied by hand. The bond strength was then evaluated by filling the chip with water and progressively increasing the inlet pressure of 100 mbar every 30 seconds with a constantly closed outlet (pressure controller MFCS, Fluigent). The maximum pressures obtained before the bond burst are shown in Fig. 1 for either a room temperature environment (ca. 25 °C) or when placed on a heating plate (Techne Prime) at 95 °C, equipped with a flat adapter.23 In the latter case, in order to evaluate the effect of the pressure applied during the bonding, we also tested 20 bar (equivalent to local pressures applicable by a tweezer tip).
1 ratio, by replica molding of a brass micromilled mold), COC (Topas 6013, by hot embossing of the same mold), PMMA (OTMT), PC (Radiospares) (by direct material micromilling), glass slide (ROTH, with laser engraver C180II from GCC LaserPro) and stereolithographic 3D printer resin (DS2000, by DWS). The PCR sealing tape (ThermalSeal RTS™, Excel Scientific) having a thickness of 100 μm, a contact angle of 101.9 ± 1.1° and a roughness of Ra = 131.9 ± 3.4 nm (see ESI† Fig. S1a), was cut with a size of 20 × 35 mm and manually placed over the pattern, pressing directly with fingers or by simple tweezers. We estimated the local force applied manually to be between 1 and 5 kg, corresponding to 1–5 bars by direct finger pressing (surface of ∼1 cm2) or 10–50 bars with a tweezer tip (surface of ∼1 mm2). However, for a user-independent characterization of the bond strength, the protocol was performed by applying pressure with a hydraulic press at 3 bar, corresponding to the pressure range applied by hand. The bond strength was then evaluated by filling the chip with water and progressively increasing the inlet pressure of 100 mbar every 30 seconds with a constantly closed outlet (pressure controller MFCS, Fluigent). The maximum pressures obtained before the bond burst are shown in Fig. 1 for either a room temperature environment (ca. 25 °C) or when placed on a heating plate (Techne Prime) at 95 °C, equipped with a flat adapter.23 In the latter case, in order to evaluate the effect of the pressure applied during the bonding, we also tested 20 bar (equivalent to local pressures applicable by a tweezer tip).
As observed, at room temperature the bonding with all thermoplastics and 3D resin resisted at pressures over 5 bar (which represents the highest pressure that we could apply), while PDMS and glass presented maximum bursting pressures of 4.0 and 2.6 bar, respectively. In any case, this is comparable to the bond strengths that can be obtained by oxygen plasma for PDMS–PDMS (1.4–3.4 bar) or PDMS–glass (2.7–4.8 bar).2
The same tests were repeated under high temperature conditions (95 °C), resulting in a decrease in the bursting pressures of all the materials, but which should be nonetheless sufficient for most microfluidic applications (1.6–1.8 bar for all thermoplastics and PDMS). Additionally, we observed that it is possible to increase the bond strength for most materials by increasing the bonding pressure (20 bar), allowing maximum pressures over 2 bar for PDMS and PMMA and even beyond 3 bar in the case of COC (see Fig. 1). The lower values obtained in the case of the 3D printed device at 95 °C might be due to the nature of the printed material itself, which is originally designed for the production of transparent medical prostheses with working temperatures between 20 °C and 60 °C (DigitalWAX DL-260).
It is important to underline that the bonding obtained with any of the tested pressures can be very easily removed with manual pulling forces (e.g. with tweezers), leaving no visible trace of the adhesive. The device can then be immediately resealed for further use or for the liquid content collection.
Since COC is a widely used thermoplastic material in microfluidics due to its remarkable properties, such as compatibility with biological applications,24,25 chemical resistance26,27 and high transparency,28 we selected it for forthcoming evaluations.
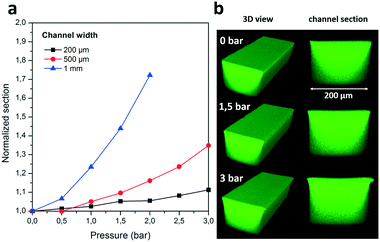
The integrity and stability of the bonding was further evaluated by confocal microscopy (TCS SP8, Leica) by filling COC channels of different widths (200, 500, and 1000 μm) and same depth (200 μm) with a fluorescein solution (3 μM in water, by Sigma-Aldrich) and applying pressure during the measurement. This allowed us to evaluate the total area of the channel section in a wide range of pressures as indicated in Fig. 2a, where the initial area is normalized as 1. As shown, the smallest channel presents the highest stability, with an increase in the total area of the section below 11% for an applied pressure of 3 bar. For larger widths the section increase is proportionally higher, although still below 10% for pressures up to 1.5 bar for 500 μm wide channels and up to 0.5 bar for 1 mm. Therefore, the maximum pressure applicable must be chosen considering the geometry of the microfluidic channels. However, for most microfluidic applications in which working pressures are below 1 bar and channel sizes in the range of tens to hundreds of microns, this adhesive tape solution seems to be well adapted. Furthermore, confocal imaging shows that the wall formed by the adhesive tape tends to stay flat for all pressures and the deformation takes place only at the interface between the tape and the chip material (Fig. 2b). Unbonding of this interface is the source of rupture if the pressure is increased to the maximum values shown in Fig. 1. We hypothesize that this might explain the lower bond strength observed for glass, as the aqueous solution could have an easier access to the interface due to higher wettability of the material. However, in all cases the aspect ratio of the section is highly preserved even in the upper pressure range. Further, any possible deformation at high pressure is homogeneously distributed along the length of the channel, as seen in Fig. 2b, avoiding the formation of Saffman–Taylor fingers that would implicate the formation of dead-end channels and stagnant areas. Finally, we evaluated the reversibility of this deformation by performing pressure cycles in which increasing pressures were alternated with zero pressure conditions (for the 200 μm-wide channel). The deformation was seen to be partly reversible, with a permanent section increase below 5% for pressures up to 2 bars (see ESI† Fig. S2).
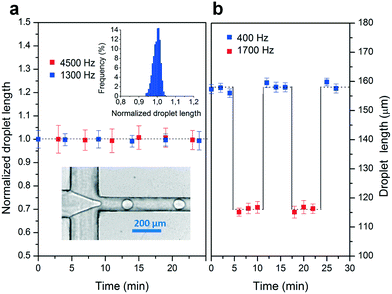
In order to evaluate the geometrical stability over a long experimental time with a particularly pressure-demanding application,29 tests were performed by high-frequency generation of monodisperse water droplets in oil (FC-40 oil, 3M) containing 1% (w/w) Krytox-PEG surfactant (Fluoro-Surfactant, by Ran Biotechnologies). In detail, the COC device was designed with a flow focusing geometrical configuration sealed with the adhesive (lower insert of Fig. 3a: oil and water inlets 200 μm wide, droplet generation channel 100 μm wide and 50 μm high). Since the droplet size depends on the channel geometry, at first, it was constantly evaluated for 25 minutes during the generation of either 1300 (inlet oil flow rate 2.4 μL s−1 and water 0.4 μL s−1, flows set by Cetoni syringe pumps) or 4500 droplets per second (inlet oil flow rate 6 μL s−1, water 0.9 μL s−1) by their recording at a frequency of 7400 Hz (Nikon Ti microscope, Phantom v4.2 camera by Vision Research) followed by image processing (user-customized LabView program). These frequencies correspond to a range of pressures in the generation channel between 0.4 and 1 bar, covering the range of values for the inlet pressure typically used for droplet generation by flow-focusing.30 As shown in Fig. 3a, the droplet size was highly stable and reproducible during all the experimental time, confirming the absence of any deformation in the geometry of the channel. Furthermore, this stability is maintained even in the case of longer experimental times, as indicated in ESI† Fig. S3. The results show in all cases a Gaussian distribution (Fig. 3a upper insert) of the droplet size with a relatively low coefficient of variation below 2.8% at 1300 Hz and below 4.8% at 4500 Hz, indicating again a highly preserved geometry. Next, the stability of the bonding was further tested with pressure cycles in which the flow rate of the aqueous phase was periodically switched between two values keeping a constant oil flow rate (4 μL s−1). The two frequency values tested (400 and 1700 Hz) were chosen to represent the typical working conditions for flow focusing, in a range between 500 and 2000 Hz.30,31 This resulted in monodisperse droplets generated at two different frequencies (400 Hz water at 0.4 μL s−1 and 1700 Hz water at 0.8 μL s−1) and two different sizes that showed an equally high stability over time. In particular, we do not observe neither deviations in the droplet length due to plastic deformation of the device, nor any slow transient behavior that would suggest elastic deformation of the adhesive tape after each change in experimental conditions.
The chemical resistance of the sealing and adhesive in the presence of organic solvents was also evaluated by sealing glass channels containing the chemical solution (see ESI† Fig. S4 for details). The integrity was not visibly affected by the presence of polar solvents (ethanol, dimethyl sulfoxide, acetonitrile), although it was compromised by non-polar solvents (toluene, cyclohexane) after 24 hours of evaluation periods. We further investigated the possible use of the adhesive for microchip capillary electrophoresis applications. In this case a straight COC channel having a cross section of 200 × 200 μm and a length of 3 cm was sealed with the adhesive, and the electroosmotic mobility (μeo) inside was obtained according to the current monitoring method32 in the presence of phosphate buffer pH 7.5. A HVS448 1500 V power supply (LabSmith, Livermore, CA) was employed to provide high voltage and current measurements. A cathodic electroosmotic flow was observed, and the obtained values (μeo = 1.69 × 10−4 ± 0.29 cm2 V−1 s−1) were comparable to the case of whole COC chips (μeo = 2.76 × 10−4 ± 0.14 cm2 V−1 s−1),33 evidencing the absence of a dramatic impact of the adhesive tape regarding electrokinetic behavior.
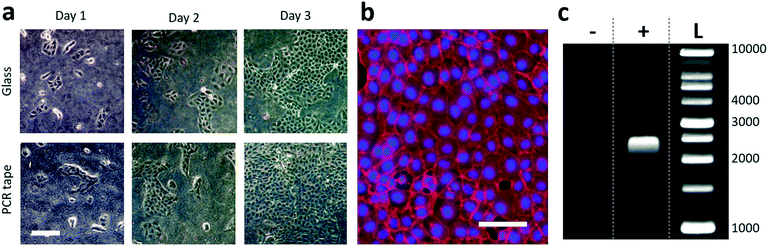
Finally, the tape solution proposed here has been tested for compatibility with both cell culture and molecular biology. The former was performed by closing a PDMS chamber (9 × 2 mm, 60 μm high, 1.2 μL internal volume, connected to four external well reservoirs, see ESI† Fig. S5) with either a standard glass slide or the adhesive tape. PDMS was chosen here because of its gas permeability, which is crucial for cell culture. Both chips were oxygen plasma treated for 30 seconds, filled with phosphate buffered saline (PBS) containing 0.01 mg mL−1 poly-D-lysine (Sigma-Aldrich) for promoting the cell adhesion, and incubated for 2 hours at room temperature. The chambers were sequentially rinsed with PBS and with cell culture medium (DMEM GlutaMax high glucose supplemented with 10% fetal bovine serum (v/v), Thermo Fisher Scientific). Next, 3 μL of medium containing Madin–Darby canine kidney (MDCK) cells at 6 × 105 cells per mL was injected in the chips prior to 10 min incubation in a humidified CO2 incubator (37 °C, 5% CO2) for cell attachment to the surface (adhesive tape or glass). After filling each reservoir with 50 μL of culture medium, the chips were enclosed in a plastic box containing 1 mg mL−1 ethylenediaminetetraacetic acid (EDTA, Sigma-Aldrich) to avoid evaporation and placed in the incubator for the following days. Phase-contrast images of cell growth were taken after 24, 48 and 72 h as shown in Fig. 4a. As observed, the cells presented an apparently equivalent adhesion to the surface of both glass and the adhesive tape, resulting in comparable cell growth rates and spreading. Full confluency was reached at day 4; at this point the cells had been fixed by incubation with 4% paraformaldehyde (Electron Microscopy Sciences) in PBS for 10 min, permeabilized by incubation with 0.2% Triton-X (Life Technologies) in PBS for 30 min, and stained for nucleus and actin by incubation with 1 μg mL−1 Hoechst 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 258 (Sigma-Aldrich) and 1/300 volume of Alexa Fluor® 647 Phalloidin (Thermo Fisher Scientific) in PBS, respectively, for 2 h at room temperature. Fig. 4b shows a fluorescence image (Nikon Ti Microscope, CoolSNAP HQ2) of the cells grown on the surface of the adhesive, presenting a healthy configuration. Furthermore, this not only shows the possibility of cell culture on the adhesive tape opportunely treated, but also the ability to perform high quality fluorescence imaging through it with no loss of quality, auto-fluorescence artifacts or light absorption (see ESI† Fig. S1b for the UV-visible absorption spectrum of the tape).
258 (Sigma-Aldrich) and 1/300 volume of Alexa Fluor® 647 Phalloidin (Thermo Fisher Scientific) in PBS, respectively, for 2 h at room temperature. Fig. 4b shows a fluorescence image (Nikon Ti Microscope, CoolSNAP HQ2) of the cells grown on the surface of the adhesive, presenting a healthy configuration. Furthermore, this not only shows the possibility of cell culture on the adhesive tape opportunely treated, but also the ability to perform high quality fluorescence imaging through it with no loss of quality, auto-fluorescence artifacts or light absorption (see ESI† Fig. S1b for the UV-visible absorption spectrum of the tape).
The compatibility of the method with molecular biology applications in which the reagent solution is in direct contact with the adhesive was then tested. An on-chip PCR was performed to verify the stability of the adhesive for a high number of fast temperature cycles as well as the viability of the biological protocol. A solution containing the template DNA molecules (3 ng μL−1, 2500 bp), enzymes (Phusion High-Fidelity DNA polymerase, ThermoFischer), dNTP mix and PCR primers were prepared and loaded inside a tape-sealed COC chip containing a straight channel (300 μm width, 300 μm depth) and a middle circular chamber (6 mm diameter, same depth) with a total volume of 15 μL. The PCR cycle parameters were: denaturation 98 °C 30 s, annealing 58 °C 10 s and extension 72 °C 30 s for a total of 35 cycles. The solution was recovered at the end of the process by pipetting and migrated in an agarose gel (SYBR(R) Green I stain) together with the products of a negative control performed with no initial DNA template, following the same protocol. The results are shown in Fig. 4c. The presence of PCR products, as indicated by the obtained positive bands, confirm the viability of enzymatic reactions with high temperature cycling and with the reagents in direct contact with the adhesive tape.
Conclusions
The PCR adhesive tape evaluated in these tests shows remarkable properties for the easy, low cost and reversible bonding of chips made with a wide range of transparent materials commonly used in the microfluidics community. Besides requiring no heating, a strong bonding can be obtained with the local forces typically obtained with a manual pressing. This is therefore an interesting alternative to more costly and complex bonding procedures that can only be performed in clean room-like facilities, making it suitable and convenient even for educational purposes. On the other hand, as demonstrated by the experiments performed with a press, the technique can also be implemented in manufacturing processes, and in this case manual bonding can facilitate prototyping and is not a mandatory step. The bonding remains highly resistant even for high temperature applications (95 °C), allowing in particular PCR cycling procedures that can lead to important vapor pressures inside the microfluidic channels. Moreover, the aspect ratio of the channel section is highly preserved, which, coupled to the low surface roughness of the adhesive, allows precise and demanding microfluidic procedures such as monodisperse droplet generation. Its optical properties in terms of transparency and low auto-fluorescence are also particularly interesting to include a window with high optical quality, into devices prepared by printed resins that usually present a low optical transmittance and/or a rough surface, as well as for general biological imaging. These optical properties should make possible the high quality imaging of biological processes such as cell culture or enzymatic reactions, which can be performed in direct contact with the surface of the tape. This is further reinforced by the fact that the flat configuration of the adhesive is well preserved even under pressure conditions. Finally, the bonding reversibility allows an easy adhesive removal and the possibility to reuse the chip after rinsing or to collect the sample inside (e.g. cultured cells) for further analysis. Therefore, we believe that this adhesive tape could be a versatile and simple bonding solution for a wide range of common microfluidic applications in both specialized and non-technical environments.Acknowledgements
This work is supported by the French National Research Agency (ANR) as part of the “Investissements d'Avenir” program (reference: ANR 10-NANO 0207), ARC foundation for young researcher fellowship (D. F.) and ERC Advanced Grant CellO (FP7-IDEAS-ERC-321107). MS was supported by a Curie Institute International PhD fellowship. We are also grateful to Fahima Faqir and Madison Oger (Institut Curie) for discussions on PCR reactions.References
- Y. Temiz, R. D. Lovchik, G. V. Kaigala and E. Delamarche, Microelectron. Eng., 2015, 132, 156–175 CrossRef CAS.
- S. Bhattacharya, A. Datta, J. M. Berg and S. Gangopadhyay, J. Microelectromech. Syst., 2005, 14, 590–597 CrossRef CAS.
- K. Haubert, T. Drier and D. Beebe, Lab Chip, 2006, 6, 1548 RSC.
- E. Ouellet, C. W. T. Yang, T. Lin, L. L. Yang and E. T. Lagally, Langmuir, 2010, 26, 11609–11614 CrossRef CAS PubMed.
- N. Y. Lee and B. H. Chung, Langmuir, 2009, 25, 3861–3866 CrossRef CAS PubMed.
- C. W. Beh, W. Zhou and T.-H. Wang, Lab Chip, 2012, 12, 4120–4127 RSC.
- C. W. Tsao and D. L. DeVoe, Microfluid. Nanofluid., 2009, 6, 1–16 CrossRef CAS.
- S. Miserere, G. Mottet, V. Taniga, S. Descroix, J.-L. Viovy and L. Malaquin, Lab Chip, 2012, 12, 1849 RSC.
- N. Keller, T. M. Nargang, M. Runck, F. Kotz, A. Striegel, K. Sachsenheimer, D. Klemm, K. Länge, M. Worgull, C. Richter, D. Helmer and B. E. Rapp, Lab Chip, 2016, 16, 1561–1564 RSC.
- D. J. Harrison, A. Manz, Z. Fan, H. Luedi and H. M. Widmer, Anal. Chem., 1992, 64, 1926–1932 CrossRef CAS.
- Z.-J. Jia, Q. Fang and Z.-L. Fang, Anal. Chem., 2004, 76, 5597–5602 CrossRef CAS PubMed.
- S. Waheed, J. M. Cabot, N. P. Macdonald, T. Lewis, R. M. Guijt, B. Paull and M. C. Breadmore, Lab Chip, 2016, 16, 1993–2013 RSC.
- D. Bartolo, G. Degré, P. Nghe and V. Studer, Lab Chip, 2008, 8, 274–279 RSC.
- S. Silvestrini, D. Ferraro, T. Tóth, M. Pierno, T. Carofiglio, G. Mistura and M. Maggini, Lab Chip, 2012, 12, 4041 RSC.
- L. Yu, F. E. H. Tay, G. Xu, B. Chen, M. Avram and C. Iliescu, J. Phys.: Conf. Ser., 2006, 34, 776 CrossRef CAS.
- H. Wu, B. Huang and R. N. Zare, Lab Chip, 2005, 5, 1393–1398 RSC.
- R. W. R. L. Gajasinghe, S. U. Senveli, S. Rawal, A. Williams, A. Zheng, R. H. Datar, R. J. Cote and O. Tigli, J. Micromech. Microeng., 2014, 24, 75010 CrossRef.
- L. Tang and N. Y. Lee, Lab Chip, 2010, 10, 1274 RSC.
- P. K. Yuen and V. N. Goral, Lab Chip, 2010, 10, 384–387 RSC.
- J. Saez, L. Basabe-Desmonts and F. Benito-Lopez, Microfluid. Nanofluid., 2016, 20, 116 CrossRef.
- J. Kim, R. Surapaneni and B. K. Gale, Lab Chip, 2009, 9, 1290 RSC.
- C. S. Thompson and A. R. Abate, Lab Chip, 2013, 13, 632 RSC.
- D. Ferraro, J. Champ, B. Teste, M. Serra, L. Malaquin, J.-L. Viovy, P. de Cremoux and S. Descroix, Sci. Rep., 2016, 6, 25540 CrossRef CAS PubMed.
- P. S. Nunes, P. D. Ohlsson, O. Ordeig and J. P. Kutter, Microfluid. Nanofluid., 2010, 9, 145–161 CrossRef CAS.
- K. Perez-Toralla, G. Mottet, E. T. Guneri, J. Champ, F.-C. Bidard, J.-Y. Pierga, J. Klijanienko, I. Draskovic, L. Malaquin, J.-L. Viovy and S. Descroix, Lab Chip, 2015, 15, 811–822 RSC.
- K. W. Ro, J. Liu and D. R. Knapp, J. Chromatogr. A, 2006, 1111, 40–47 CrossRef CAS PubMed.
- A. Bruchet, V. Taniga, S. Descroix, L. Malaquin, F. Goutelard and C. Mariet, Talanta, 2013, 116, 488–494 CrossRef CAS PubMed.
- A. Piruska, I. Nikcevic, S. H. Lee, C. Ahn, W. R. Heineman, P. A. Limbach and C. J. Seliskar, Lab Chip, 2005, 5, 1348 RSC.
- E. Piccin, D. Ferraro, P. Sartori, E. Chiarello, M. Pierno and G. Mistura, Sens. Actuators, B, 2014, 196, 525–531 CrossRef CAS.
- T. Ward, M. Faivre, M. Abkarian and H. A. Stone, Electrophoresis, 2005, 26, 3716–3724 CrossRef CAS PubMed.
- M. T. Guo, A. Rotem, J. A. Heyman and D. A. Weitz, Lab Chip, 2012, 12, 2146 RSC.
- X. Huang, M. J. Gordon and R. N. Zare, Anal. Chem., 1988, 60, 1837–1838 CrossRef CAS.
- K. Perez-Toralla, J. Champ, M. R. Mohamadi, O. Braun, L. Malaquin, J.-L. Viovy and S. Descroix, Lab Chip, 2013, 13, 4409–4418 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6lc01319h |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2017 |